Abstract
Background
Atypical bovine spongiform encephalopathies (BSEs) are recently recognized prion diseases of cattle. Atypical BSEs are rare; approximately 30 cases have been identified worldwide. We tested prion gene (PRNP) haplotypes for an association with atypical BSE.
Methodology/Principle Findings
Haplotype tagging polymorphisms that characterize PRNP haplotypes from the promoter region through the three prime untranslated region of exon 3 (25.2 kb) were used to determine PRNP haplotypes of six available atypical BSE cases from Canada, France and the United States. One or two copies of a distinct PRNP haplotype were identified in five of the six cases (p = 1.3×10−4, two-tailed Fisher's exact test; CI95% 0.263–0.901, difference between proportions). The haplotype spans a portion of PRNP that includes part of intron 2, the entire coding region of exon 3 and part of the three prime untranslated region of exon 3 (13 kb).
Conclusions/Significance
This result suggests that a genetic determinant in or near PRNP may influence susceptibility of cattle to atypical BSE.
Introduction
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are infectious, invariably fatal neurodegenerative disorders that occur in humans, ruminants, cats, and mink [1]. TSEs are unique in their ability to manifest through acquired, inherited, and sporadic routes [1]. Classical bovine spongiform encephalopathy (BSE) is an acquired cattle TSE of unknown origin that spreads through the consumption of meat and bone meal contaminated with the infectious prion agent [2]. Classical BSE is accepted as the probable cause of the human TSE variant Creutzfeldt-Jakob Disease (CJD) [3], [4]. Two BSEs distinct from classical BSE, so called “atypical BSEs” (H-type and L-type) have recently been identified in Asian, North American and European cattle [2]. Approximately, 30 atypical BSEs have been identified worldwide and their etiology is unclear.
Variation in the prion gene (PRNP) correlates with TSE susceptibility in some mammals including cattle [1], [5]–[7]. The deletion alleles of two bovine PRNP insertion/deletion polymorphisms, one within the promoter region and the other in intron 1, associate with classical BSE susceptibility [5]–[7]. These same alleles do not correlate with atypical BSE susceptibility [8]. In 2006, a United States atypical BSE case was identified and subsequently found to have a PRNP nonsynonymous polymorphism (E211K) that is homologous to the human PRNP E200K polymorphism (observation by J.A.R). The human K200 allele is a highly-penetrant risk factor for genetic CJD [9]. To date, the K211 allele has not been observed in other atypical BSE cases or reported in healthy cattle [10], [11]. Thus, while the K211 allele may have been a genetic cause for one case of atypical BSE, it has not accounted for the majority of atypical cases. Consequently, any association of PRNP alleles with atypical BSE was largely unknown prior to this study.
PRNP variation in cattle is complex. Bovine PRNP polymorphism alleles reflect a region of high linkage disequilibrium (LD) from the promoter through a portion of intron two, and a region of low LD from intron two past the three prime untranslated region. This genetic architecture is present across populations of Bos taurus breeds and a similar trend has been observed in a small sampling of Bos indicus influenced breeds [11]. A set of 19 haplotype tagging polymorphisms (htSNPS) was previously developed that accounts for the genetic architecture of PRNP and characterizes haplotype diversity within and across PRNP [11]. In this study, we used the htSNPs to test PRNP haplotypes for an association with atypical BSE and report the association of a relatively uncommon PRNP haplotype with atypical BSE.
Results and Discussion
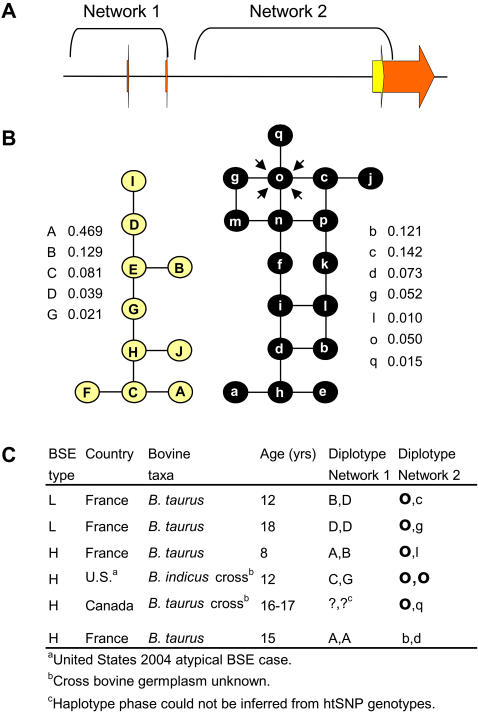
The 19 PRNP htSNPs were used to determine PRNP haplotypes of six available atypical BSE cases that originated from Canada, France and the United States. The haplotypes were phased in previously defined PRNP regions of high and low LD (Fig. 1A; network 1 spans the high LD region, network 2 spans the low LD region). Additionally, the entire prion protein (PrP) coding region was sequenced for each of the six atypical BSE cases. None of the cases contained previously unknown SNP alleles in the PrP coding region or the K211 allele. However, one or two copies of a distinct haplotype were identified by haplotype reconstructions in five of the six cases. The haplotype spans a portion of intron 2, the entire coding region, and a portion of the 3′ UTR of PRNP (13 kb), (haplotype “o”, Fig. 1B and 1C, Table 1).
Figure 1. Prion haplotypes of atypical BSE cases.
(A) Physical map of bovine PRNP. Orange and yellow arrows represent untranslated and protein coding regions, respectively. PRNP regions spanned by prion haplotypes are indicated by brackets labeled network 1 and network 2. (B) PRNP haplotype relationships and frequencies in U.S. cattle. Haplotypes in network 1 are represented as yellow circles, are defined by 9 htSNPs and span a portion of the PRNP promoter, exon 1, intron 1, exon 2, and a small portion of intron 2 (6.3 kb). Haplotypes in network 2 are represented as black circles, are defined by 10 htSNPs and span most of intron 2, the entire coding region, and a portion of the 3′ UTR of PRNP (13 kb). Numbers represent the frequencies of PRNP haplotypes in the control group of U.S. cattle. (C) Atypical BSE case information. BSE type, country of origin, bovine taxa, age, and PRNP diplotyes of the atypical BSE cases.
Table 1. PRNP haplotype sequences in Networks 1 and 2.
| Haplotype tagging polymorphisms | |||||||||||||||||||
| Network 1 | Network 2 | ||||||||||||||||||
| Haplotype | tattt-(T,G)-gtctca | taccc-(C,T)-aaatg | gcttc-(C,T)-tatca | atcta-(A,G)-ttcac | cgact-(C,T)-acccg | tgggc(I,Z)-tggctb | ggagc-(G,A)-tccgt | agagg-(C,T)-ggccc | acaga-(A,T)-gataa | ggaat-(G,A)-tgtat | ttagg-(T,C)-ggcatc | tcttt-(T,C)-ttttt | tattc-(A,G)-gttac | tcttg-(G,C)-ggggg | tatag-(C,G)-tcaaa | gagtc-(G,A)-gacac | aaacc-(C,T)-agtaa | gtcaa-(C,T)-atcac | actta-(C,T)-gggya |
| 449d | 1392 | 1576 | 1701 | 4136 | 4450e | 4732 | 4776 | 6811 | 8631 | 9162 | 9786 | 13793 | 13861 | 13925 | 17284 | 20720 | 20957 | 21680 | |
| A | T | C | C | G | C | Z | G | C | A | —f | — | — | — | — | — | — | — | — | — |
| B | G | C | C | A | C | I | A | T | A | — | — | — | — | — | — | — | — | — | — |
| C | T | C | C | A | C | Z | G | C | A | — | — | — | — | — | — | — | — | — | — |
| D | G | C | C | A | T | I | A | C | A | — | — | — | — | — | — | — | — | — | — |
| E | G | C | C | A | C | I | A | C | A | — | — | — | — | — | — | — | — | — | — |
| F | T | C | C | A | C | Z | G | C | T | — | — | — | — | — | — | — | — | — | — |
| G | G | C | C | A | C | I | G | C | A | — | — | — | — | — | — | — | — | — | — |
| H | T | C | C | A | C | I | G | C | A | — | — | — | — | — | — | — | — | — | — |
| I | G | T | C | A | T | I | A | C | A | — | — | — | — | — | — | — | — | — | — |
| J | T | C | T | A | C | I | G | C | A | — | — | — | — | — | — | — | — | — | — |
| a | — | — | — | — | — | — | — | — | — | A | T | T | G | C | G | G | T | C | T |
| b | — | — | — | — | — | — | — | — | — | A | T | T | G | C | C | G | C | C | C |
| c | — | — | — | — | — | — | — | — | — | G | T | C | A | G | C | G | C | C | C |
| d | — | — | — | — | — | — | — | — | — | A | T | T | G | C | C | G | C | C | T |
| e | — | — | — | — | — | — | — | — | — | A | T | T | G | C | G | G | C | T | T |
| f | — | — | — | — | — | — | — | — | — | G | T | T | A | C | C | G | C | C | T |
| g | — | — | — | — | — | — | — | — | — | G | C | C | A | G | C | G | C | C | T |
| h | — | — | — | — | — | — | — | — | — | A | T | T | G | C | G | G | C | C | T |
| i | — | — | — | — | — | — | — | — | — | A | T | T | A | C | C | G | C | C | T |
| j | — | — | — | — | — | — | — | — | — | G | T | C | A | G | C | A | C | C | C |
| k | — | — | — | — | — | — | — | — | — | A | T | T | A | G | C | G | C | C | C |
| l | — | — | — | — | — | — | — | — | — | A | T | T | A | C | C | G | C | C | C |
| m | — | — | — | — | — | — | — | — | — | G | C | T | A | G | C | G | C | C | T |
| n | — | — | — | — | — | — | — | — | — | G | T | T | A | G | C | G | C | C | T |
| o | — | — | — | — | — | — | — | — | — | G | T | C | A | G | C | G | C | C | T |
| p g | — | — | — | — | — | — | — | — | — | G | T | T | A | G | C | G | C | C | C |
| q | — | — | — | — | — | — | — | — | — | G | T | C | A | C | C | G | C | C | T |
Haplotype tagging polymorphism is enclosed in parenthesis. Five nucleotides of flanking sequence on the 5′ and 3′ ends are included from bottom to top, respectively.
Twelve-base InDel, I = GGGGGCCGCGGC, Z = deletion.
One or two G nucleotides immediately adjacent (5′ end) to the haplotype tagging polymorphism have been observed in U.S. cattle.
Nucleotide position in GenBank accession # DQ457195.
The insertion polymorphism allele spans nucleotide 4450 through 4461.
Not applicable.
Inferred haplotype.
The frequency of the implicated haplotype in atypical BSE cases was compared to its frequency in a control group of 114 diverse DNA samples representing 21 breeds of U.S. beef and dairy cattle, since unaffected controls from the farms where the atypical BSE cases originated are not available, nor are diversity panels of beef and dairy cattle in Canada and France. However, the control group of U.S. cattle represents germplasm that is collectively found in Canada, France, and the United States, and current evidence from the international bovine HapMap project indicates that diversity within Bos taurus breeds is similar between countries (personal communication from T.P.L.S.). Therefore, we used the group of U.S. cattle as a surrogate control in this study which involves natural occurrences of atypical BSE cases from three countries on two different continents. The implicated haplotype was observed in both Bos taurus and Bos indicus individuals in the control group and had a frequency of 0.050, ten-fold less than the atypical BSE-cases (frequency = 0.50). A Fisher's exact two-tailed test showed a significant association of the haplotype with atypical BSE (p = 1.3×10−4), as did the difference between proportions (CI95% 0.263–0.901).
This result suggests that a genetic determinant in or near PRNP may influence susceptibility of cattle to atypical BSE. The causative allele(s) remains to be identified and probably occurs on the background of the implicated PRNP haplotype. Complete sequencing of PRNP from atypical BSE cases and BSE negative controls that both have the implicated haplotype may reveal PRNP alleles with predictive power for atypical BSE. The implicated haplotype itself does not effectively predict atypical BSE because of its frequency in healthy cattle. However, our results combined with the discovery of the PRNP K211 allele suggest that atypical BSE may be managed through the identification of cattle with known genetic risk factors for the disease and their removal from livestock populations.
Materials and Methods
Composition of atypical BSE group
Atypical BSE cases were selected for this study solely on the basis of available DNA for PRNP sequencing and genotyping. DNA samples were obtained from six unrelated BSE cases confirmed as atypical H or L type BSE by Western blot profile (high or low molecular mass of unglycosylated protease-resistant prion protein (PrPres) [12]–[14]. Two atypical L-type and two atypical H-type BSE cases originated in France. Two additional atypical H-type BSE cases originated from Canada and the United States.
Composition of cattle control group
Samples from two cattle DNA diversity panels were used to construct the cattle control group; the U.S. Meat Animal Research Center (USMARC) Beef Cattle Discovery Panel 2.1 (MBCDP2.1) [15] and the USMARC Dairy Cattle Panel (MDCP1.5) [11], [16]. Breeds in this group include Angus (n = 8) Hereford (n = 8), Limousin (n = 8), Simmental (n = 7), Charolais (n = 6), Beefmaster (n = 5), Red Angus (n = 6), Gelbvieh (n = 6), Brangus (n = 5), Salers (n = 5), Brahman (n = 6), Shorthorn ( n = 5), Maine-Anjou (n = 5), Longhorn (n = 4), St. Gertrudis (n = 4), Chianina (n = 4), Holstein (n = 8), Jersey (n = 7), Guernsey (n = 3), Aryshire (n = 2), and Brown Swiss (n = 2). A total of 21 breeds and 114 individuals are represented in the group.
PRNP amplification and sequence-based genotyping of htSNPs
Twelve segments of PRNP were amplified for sequence-based genotyping of 19 htSNPs (Table S1). In addition, the complete prion protein coding region was sequenced. All but two PRNP segments were amplified with the following reagents (per 55 uL reaction), 1.25 units of Thermo-Start DNA Polymerase, 2.3 mM MgCl2, 0.181 mM dNTPs, 0.4 uM forward and reverse amplification primer, and 50 ng genomic DNA. Two segments were amplified with identical concentrations of Taq, dNTPs, primers, and genomic DNA as described above. However, one segment, BTAPRNPDS13a2, was amplified with 1.36 mM MgCl2 and 3% DMSO and the other, segment BTAPRNPDS13b, was amplified with 1.36 mM MgCl2 and 2% DMSO. PCR conditions for the 12 segments were the following: 94°C for 15 min, 40 cycles of 94°C for 20 sec, 58°C for 30 sec (excluding BTAPRNPDS13a2), 72°C for 60 sec, and a final incubation at 72°C for 3 minutes. The primer extension temperature for segment BTAPRNPDS13a2 was conducted at 53°C for 30 sec. Following an Exonuclease I digestion [17], the amplicons were sequenced with BigDye terminator chemistry on an ABI 3730 capillary sequencer (PE Applied Biosystems, Foster City, California). All sequencing primers listed in Table S1 were used in duplicate or quadruplicate for each atypical BSE sample to obtain multiple genotypes of each htSNP.
SNP genotyping, haplotype phasing and statistical testing
PRNP sequences were processed for polymorphism detection and genotyping with Phred, Phrap, Polyphred, and Consed software [18]. Haplotype phase was determined with Phase (version 2.1) [19], [20]. The frequencies of PRNP haplotype “o” in the atypical BSE case group and the control group were tested for significance with a Fisher's exact two-tailed test in WinPepi (version 4.5) [21]. The 95% confidence interval for the difference between the frequency proportions with continuity correction was also calculated in WinPepi.
Supporting Information
Oligonucleotides for amplficiation and sequence genotyping of PRNP htSNPs and the complete PrP coding region. This table lists the oligonucleotides used for amplification and sequence genotyping of PRNP htSNPs and the complete PrP coding region.
(1.66 MB XLS)
Acknowledgments
The authors thank Gennie Schuller-Chavez for outstanding technical assistance and Joan Rosch for secretarial assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the USDA National Research Initiative, Competitive Grant No. 2005-35212-15890 and the Agricultural Research Service.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P, McShane LM, Zanusso G, Detwiler L. On the question of sporadic or atypical bovine spongiform encephalopathy and Creutzfeldt-Jakob disease. Emerg Infect Dis. 2006;12:1816–1821. doi: 10.3201/eid1212.060965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, et al. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 4.Hill AF, Desbruslais M, Joiner S, Sidle KC, Gowland I, et al. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 5.Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, et al. Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics. 2004;5:19–25. doi: 10.1007/s10048-003-0171-y. [DOI] [PubMed] [Google Scholar]
- 6.Juling K, Schwarzenbacher H, Williams JL, Fries R. A major genetic component of BSE susceptibility. BMC Biol. 2006;4:33. doi: 10.1186/1741-7007-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haase B, Doherr MG, Seuberlich T, Drögemüller C, Dolf G, et al. PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genet. 2007;8:15. doi: 10.1186/1471-2156-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunelle BW, Hamir AN, Baron T, Biacabe AG, Richt JA, et al. Polymorphisms of the prion gene promoter region that influence classical bovine spongiform encephalopathy susceptibility are not applicable to other transmissible spongiform encephalopathies in cattle. J Anim Sci. 2007;85:3142–3147. doi: 10.2527/jas.2007-0208. [DOI] [PubMed] [Google Scholar]
- 9.Kovács GG, Puopolo M, Ladogana A, Pocchiari M, Budka H, et al. Genetic prion disease: the EUROCJD experience. Hum Genet. 2005;118:166–174. doi: 10.1007/s00439-005-0020-1. [DOI] [PubMed] [Google Scholar]
- 10.Heaton MP, Leymaster KA, Freking BA, Hawk DA, Smith TPL, et al. Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer. Mamm Genome. 2003;14:765–777. doi: 10.1007/s00335-003-2283-y. [DOI] [PubMed] [Google Scholar]
- 11.Clawson ML, Heaton MP, Keele JW, Smith TPL, Harhay GP, et al. Prion gene haplotypes of U.S. cattle. BMC Genet. 2006;7:51. doi: 10.1186/1471-2156-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron T, Biacabe A-G, Arsac J-N, Benestad S, Groschup MH. Atypical transmissible spongiform encephalopathies (TSEs) in ruminants. Vaccine. 2006;25:5625–5630. doi: 10.1016/j.vaccine.2006.10.058. [DOI] [PubMed] [Google Scholar]
- 13.Biacabe A-G, Laplanche J-L, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004;5:110–115. doi: 10.1038/sj.embor.7400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richt JA, Kunkle RA, Alt D, Nicholson EM, Hamir AN, et al. Identification and characterization of two bovine spongiform encephalopathy cases diagnosed in the United States. J Vet Diagn Invest. 2007;19:142–154. doi: 10.1177/104063870701900202. [DOI] [PubMed] [Google Scholar]
- 15.Heaton MP, Chitko-McKown CG, Grosse WM, Keele JW, Keen JE, et al. Interleukin-8 haplotype structure from nucleotide sequence variation in commercial populations of U.S. beef cattle. Mamm Genome. 2001;12:219–226. doi: 10.1007/s003350010269. [DOI] [PubMed] [Google Scholar]
- 16.Heaton MP, Keen JE, Clawson ML, Harhay GP, Bauer N, et al. Use of bovine single nucleotide polymorphism markers to verify sample tracking in beef processing. J Am Vet Med Assoc. 2005;226:1311–1314. doi: 10.2460/javma.2005.226.1311. [DOI] [PubMed] [Google Scholar]
- 17.Smith TP, Godtel RA, Lee RT. PCR-based setup for high-throughput cDNA library sequencing on the ABI 3700 automated DNA sequencer. Biotechniques. 2000;29:698–700. doi: 10.2144/00294bm05. [DOI] [PubMed] [Google Scholar]
- 18.Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- 19.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J of Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J of Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abramson JH. WINPEPI (PEPI-for-Windows): computer programs for epidemiologists. Epidemiol Perspect Innov. 2004;1:6. doi: 10.1186/1742-5573-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides for amplficiation and sequence genotyping of PRNP htSNPs and the complete PrP coding region. This table lists the oligonucleotides used for amplification and sequence genotyping of PRNP htSNPs and the complete PrP coding region.
(1.66 MB XLS)