Abstract
Salmonella infection triggers activation of innate immune cells through the interaction of bacterial products with Toll-like Receptors (TLRs). Toll/IL-1 domain-containing adaptor protein (TIRAP) is an adaptor protein involved in downstream signaling through TLR-1, TLR-2, TLR-4, and TLR-6. We examined the role of TIRAP during infection with attenuated Salmonella. Surprisingly, TIRAP-deficient mice were fully capable of resolving primary infection with Salmonella and actually exhibited accelerated clearance of bacteria at a late stage of the infection. Consistent with enhanced bacterial clearance, TIRAP-deficient mice resolved bacterial-associated splenic inflammation more rapidly than wild-type (Wt) mice and splenocytes from Salmonella-infected TIRAP-deficient mice produced more IFN-γ upon in vitro re-stimulation. Upon secondary challenge, TIRAP-deficient and Wt mice displayed a similar level of protective immunity against virulent Salmonella. Together these data indicate that TIRAP-mediated signaling is completely dispensable for clearance of Salmonella infection, and actually has a mild deleterious effect upon the resolution of primary infection.
Keywords: Bacterial Infection, Toll-Like Receptors, CD4 T cells
Introduction
Human typhoid is caused by infection with Salmonella enterica serovar typhi and remains a considerable health care problem in many parts of the developing world. Salmonella enterica serovar typhimurium (hereafter referred to as S. typhimurium) infection of inbred mouse strains provides the best model to study human typhoid in a laboratory setting [1]. Murine S. typhimurium infection causes rapid activation of innate and adaptive immune responses [2,3]. Understanding the processes by which Salmonella infection activates cells of the innate and adaptive immune system is critical to the rational design of novel therapeutics and vaccines against typhoid.
Toll-like receptors (TLRs) are expressed by many eukaryotic cells and represent an important means by which a host can identify pathogenic microbes and produce a rapid inflammatory response [4]. Salmonella express several microbial signatures that have the potential to activate TLRs in vivo, including lipopolysaccharide (LPS) [5,6], flagellin [7,8], and lipoproteins [9]. Although each of these bacterial products has been shown to activate innate immune cells in vitro, it is not yet clear which of these products is most important in eliciting an inflammatory response during Salmonella infection, or whether there exists significant redundancy in this process. One recent report demonstrated that TLRs are temporarily activated in response to Salmonella infection, suggesting that different TLRs contribute to innate activation at different stages following infection [9].
Activation of TLRs encourages dimerization of the receptor, leading to the recruitment of cytoplasmic adaptor proteins such as MyD88 and TIRAP to the signaling complex [10]. The importance of adaptor protein recruitment for TLR activation during Salmonella infection has been elegantly described. C3H/HeJ mice have a point mutation in the TIR domain of TLR4 preventing the recruitment of adaptor proteins and the ability to respond to Salmonella LPS [5,6]. An alternate group of cytoplasmic adaptor proteins, Toll/IL1R domain containing adaptor inducing IFN-β (TRIF) and TRIF-related adaptor molecule (TRAM), are also involved in MyD88-independent TLR signaling [10]. Recruitment of these adaptor proteins to the TLR signaling complex leads to NF-kB and p38 activation, and is required for optimal production of inflammatory cytokines in response to most TLR ligands [11-14].
Although MyD88 plays a major role in signaling through most TLRs, TIRAP is thought to be required for signaling through TLR1/2, TLR2/6, and TLR4 but dispensable for signaling through TLRs 3, 5, 7, and 9 [14,15].
MyD88 signaling is required for the activation of NADPH Oxidase activity in Salmonella-infected phagocytes [16], and MyD88-deficient mice display enhanced susceptibility to Salmonella infection [9]. These data demonstrate the importance of TLR signaling for bacterial clearance after Salmonella infection. However, it remains unclear which TLRs are critical to innate immune activation and whether TIRAP contributes significantly to innate responses to Salmonella.
As noted above, the importance of TLR4 signaling to resolution of Salmonella infection has been clearly documented [5,6]. The contribution of other TLRs during Salmonella infection is less clear. TLR5 is activated by Salmonella flagellin [7,8] and it was previously thought that a mutation in TLR5 might be responsible for the increased susceptibility to Salmonella displayed by MOLF/Ei mice [17]. However, recent reports point to an alternative deficiency in MOLF/Ei mice [18], and indicate that TLR5 deficiency has only a modest effect on Salmonella clearance [19,20]. Although macrophage expression of TLR2 is increased upon Salmonella infection, and signaling through TLR2 may be required for optimal macrophage activation, TLR2-deficient mice display only mildly increased susceptibility to Salmonella [9]. Therefore, the involvement of TLRs other than TLR2, 4, or 5, during Salmonella infection is suggested by the increased susceptibility of MyD88-deficient mice, but currently remains undefined.
The restricted nature of TIRAP involvement in TLR1/2, TLR2/6, and TLR4 signaling suggested that TIRAP deficiency could help elucidate the involvement of this group of TLRs during Salmonella infection. A recent study demonstrated that TIRAP expression in the lung is required for host resistance to Klebsiella pneumoniae, but not Pseudomonas aeruginosa infection [21]. Thus, different bacterial infections appear to preferentially target a limited range of TLRs in order to activate innate inflammatory responses. Although previous studies have confirmed a role for MyD88, TLR4 and TLR5 during the innate immune response to Salmonella, the role of the TIRAP has not been directly examined. Given the association between TIRAP and TLR4 signaling, it might be anticipated that TIRAP plays an important role in innate immune protection against Salmonella. However, our data demonstrate that TIRAP-deficient mice have no deficiency in the resolution of primary Salmonella infection, and in fact display a mild acceleration in bacterial clearance. Furthermore, TIRAP-deficient mice more rapidly resolved the splenic inflammation accompanying Salmonella infection, developed increased T cell IFN-γ production, and were protected from secondary infection with virulent Salmonella. Together, these data suggest a paradoxical detrimental role for TIRAP signaling during primary bacterial infection.
Materials and Methods
Mouse strains
TIRAP-deficient mice [15] were generously provided by Dr. Medzhitov, Yale University, and were backcrossed to C57BL/6 for 8-10 generations in our animal facility. F10 C57BL/6 TIRAP-deficient mice were screened by PCR for TIRAP deficiency and maintained by intercrossing homozygous pairs. SM1 Rag-deficient TCR transgenic mice expressing CD90.1 and CD45.1 alleles have been described [22,23]. C57BL/6 mice were purchased from NCI-Frederick, (Frederick, MD) and were used at 8-16 weeks of age. All mice were housed in specific pathogen-free conditions and cared for in accordance to Research Animal Resources (RAR) practices at the University of Minnesota.
Salmonella infection and bacterial colonization in vivo
S. typhimurium BRD509 and SL1344 strains were grown overnight in LB broth without shaking and diluted in PBS after determining bacterial concentrations using a spectrophotometer. Mice were infected intravenously with 5×105 bacteria via the lateral tail vein, and monitored daily for signs of infection. In all infection experiments, the estimated bacterial dose was confirmed by plating serial dilutions onto MacConkey agar plates, and incubating overnight at 37°C. To determine bacterial colonization in vivo, spleens from infected mice were removed and homogenized in PBS and serial dilutions were plated on MacConkey agar plates. Plates were incubated overnight at 37°C and bacterial counts determined for each individual organ.
IFN-γ Elisa
Spleens were harvested from infected Wt and TIRAP-deficient mice and a single cell suspension generated. Spleen cells were placed in 96-well plates and incubated for 48 hours, with and without the addition of Heat-Killed Salmonella typhimurium (HKST). Supernatants were harvested and transferred to protein-binding plates previously coated with purified antibody specific for IFN-γ (eBioscience, San Diego, CA). IFN-γ production was visualized using a biotin-conjugated antibody for IFN-γ (eBioscience) extravidin peroxidase (Sigma, St. Louis, MO), and O-Phenylenediamine substrate (Sigma). Color change was determined using a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA).
Salmonella-specific antibody
High protein binding plates were prepared by adding HKST diluted in carbonate buffer and incubating overnight at 4°C. Free binding sites were blocked by adding 10% FCS/PBS and plates were washed 4× in PBS/Tween. Blood was obtained from mice by retro-orbital bleeding and serum recovered and stored at -20°C. Serum samples were thawed, diluted in FCS/PBS and incubated on plates for one hour at 37°C. After further washing, biotin-conjugated antibody for IgG2a (eBioscience), extravidin peroxidase, and O-Phenylenediamine substrate were added sequentially to the plates. Color change was determined using a SpectraMax M2 microplate reader.
Adoptive Transfer of SM1 cells
Spleen and lymph nodes (inguinal, axillary, brachial, cervical, and mesenteric) were harvested from SM1 Rag-deficient, congenic TCR transgenic mice [22] and a single cell suspension generated. An aliquot of cells was stained using antibodies specific for to CD4, Vβ 2, CD90.1, or CD45.1 (eBioscience), and the percentage of SM1 cells determined by flow cytometry using a FACSCanto (BD Biosciences, San Jose, CA). In some experiments SM1 cells were incubated with CFSE for 10 minutes at 37°C immediately prior to adoptive transfer. For transfer, 1-5 ×106 SM1 cells were injected intravenously into a recipient C57BL/6 or TIRAP-deficient mouse via the lateral tail vein.
Flow Cytometric Analysis
Spleens were harvested and a single cell suspension re-suspended in Fc Block (24G2 supernatant plus rat and mouse serum) containing a variety of surface staining antibodies and incubated for 30 minutes at 4°C. Surface staining antibodies included, Fluoroscein isothiocyanate- (FITC-), phycoerytherin- (PE-), CyChrome-, PE-Cy5-, or allophycocyanin-conjugated antibodies specific for CD4, CD11a, CD45.1, and CD90.1. After staining, cells were fixed and data acquired using a FACSCanto. Data analysis was accomplished using FlowJo software (TreeStar, San-Carlos, CA).
Tracking SM1 cells in vivo
After Salmonella infection, spleen cells were harvested at different time points into Eagle’s Hanks Amino Acids Medium (EHAA) (Biofluids, Rockville, MD) containing 2% fetal bovine serum and 5mM EDTA. Cells were stained as described above and the percentage of SM1 cells per spleen was determined by flow cytometry.
Results
TIRAP-deficient mice display accelerated clearance of primary Salmonella infection
In order to examine the role of TIRAP during Salmonella infection, TIRAP-deficient C57BL/6 and Wild-type (Wt) mice were infected intravenously with 5×105 attenuated Salmonella (BRD509) and the number of bacteria in the spleen monitored over time. In both Wt and TIRAP-deficient mice, the number of bacteria expanded between 1 and 3 weeks after infection, began to reduce around 3 weeks post-infection, and was finally below the limit of detection 7 weeks after infection (Fig. 1A). Interestingly, the number of bacteria at 5 weeks post-infection was significantly lower in TIRAP-deficient versus Wt mice (Fig. 1A). We therefore specifically re-examined bacterial burdens at late time points following infection of TIRAP-deficient and Wt mice. Bacterial burdens were somewhat higher overall in this experiment but, as noted in the previous experiment, TIRAP-deficient mice had significantly lower bacterial burdens than Wt mice at later time points after infection (Fig. 1B). These data demonstrate first of all that TIRAP is not required for the resolution of Salmonella infection, and secondly that bacterial clearance occurs more rapidly in TIRAP-deficient mice.
Figure 1. TIRAP is detrimental to the clearance of Salmonella infection.
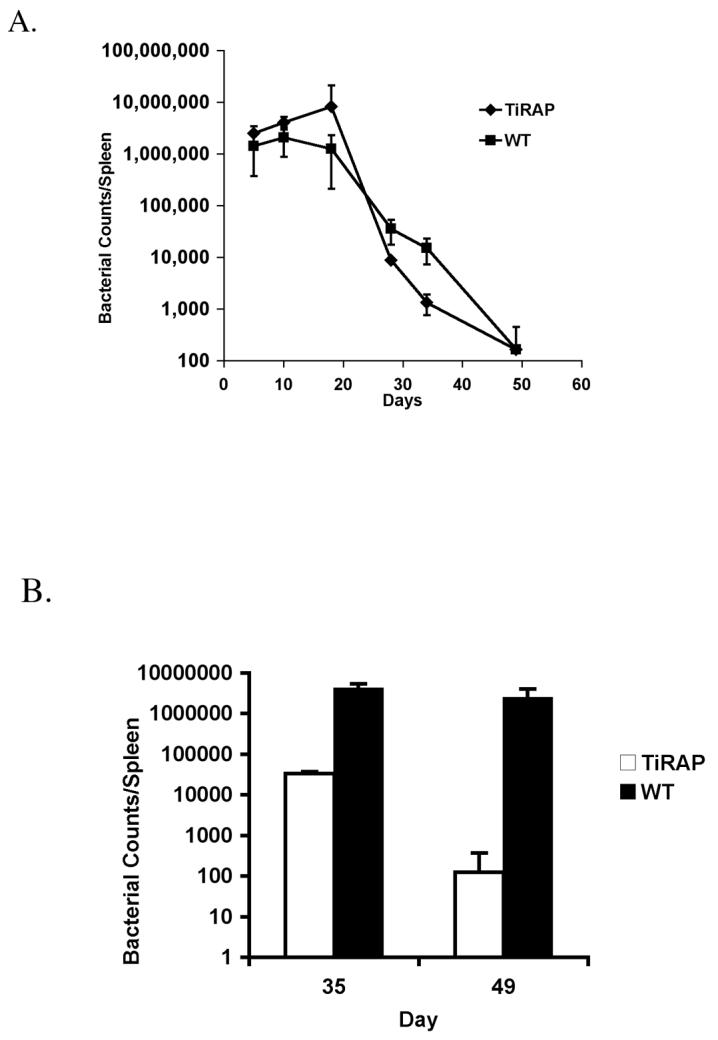
C57BL/6 Wt and TIRAP-/- mice were infected intravenously with 5×105 attenuated Salmonella, BRD509. (A) Spleens from infected mice were recovered at regular intervals after infection and the number of bacteria determined by plating serial dilutions. Data show the number of bacteria per spleen and are representative of at least 3 mice per group. (B) Spleens from infected mice were recovered 35 and 49 days after infection and the number of bacteria determined by plating serial dilutions. Data show the number of bacteria per spleen and are representative of at least 3 mice per group.
TIRAP-deficient mice make more IFN-γ and resolve splenic inflammation more rapidly than Wt mice
The onset of Salmonella clearance around 3-weeks post-infection is mediated by an adaptive immune response that requires the production of IFN-γ [24,25]. We therefore examined the production of IFN-γ by splenocytes from infected TIRAP-deficient and Wt mice, following in vitro re-stimulation with Salmonella antigens. Splenocytes from TIRAP-deficient mice produced more IFN-γ in response to stimulation than splenocytes from Wt mice (Fig. 2A), correlating with the enhanced clearance of bacteria in vivo. Next, we examined the cellular composition of the spleen of TIRAP-deficient and Wt mice at a late stage after infection. The spleens of TIRAP-deficient mice were smaller than Wt mice at this stage (data not shown), consistent with early bacterial clearance. Indeed, the spleen of TIRAP-deficient mice contained a higher percentage of B cells, CD8 T cells, and CD4 T cells, and a much lower percentage of NK1.1+ and CD11b+ than were detected in infected Wt spleens (Fig. 2B). These data suggest that the inflammatory infiltration of NK cells and macrophages is more rapidly resolved in TIRAP-deficient mice, consistent with rapid bacterial clearance.
Figure 2. Enhanced IFN-γ production and resolution of splenic infiltration in TIRAP-deficient mice.
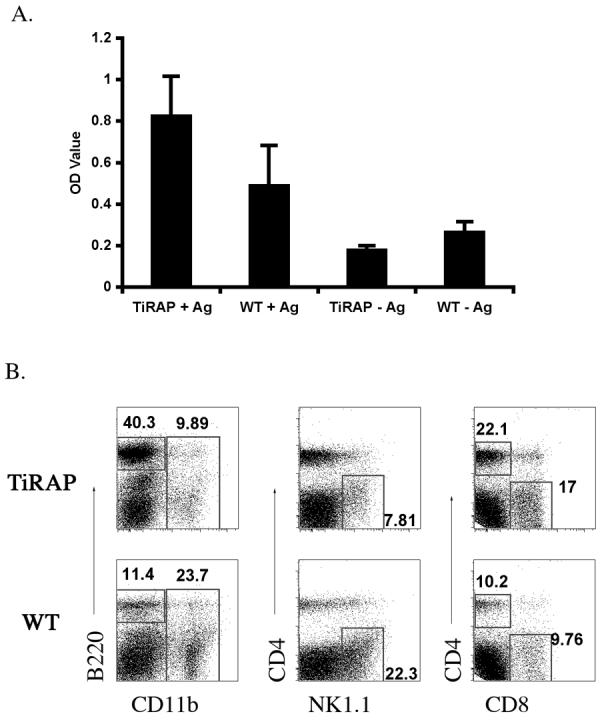
C57BL/6 Wt and TIRAP-/- mice were infected intravenously with 5×105 attenuated Salmonella, BRD509. (A) Spleens from infected mice were harvested 35 days later and cultured in vitro in medium alone (-Ag) or with added HKST (+Ag). Supernatants were harvested 72 hours later and IFN-γ production measured by ELISA. Data shows the mean OD ± SD for 3 mice per group. (B) Spleens from infected mice were recovered 35 days after infection and stained with antibodies specific for cell-specific surface markers. Data show FACS plots for individual mice and are representative of 3 mice per group. Numbers show the percentage of cells within each boxed gate.
Salmonella-specific CD4 T cells expand and persist normally in TIRAP-deficient mice
Given the difference in bacterial clearance in TIRAP-deficient and Wt mice and the role of CD4 T cell in Salmonella immunity, we examined the development of Salmonella-specific CD4 T cell responses in TIRAP-deficient and Wt mice. TIRAP-deficient and Wt mice were adoptively transferred with SM1 cells and spleens harvested on day 3, 7, and 18 following Salmonella infection or immunization with HKST. Three days after infection or HKST immunization SM1 T cells expanded to a similar degree (Fig. 3-Day3), and had undergone a similar number of cell divisions (Fig. 4-Day3), in both TIRAP-deficient and Wt mice. These data indicate that TIRAP-dependent inflammatory responses are not required for optimal flagellin-specific CD4 T cell expansion. As previously reported [26], SM1 T cells contracted and failed to enter the memory pool in Salmonella-infected Wt mice, but were clearly detected in the memory CD4 population of HKST-immunized mice, 7 and 18 days after immunization (Fig. 3 and 4). CD4 contraction and memory development was therefore identical in TIRAP-deficient mice, indicating that TIRAP does not play a significant role in flagellin-specific CD4 T cell activation in response to Salmonella.
Figure 3. Normal expansion and development of Salmonella-specific memory cells in a TIRAP-deficient environment.
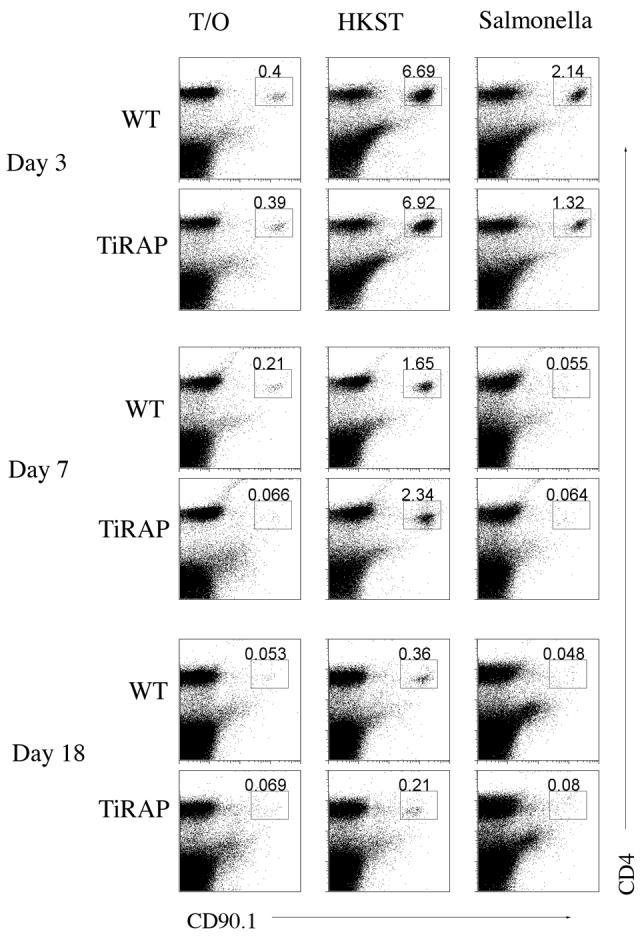
Wt and TIRAP-/- mice were adoptively transferred with SM1 T cells and infected intravenously the following day with 5×105 attenuated Salmonella, BRD509 or immunized intravenously with 1×108 HKST. Spleens were harvested at various time points later and stained using antibodies specific for CD4 and CD90.1. FACS plots show the percentage of SM1 T cells in the spleen of uninfected (Transfer Only), HKST immunized (HKST), and infected (Salmonella) mice. Numbers are the percentage of SM1 cells among all live gated spleen cells. Data are representative of 3 mice per group and 2 individual experiments.
Figure 4. Normal activation and cell division of Salmonella-specific T cells in a TIRAP-deficient environment.
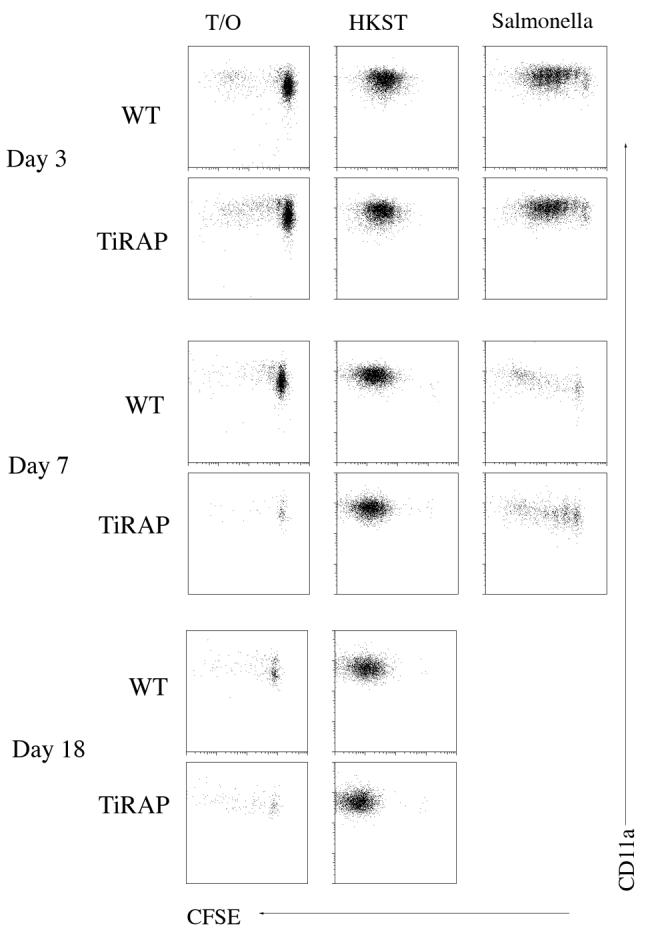
Wt and TIRAP-/- mice were adoptively transferred with CFSE-dyed SM1 T cells and infected intravenously the following day with 5×105 attenuated Salmonella, BRD509 or immunized intravenously with 1×108 HKST. Spleens were harvested at various time points later and stained using antibodies specific for CD4 and CD90.1 to identify SM1 cells, and CD11a to identify activated T cells. FACS plots show CFSE-dye dilution and CD11a expression on gated SM1 T cells in the spleen of uninfected (Transfer Only), HKST immunized (HKST), and infected (Salmonella) mice. There were too few SM1 cells at day 18 in infected mice to gather sufficient data for analysis. Data are representative of 3 mice per group and 2 individual experiments.
Vaccinated TIRAP-deficient mice resist secondary infection with virulent Salmonella
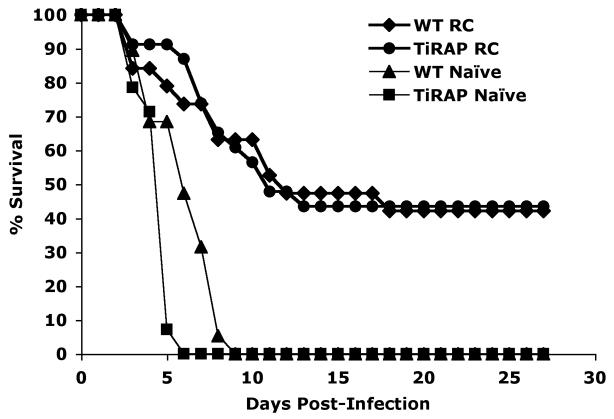
In addition to CD4 T cell responses, infection of TIRAP-deficient mice generated an elevated anti-Salmonella IgG2a antibody titer similar to Wt mice (Fig. 5). Thus, after bacterial clearance TIRAP-deficient mice had generated a robust humoral and cellular immune response to Salmonella. One of the hallmarks of exposure to live attenuated Salmonella is the acquisition of protective immunity to subsequent challenge with virulent Salmonella [27]. We examined whether TIRAP is required during secondary infection by re-infecting Wt and TIRAP-deficient mice with virulent Salmonella, 50 days following administration of BRD509. Prior to infection a small cohort of Wt and TIRAP-deficient mice were sacrificed to confirm that primary clearance of BRD509 had occurred in both groups (data not shown). As expected, Wt and TIRAP-deficient mice succumbed rapidly to infection with virulent Salmonella with TIRAP-deficient mice being slightly more susceptible than Wt mice (Fig. 6). At this relatively high challenge dose a percentage of previously immunized Wt and TIRAP-deficient mice also succumbed to infection. However, both Wt and TIRAP-deficient mice displayed increased resistance to virulent infection as indicated by a delay in time of death and ultimate survival of around 40% of infected mice (Fig. 6). These data demonstrate that TIRAP is not required for the acquisition of protective immunity to virulent Salmonella.
Figure 5. TIRAP-deficient mice develop antibody responses after Salmonella infection.
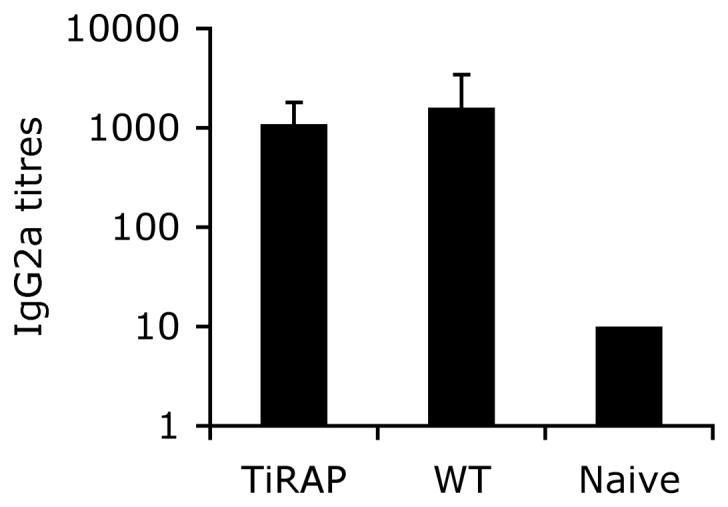
Serum was obtained from infected Wt or TIRAP-/- mice, 3 months after intravenous infection with 5×105 BRD509, and from naïve Wt mice. IgG2a responses to Salmonella were determined by ELISA. Data show mean Salmonella-specific IgG2a titers ± SD of 8-9 mice per group.
Figure 6. Development of protective immunity against virulent Salmonella in TIRAP-deficient mice.
C57BL/6 Wt and TIRAP-/- mice were infected intravenously with 5×105 attenuated Salmonella, BRD509 and 50 days later naïve or vaccinated mice were challenged intravenously with 1×105 virulent Salmonella SL1344. Cages were monitored daily for death or moribund mice. Data show percentage survival of mice using combined data from 2 individual experiments and at least 10 mice per group.
Discussion
TLR signaling deficiency has a long association with the mouse model of Salmonella infection [28], and MyD88-deficient mice certainly display increased susceptibility to infection [9]. TIRAP is an adaptor molecule that participates in MyD88-dependent TLR activation through TLR1/2, TLR2/6 and TLR4, but is not thought to be required for signaling through other TLRs [10]. Indeed, TIRAP deficient mice display normal inflammatory responses to ligands for TLR5, 7, and 9 but have defects in activation through TLR1, 2, 4 and 6 [14,15]. Thus TIRAP-deficient mice can potentially be used to determine whether certain groupings of TLRs are important for host defense. Recent reports have demonstrated that TIRAP is required for innate pulmonary defense against E. coli and Klebsiella pneumoniae, but not Pseudomonas aeruginosa [21,29]. Thus, certain pathogens appear predisposed to induce TIRAP-dependent signaling while others are not.
Our data demonstrate that TIRAP is not required for primary clearance of Salmonella infection in the mouse model, although these mice did display slightly increased susceptibility when challenged with virulent Salmonella. These data are surprising, particularly given the acknowledged importance of TIRAP to TLR4 signaling [15], the increased susceptibility of TLR4-deficient mice to Salmonella infection [5,6], and the requirement for TIRAP in defense against the closely related organism E. coli [29]. However, our data are consistent with a model where MyD88-independent signaling through TLR4 is sufficient to induce the required innate immune response to Salmonella in the absence of TIRAP. Indeed, TLR4 signaling is known to mediate dendritic cell maturation in the absence of TIRAP, although B cell proliferation and cytokine production are considerably diminished [14,15]. Furthermore, MyD88-independent activation is known to cause production of IFN-β, and it is possible that this cytokine contributes directly to Salmonella clearance in the absence of TIRAP. Alternatively, our data might imply that during Salmonella infection TIRAP-independent TLRs such as TLR5 and TLR9 are fully sufficient to induce maximal innate responses in vivo. Indeed, experiments examining epithelial cell interactions with Salmonella indicate that TLR5 is the major receptor involved in innate activation [30]. However, such a model would not explain the increased susceptibility of TLR4-deficient mice to Salmonella infection [5,6]. These explanations are not mutually exclusive, and we would suggest that a degree of compensation via other TLRs, plus a role for MyD88-independent TLR4 signaling are likely to explain the lack of a requirement for TIRAP during Salmonella infection. Whatever the explanation, our data demonstrate a surprising lack of involvement for a major TLR4 adaptor protein in innate immunity to Salmonella infection. However, our experiments cannot rule out a role for TIRAP when challenging with a lower infectious dose, or by a different route.
Our experiments actually uncovered more rapid bacterial clearance, resolution of splenic NK1.1 and CD11b cell infiltrates, and enhanced Salmonella-specific IFN-γ responses in TIRAP-deficient mice. This mildly enhanced response was only detected towards the late stage of bacterial clearance when adaptive immune responses mediate protective immunity [24,25]. However, it should also be noted that this small increase in IFN-γ production in TIRAP-deficient mice also correlates with a higher percentage of CD4 T cells in the spleen of these mice and may therefore simply be indicative of a higher frequency of Salmonella-specific Th1 cells in the in vitro culture system.
Although together our data suggested a mildly enhanced adaptive response to Salmonella in TIRAP-deficient mice, we detected no difference in Salmonella flagellin-specific CD4 T cell expansion, cell division, or memory frequency in TIRAP versus Wt mice. As our transferred T cells are TIRAP-sufficient, it is possible that T cell intrinsic TIRAP signaling is somehow detrimental to the development of IFN-γ -producing T cells. Indeed, recent data suggest an important contributory role for TLR activation in CD4 T cell activation [31]. However, we think it more likely that TIRAP deficiency simply forces innate immune activation through alternative TIRAP-independent TLRs and that these TLRs are slightly more efficient in generating an inflammatory response to clear bacteria. Thus, in Wt mice, TIRAP signaling probably does not contribute to bacterial clearance but may actually be a liability, hindering innate immune activation through more efficient TLRs. However, given the reported susceptibility of TIRAP-deficient mice to E. coli and Klebsiella pneumoniae [21,29], it seems clear that TIRAP is actively involved in signaling processes that contribute to clearance in other bacterial infections.
Prior exposure to attenuated Salmonella can confer enhanced immunity to challenge with virulent Salmonella [32]. Our data demonstrate that TIRAP is not required for the development of vaccine-induced immunity to Salmonella infection. Again these data are surprising given the proposed role of TLR4 in Salmonella immunity and the importance of TIRAP to TLR4 signaling. Together, our data demonstrate that TIRAP is not required for primary or vaccine-induced immunity to Salmonella infection, and indicate that TIRAP-independent pathways are fully sufficient, and may actually be more efficient in mediating bacterial clearance in this infection model.
Acknowledgements
We would like to thank Joseph Foley and Michael Jarcho for technical assistance. This work was supported by grants from National Institutes of Health (AI056172 and AI055743 to S.J.M).
Abbreviations
- HKST
Heat-killed Salmonella typhimurium
- Wt
Wild type
- TIRAP
Toll/IL-1R Domain-Containing Adaptor Protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of salmonella infections: enteritis versus typhoid fever. Micrtobes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- [2].Wick MJ. Living in the danger zone: innate immunity to Salmonella. Curr Opin Microbiol. 2004;7:51–7. doi: 10.1016/j.mib.2003.12.008. [DOI] [PubMed] [Google Scholar]
- [3].Srinivasan A, McSorley SJ. Activation of Salmonella-specific immune responses in the intestinal mucosa. Arch Immunol Ther Exp (Warsz) 2006;54:25–31. doi: 10.1007/s00005-006-0003-5. [DOI] [PubMed] [Google Scholar]
- [4].Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- [5].Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- [6].Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–52. [PubMed] [Google Scholar]
- [7].Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- [8].Gewirtz AT, Nava TA, Lyons S, Godowski PJ, Madera JL. Cutting Edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- [9].Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A. Toll-like receptors are temporally involved in host defense. J Immunol. 2004;172:4463–9. doi: 10.4049/jimmunol.172.7.4463. [DOI] [PubMed] [Google Scholar]
- [10].West AP, Koblansky AA, Ghosh S. Recognition and Signaling by Toll-Like Receptors. Annu Rev Cell Dev Biol. 2006;22:409–37. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- [11].Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18- mediated function. Immunity. 1998;9:143–50. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- [12].Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–22. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- [13].Horng T, Barton GM, Medzhitov R. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2:835–41. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- [14].Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–9. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- [15].Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–33. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- [16].Laroux FS, Romero X, Wetzler L, Engel P, Terhorst C. Cutting edge: MyD88 controls phagocyte NADPH oxidase function and killing of gram-negative bacteria. J Immunol. 2005;175:5596–600. doi: 10.4049/jimmunol.175.9.5596. [DOI] [PubMed] [Google Scholar]
- [17].Sebastiani G, Leveque G, Lariviere L, Laroche L, Skamene E, Gros P, et al. Cloning and characterization of the murine toll-like receptor 5 (Tlr5) gene: sequence and mRNA expression studies in Salmonella-susceptible MOLF/Ei mice. Genomics. 2000;64:230–40. doi: 10.1006/geno.2000.6115. [DOI] [PubMed] [Google Scholar]
- [18].Angers I, Sancho-Shimizu V, Descoteaux A, Gewirtz AT, Malo D. Tlr5 is not primarily associated with susceptibility to Salmonella Typhimurium infection in MOLF/Ei mice. Mamm Genome. 2006;17:385–97. doi: 10.1007/s00335-005-0132-x. [DOI] [PubMed] [Google Scholar]
- [19].Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–74. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- [20].Feuillet V, Medjane S, Mondor I, Demaria O, Pagni PP, Galan JE, et al. Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc Natl Acad Sci U S A. 2006;103:12487–92. doi: 10.1073/pnas.0605200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jeyaseelan S, Young SK, Yamamoto M, Arndt PG, Akira S, Kolls JK, et al. Toll/IL-1R domain-containing adaptor protein (TIRAP) is a critical mediator of antibacterial defense in the lung against Klebsiella pneumoniae but not Pseudomonas aeruginosa. J Immunol. 2006;177:538–47. doi: 10.4049/jimmunol.177.1.538. [DOI] [PubMed] [Google Scholar]
- [22].McSorley SJ, Asch S, Costalonga M, Rieinhardt RL, Jenkins MK. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- [23].Srinivasan A, Foley J, Ravindran R, McSorley SJ. Low-dose salmonella infection evades activation of flagellin-specific CD4 T cells. J Immunol. 2004;173:4091–9. doi: 10.4049/jimmunol.173.6.4091. [DOI] [PubMed] [Google Scholar]
- [24].Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA-infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J. Immunol. 1996;156:3321–3326. [PubMed] [Google Scholar]
- [25].Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175:4603–10. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- [26].Srinivasan A, Foley J, McSorley SJ. Massive Number of Antigen-Specific CD4 T Cells during Vaccination with Live Attenuated Salmonella Causes Interclonal Competition. J Immunol. 2004;172:6884–93. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- [27].McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:3344–8. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Plant J, Glynn AA. Natural resistance to Salmonella infection, delayed hypersensitivity and Ir genes in different strains of mice. Nature. 1974;248:345–7. doi: 10.1038/248345a0. [DOI] [PubMed] [Google Scholar]
- [29].Jeyaseelan S, Manzer R, Young SK, Yamamoto M, Akira S, Mason RJ, et al. Toll-IL-1 receptor domain-containing adaptor protein is critical for early lung immune responses against Escherichia coli lipopolysaccharide and viable Escherichia coli. J Immunol. 2005;175:7484–95. doi: 10.4049/jimmunol.175.11.7484. [DOI] [PubMed] [Google Scholar]
- [30].Zeng H, Carlson AQ, Guo Y, Yu Y, Collier-Hyams LS, Madara JL, et al. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J Immunol. 2003;171:3668–74. doi: 10.4049/jimmunol.171.7.3668. [DOI] [PubMed] [Google Scholar]
- [31].Gelman AE, Larosa DF, Zhang J, Walsh PT, Choi Y, Sunyer JO, et al. The Adaptor Molecule MyD88 Activates PI-3 Kinase Signaling in CD4(+) T Cells and Enables CpG Oligodeoxynucleotide-Mediated Costimulation. Immunity. 2006;25:783–93. doi: 10.1016/j.immuni.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hoiseth SK, Stocker BAD. Aromatic-dependent salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]