Abstract
The mechanism of mRNA export is a complex issue central to cellular physiology. We characterized previously yeast Gle1p, a protein with a leucine-rich (LR) nuclear export sequence (NES) that is essential for poly(A)+ RNA export in Saccharomyces cerevisiae. To characterize elements of the vertebrate mRNA export pathway, we identified a human homologue of yeast Gle1p and analyzed its function in mammalian cells. hGLE1 encodes a predicted 75-kDa polypeptide with high sequence homology to yeast Gle1p, but hGle1p does not contain a sequence motif matching any of the previously characterized NESs. hGLE1 can complement a yeast gle1 temperature-sensitive export mutant only if a LR-NES is inserted into it. To determine whether hGle1p played a role in nuclear export, anti-hGle1p antibodies were microinjected into HeLa cells. In situ hybridization of injected cells showed that poly(A)+ RNA export was inhibited. In contrast, there was no effect on the nuclear import of a glucocorticoid receptor reporter. We conclude that hGle1p functions in poly(A)+ RNA export, and that human cells facilitate such export with a factor similar to yeast but without a recognizable LR-NES. With hGle1p localized at the nuclear pore complexes, hGle1p is positioned to act at a terminal step in the export of mature RNA messages to the cytoplasm.
The nuclear export of proteins and ribonucleoprotein (RNP) particles through the nuclear pore complex (NPC) is a facilitated and signal-dependent process (1–3). Moreover, RNA processing and transport events are tightly coupled, such that splicing, polyadenylation, and capping all affect the export process (4–9). Throughout the processing and exit pathway, RNA is bound by distinct proteins and the critical signals for export are predicted to reside on these proteins (1, 2, 10). This has been demonstrated clearly in studies of the HIV-1 Rev protein, which specifically binds unspliced viral RNA (11–13). The RNA-binding domain of Rev is distinct from a region containing a leucine-rich (LR) nuclear export sequence (NES), which is both necessary and sufficient for mediating nuclear export (14, 15). The LR-NES is recognized in the nucleoplasm by a nuclear export receptor, Crm1p/exportin (16), that is a member of a family of β nuclear transport factors (17, 18). Thus, the NES of Rev directs export of the protein and bound viral RNA coincidentally through the NPC by interaction with an exporting β.
In vertebrate cells, different RNA classes are exported by independent pathways with each RNA type (mRNA, U snRNA, tRNA, or rRNA) potentially requiring at least a subset of distinct factors (reviewed in refs. 1, 2, and 19). U snRNA and 5S rRNA export may require proteins with LR-NESs that are recognized by Crm1p/exportin (14, 16, 20). Interestingly, mRNA export may also utilize aspects of the LR-NES machinery (21). The temperature-dependent poly(A)+ RNA export defects in two β yeast mutants (pse1 and crm1/xpo1) suggest these β family members may be involved (22, 23).
Given the complexity of the mRNA maturation and export process, the combined actions of several different proteins may determine the rate of mRNA export. Candidates for mRNA export mediators include a specific subset of heterogeneous nuclear (hn) RNP proteins that shuttle between the nucleus and cytoplasm (1, 2). The shuttling hnRNP A1 and hnRNP K proteins contain both RNA-binding motifs and unique NESs (24–26). These properties position A1 and/or K to function as active carriers by association with mRNA in the nucleus, export via their NESs, and release of the mRNA cargo in the cytoplasm. The observation that mutant alleles of the gene encoding the shuttling yeast hnRNP-like Npl3p are deficient for poly(A)+RNA export supports this hypothesis (27). Interestingly, the NESs in A1 (termed M9) and K (termed KNS) are not related in primary amino acid sequence to one another, or to LR-NESs (25, 26). It is unknown how the NES signals in the hnRNP proteins are utilized during export. The nuclear import of A1 requires recognition of the M9 domain by the β family member transportin (28). Although transportin also could play a role in A1 nuclear export via the M9 domain, it is probably not sufficient. Yeast mutants of Kap104p, the transportin homologue, do not exhibit a primary defect in poly(A)+ RNA export (29), and recent microinjection studies have concluded that M9 recognition during mRNA export differs from that during protein import (30). Thus, an as yet unidentified intermediary between the NPCs and hnRNPs is required for exit from the nucleus.
We and others have used genetic approaches in the yeast Saccharomyces cerevisiae to dissect the pathway of nucleocytoplasmic transport (31, 32). The yeast protein Gle1p, also identified as Rss1p, is a LR-NES factor essential for poly(A)+ RNA export (33, 34). To characterize elements of the vertebrate mRNA export pathway and analyze potential mechanistic differences between species, we identified a human homologue of yeast Gle1p and analyzed its function in mammalian cells. Our findings suggest hGle1p plays a role in poly(A)+ RNA export from the nucleus.
MATERIALS AND METHODS
Cloning and Sequencing of hGle1p.
Residues 250–538 of yeast Gle1p were used to search the dbBEST database with blast (35). Two cDNA clones were detected with the same small blocks of homology: human heart cDNA clone A235F (accession no. T12405) and embryonic mouse carcinoma cDNA clone 84C06 (accession no. D21730). A third clone has since been deposited: mouse cDNA clone 583731 (accession no. AA1344115). Translation of A235F and 84C06 in all three reading frames revealed additional blocks of homology, indicating a possible sequencing frameshift. Clone A235 also had additional 3′ sequence information, A235R (accession no. T12406), with homology to the extreme carboxyl terminus of yeast Gle1p. Additional clones were identified by searching with the A235R sequence, including the human infant brain cDNA clone 31740 (accession no. R41973). Sequence from the 5′ end of clone 31740 (accession no. R17293) was analyzed and identified the slightly longer human infant brain clone 22734 (accession no. T75196). Clones 22734 and 31740 were obtained (Genome Systems, St. Louis) and full-length inserts were sequenced by the dideoxychain termination method by using appropriate oligonucleotides. Clone 31740 begins at amino acid no. 35 of the ORF in clone 22734.
Sequencing of Yeast gle1 Alleles.
The gle1–1, gle1–2, gle1–3, gle1–4, and gle1–5 alleles were isolated from the mutant strains onto LEU2/CEN plasmids by transforming a PstI/BspE1-linearized pSW397 (GLE1/LEU2/CEN) (33) into the strains. Leu+ transformants were selected, and the respective plasmids were isolated, checked for the presence of a full-length gene, and tested for the inability to complement a gle1 strain. Plasmid inserts were sequenced by the DyeDeoxy terminator method using an Applied Biosystems 373 Automated DNA Sequencer. The gle1–1 and gle1–2 alleles have identical nucleotide substitutions (CCC → CTC) in the codon for amino acid no. 380 resulting in P380L. The gle1–3, gle1–4, and gle1–5 alleles contain a nucleotide change (GGA → AGA) in the codon for amino acid no. 384 resulting in G382R.
Yeast Strains and Plasmids.
Yeast strain SWY1191 (gle1–4) (33) was grown in either YEP (1% yeast extract, 2% peptone) or synthetic minimal medium plus appropriate amino acids supplemented with 2% glucose. Yeast transformations were performed by the lithium acetate method. pSW398 (GLE1/LEU2/CEN) was used to generate the constructs in Fig. 3. The yeast Gle1p deletion constructs in pRS315 were generated by ligation of PCR products representing the 5′ and 3′ fragments with unique NcoI sites at the internal deletion point. The yeast–human chimeras were from subsequent insertion into the respective deletion construct of an NcoI fragment (in-frame “CAT”) produced by PCR. Site-directed mutagenesis of the human GLE1 gene was conducted as described (36). The hGle1+NES mutant results in the following amino acid changes: F500L, I502L, A503G, V504K, V505L, A506T, and S507L. DNA sequence analysis confirmed the mutations.
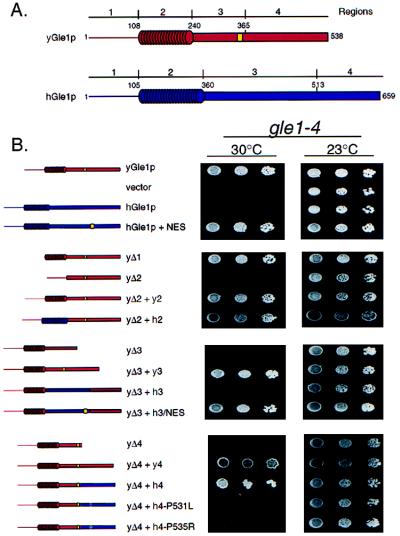
Figure 3.
Complementation analysis of yeast gle1–4 cells with hGLE1 chimeras. (A) Schematic representation of the structural regions in yeast (red) and human (blue) Gle1p proteins, with the LR-NES designated (yellow). (B) Yeast gle1–4 strains expressing the designated yeast deletion (Δ) or yeast–human chimeric Gle1p were grown at 23°C (Right) or 30°C (Left). Serial dilutions (from left to right) of equivalent cell numbers were plated on synthetic minimal medium lacking leucine. Expression of full-length yeast Gle1p supports growth at 30°C (top row), whereas vector alone does not (second row). Immunoblot analysis confirmed expression of all polypeptides (data not shown).
Production of Anti-hGle1p Antibodies and GST-hGle1p.
A 2.1-kbp PCR product generated with clone 22734 and oligonucleotides HGLE1–5′ (5′-GGGGGATCCCAATGCCGTCTGAGGGTCG-3′) and HGLE1–3′ (5′-GGGGGATCCCGAGAGGGACTGGACTA-3′) was cleaved with BamHI and inserted into pGEX-3X (Pharmacia) (pSW728). Glutathione S-transferase (GST)-hGle1p fusion protein was expressed in DH5α cells and purified by using glutathione-agarose (Sigma) as described (37). Purified GST-hGle1p protein was sent to Cocalico Biologicals, Inc. (Reamstown, PA) for production of antiserum WU1041. The hGLE1 ORF was fused in-frame to maltose-binding protein (MBP) in a similar manner by insertion into pMAL-cRI (New England Biolabs) (pSW771). MBP-hGle1p fusion protein was expressed in DH5α cells and purified by using amylose resin (New England Biolabs) according to the manufacturer’s directions. MBP-hGle1p was coupled to Affi-Gel 10 (Bio-Rad) and used to affinity-purify WU1041. Rabbit anti-MBP antibodies were produced (antiserum WU600) and affinity-purified by using MBP expressed from pMAL-cRI. Antibodies and proteins were concentrated by centrifugation in a Centricon-50 (Amicon). Total HeLa cell extracts were prepared from a 100-mm dish of cells solubilized in 1 ml cold 1% Triton X-100/PBS and analyzed by immunoblot with anti-hGle1p antibody (1:2,000) and anti-MBP antibody (1:2,000) (1 hr at room temperature) as described (37).
Microinjection and Tissue Culture.
HeLa cells were maintained in DMEM (high glucose, with l-glutamine and pyridoxine hydrochloride), 10% calf serum, 1 mM sodium pyruvate, 0.05 mM 2-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin, and grown on 12-mm circular coverslips. For expression of glucocorticoid receptor–green fluorescent protein (GR-GFP), the cells were grown in DMEM lacking phenol red with serum depleted by incubation with activated charcoal and were transiently transfected with Qiagen-purified DNA of plasmid pK7.GR.GFP (38) as described (39). Six hours after transfection, cells were rinsed with fresh medium and incubated an additional 16–20 hr before microinjection or fluorescence analysis. Dexamethasone was added at a final concentration of 10 μg/ml. Cells were microinjected by using a Narashigi micromanipulator and a constant-flow pipette with a 1:1 mixture of Texas red–dextran (70 kDa, lysine-fixable, Molecular Probes) and the respective antibody or protein mixtures. Microinjected cells were examined directly for the import of GR-GFP (transfected cells) or processed for in situ hybridization (untransfected cells).
Immunofluorescence and in Situ Hybridization.
Cells were processed by three different protocols: Fix-Triton, fixed for 30 min in cold 3% formaldehyde/buffer P (10 mM sodium phosphate, pH 7.4/150 mM NaCl/2 mM MgCl2), quenched with 0.05 M ammonium chloride/PBS for 10 min, and permeabilized by incubation for 15 min in cold 0.2% Triton X-100/buffer P; Digitonin-Fix-Triton, rinsed with PBS, with cold buffer A (20 mM Hepes-KOH, pH 7.4/110 mM potassium acetate/2 mM magnesium acetate/1 mM EGTA/2 mM DTT), then permeabilized with buffer A containing 35 μg/ml digitonin for 5 min, rinsed in buffer A alone, and then treated as above in “Fix-Triton”; Digitonin-Fix, treated as for “Digitonin-Fix-Triton” except the Triton X-100 step was eliminated. For all protocols, cells were blocked for 30 min in 2% BSA/PBS and incubated with primary antibodies diluted in block for 2 hr: rabbit anti-hGle1p (1:250, affinity-purified WU1041), mouse mAb414 (1:10, tissue culture supernatant) (40), and rabbit anti-lamin B (1:50, anti-peptide NC-6 serum) (41). Cells were washed three times with buffer P and then incubated for 1 hr at room temperature with secondary antibodies diluted in 0.1% BSA/buffer P: Cy3-labeled anti-rabbit IgG (1:500), fluorescein isothiocyanate-labeled anti-rabbit IgG (1:200), or rhodamine-labeled anti-mouse IgG (1:200) (Cappel). For in situ hybridizations, HeLa cells were fixed for 20 min in cold 3% formaldehyde in PBS, washed 3 × 5 min with cold PBS, permeabilized for 10 min in cold 0.5% Triton X-100/PBS, and washed with PBS and then 2× SSC. In situ hybridization and immunolocalization steps were conducted as described (42). Coverslips were mounted with 90% glycerol plus 1 mg/ml p-phenylenediamine. Photographs were taken with the 40× objective on an Olympus microscope with Kodak T-MAX 400 film.
RESULTS
Identification of a Human Protein with Homology to Yeast Gle1p.
To determine whether a human homologue of the yeast Gle1p was present in the available sequence databases, we conducted a series of searches with the blast program (35). Yeast Gle1p contains at least three distinct structural regions: an N-terminal, ≈100-residue region, a middle, highly charged region with predicted coiled-coil structure, and a C-terminal region that includes an essential LR-NES (33, 34). Computer analysis revealed numerous matches with the highly charged coiled-coil region of yeast Gle1p. However, none of the matches to this middle region appeared significant. In contrast, when the C-terminal 250-aa residues of yeast Gle1p were used to search the dbEST database, human and mouse sequence fragments were found that would encode proteins with high similarity to yeast Gle1p (see Materials and Methods). The complete DNA sequence was determined for a human gene in cDNA IMAGE clone 22734. The 2.2-kbp insert contains an ORF with stop codons 5′ to that for the first in-frame methionine residue and a stretch of 3′ A residues, consistent with priming at the polyadenylated tail. This indicates that the cDNA is a complete clone. The ORF of 659 aa encodes a predicted protein of ≈75 kDa (Fig. 1A).
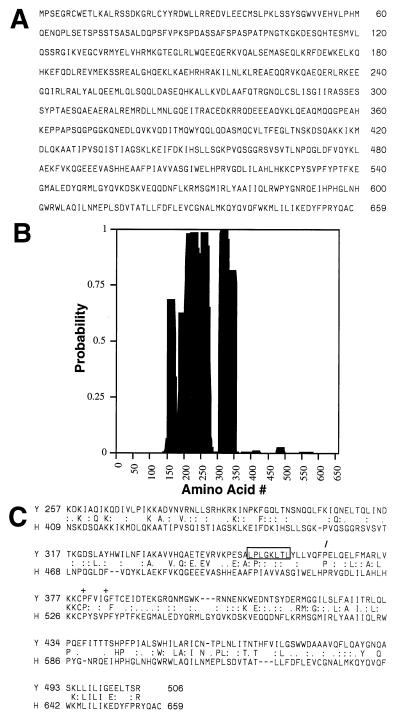
Figure 1.
Amino acid sequence of hGle1p. (A) hGle1p amino acid sequence. (B) Coiled-coil probability for hGle1p. A middle region of hGle1p shows a high probability of forming a coiled-coil structure by analysis with the coils program (48). (C) Comparison of the C-terminal regions of yeast and human Gle1p. align analysis (43) between the yeast (upper line) and human proteins (lower line) reveals significant homology. The center line designates identical (capital letter) and conserved (: or .) residues. The LR-NES in yeast Gle1p is boxed. The slash (/) designates the breakpoint for Regions 3 and 4 (Fig. 3A). The plus signs (+) mark residues changed in yeast gle1 alleles (P380G or G384R).
A GenBank search with the human protein identified yeast Gle1p with a high probability [P(N) = 2.8e-5]. Based on sequence homology as well as results described below, we designated the human protein as hGle1p. Although yeast Gle1p is smaller (538 residues), structural and sequence similarities between yeast Gle1p and hGle1p are extensive. The middle regions of both proteins are highly charged and each possesses a very high potential to form a coiled-coil structure (Fig. 1B; refs. 33 and 34). The align program (43) was used to directly compare the sequences, and the C-terminal regions (residues 250–538 of yeast and 360–659 of human) were directly matched with extensive similarity. The C-terminal regions are 54% homologous (27% identical and 27% similar residues) (Fig. 1C). Limited sequence information for mouse cDNA clones in the dbEST database (see Material and Methods) predicts proteins that are nearly identical to the C-terminal region of hGle1p. This suggests that proteins with sequence similarity to yeast Gle1p exist across species. Interestingly, the LR-NES in yeast Gle1p is not conserved in hGle1p or in the predicted mouse protein.
hGle1p Localization Is Concentrated at the NPC.
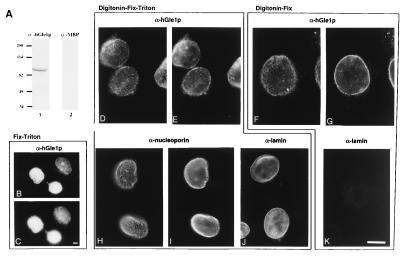
Yeast Gle1p is localized primarily at NPCs (33, 34), and we predicted that a functional homologue would have a similar distribution in human cells. To test this, polyclonal antibodies were raised against bacterially expressed GST-hGle1p. The antibodies were affinity-purified and tested by immunoblotting HeLa cell extracts and indirect immunofluorescence localization (Fig. 2). A single polypeptide migrating just greater than 82 kDa was recognized (Fig. 2A). Under different fixation conditions, the majority of the anti-hGle1p staining was localized to the nuclear envelope rim in a punctate, discontinuous pattern (e.g., Fig. 2 D and E). This staining is similar to that observed with antibodies recognizing nucleoporins (mAb414) (Fig. 2 H and I). To test whether hGle1p was cytoplasmically accessible, immunofluorescence microscopy was performed on HeLa cells in which the plasma membrane was permeabilized with digitonin (Fig. 2 F and G). Under these conditions, the nuclear envelope and NPCs remained intact restricting antibody access to the nucleus, as shown by the lack of anti-lamin B staining in similarly treated cells (Fig. 2K). Anti-lamin nuclear access was observed only with additional Triton X-100 treatment (Fig. 2J). In contrast, anti-hGle1p antibodies had access to hGle1p in cells treated with digitonin alone, reflected by punctate nuclear envelope labeling (Fig. 2G). These results suggest that a fraction of hGle1p is cytoplasmically accessible.
Figure 2.
hGle1p localizes to the NPC and is cytoplasmically accessible. (A) Immunoblot analysis with whole-cell HeLa extracts and affinity-purified rabbit antibodies: anti-hGle1p (lane 1) or anti-MBP (lane 2). Molecular mass markers are in kDa. (B–K) Intracellular distribution of hGle1p. HeLa cells were processed for labeling with primary antibodies as designated: Fix-Triton (B and C), Digitonin-Fix-Triton (D, E, and H–J), or Digitonin-Fix (F, G, and K). hGle1p localization was observed with the affinity-purified rabbit anti-hGle1p antibody (B–G), nucleoporin staining (H and I) with mAb414 (40), and lamin staining with a rabbit anti-lamin B serum (J and K) (41). Pairs of cells (B–C, D–E, H–I, and F–G) were photographed in two different focal planes for the punctate (C, D, H, and F) and nuclear rim (B, E, G, and I) staining. (Bars = 10 μm.)
hGle1p Requires a LR-NES to Function in Yeast.
To test hGle1p for complementation of the temperature-sensitive yeast gle1–4 mutant, a plasmid expressing full-length hGLE1 under control of the yeast GLE1 promoter was transformed into yeast gle1–4 cells and growth was assayed at the nonpermissive temperature of 30°C (Fig. 3). No colonies were observed even though immunoblotting confirmed that hGle1p was expressed (data not shown). Thus, full-length hGle1p did not compensate functionally for yeast Gle1p. We further tested for complementation with a panel of chimeric yeast–human proteins. The yeast protein was divided into four regions as diagrammed (Fig. 3): Region 1, the N-terminal 108 residues; Region 2, the middle coiled-coil region; Region 3, the first half of the C-terminal region harboring the LR-NES; and Region 4, the second half of the C-terminal region. Internal in-frame deletions (Δ) of each individual region from yeast Gle1p showed that only Region 1 was dispensable. In contrast Regions 2, 3, and 4 all were required because expression of the yGle1p-Δ2, yGle1p-Δ3, and yGle1p-Δ4 deleted proteins did not complement the gle1–4 mutant. To test for the ability of the individual human regions to restore function of the deleted yeast proteins, sequence encoding the corresponding human (h) region was inserted in-frame into each respective yeast-deletion construct (Fig. 3). As controls, the yeast (y) regions were placed back into the deletion constructs in an identical manner. Both the Δ2 + h2 and Δ4 + h4 chimeras allowed growth at 30°C, suggesting functional complementation by the region from hGle1p.
Interestingly, Region 3 from hGle1p was not functional in the yeast–human Δ3 + h3 chimera. To determine whether this was because of the absence of a LR-NES, the human sequence aligning with the yeast LR-NES was changed to encode a LR-NES, LPLGKLTL (residues 500–507 in hGle1p). The Δ3 + h3/NES chimera complemented the temperature-sensitive growth of the gle1–4 cells at 30°C (Fig. 3B). Moreover, when the LR-NES was incorporated into full-length hGle1p (hGle1p+NES), growth of gle1–4 cells also now was observed. However, hGle1p+NES did not complement the lethal phenotype of the gle1 null mutant (data not shown). This suggests that the complementation either requires interaction with the mutant yeast protein or that the level of activity of hGle1p+NES is not high enough for full function and viability. Overall, hGle1p is functional in yeast when supplied with a LR-NES.
In the genetic screen that identified yeast GLE1, five conditional alleles were characterized (33, 44). To delineate the structural basis for the mutant phenotype, the full-length genes for the five gle1 alleles were sequenced. Three of the alleles contain a single mutation that results in a substitution of a leucine residue for proline at residue 380. The other two alleles are a result of a change in the codon for glycine at position 384 to that for arginine. Both of these mutations reside in Region 4 and correspond to a highly conserved block of hGle1p sequence (Fig. 1C). If hGle1p function is similar to yeast Gle1p, we reasoned that similar mutations in Region 4 of hGle1p should abolish function. Two different Δ4 + h4 mutant chimeras were generated at the matching residues in the human Region 4: one that resulted in a substitution of a leucine for proline at position 529, and the other an arginine for proline at position 533. Expression of either mutant chimera in the gle1–4 cells did not allow growth at 30°C (Fig. 3B). Therefore, hGle1p function is sensitive to the same point mutations as the yeast protein, and this further supports their functional conservation.
Anti-hGle1p Antibodies Specifically Inhibit Poly(A)+ Export in Human Cells.
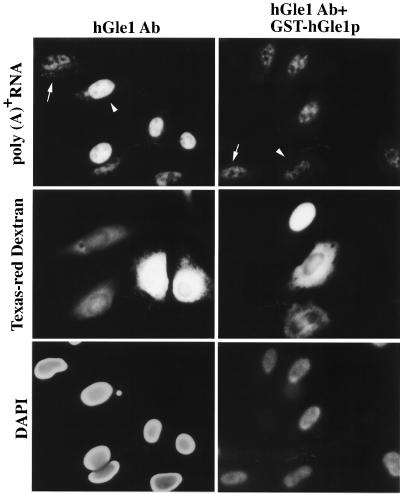
We previously suggested a role for yeast Gle1p in mRNA export based on the penetrant and specific poly(A)+ RNA export defects observed in gle1 mutant cells (33). To determine whether hGle1p plays a similar role in vertebrate mRNA export, we examined the effect on nuclear transport of antibodies raised against hGle1p. HeLa cells were co-microinjected in the cytoplasm with Texas red–dextran and affinity-purified antibodies recognizing hGle1p (Fig. 4 Left). In a given field, only a subset of the cells were injected as designated by the Texas red staining (Fig. 4 Middle). After incubation at 37°C, the localization of poly(A)+ RNA was visualized by in situ hybridization (Fig. 4 Top). Low levels of poly(A)+ RNA staining were detected in both the cytoplasm and nucleus of uninjected cells (Fig. 4 Top Left, arrow). In contrast, 6–8 hr after injection of the anti-hGle1p antibody, the cells began to show an altered pattern of poly(A)+ RNA staining (data not shown). By 12 hr the effects were marked: the poly(A)+ RNA staining disappeared from the cytoplasm and the intranuclear staining intensity significantly increased in anti-hGle1p-injected cells (Fig. 4 Top Left, arrowhead). As a control for antibody specificity, inhibition of export was relieved by coinjection of the anti-hGle1p with recombinant GST-hGle1p (Fig. 4 Right). In addition, export was not blocked by injection of affinity-purified antibodies recognizing MBP (which do not recognize any polypeptides in HeLa cells; Fig. 2A) (data not shown).
Figure 4.
Injection of anti-hGle1p antibodies inhibits poly(A)+ RNA export in HeLa cells. The distribution of poly(A)+ RNA was examined in HeLa cells co-microinjected with Texas red–dextran and either affinity-purified rabbit anti-hGle1p antibody alone (2.7 mg/ml, Left) or anti-hGle1p premixed with purified GST-hGle1p (2.7 mg/ml IgG and 2.25 mg/ml recombinant, Right). The anti-hGle1p antibody is monospecific for hGle1p (see Fig. 2A). Only a subset of the cells in a given field were injected, and all the cells were cytoplasmically injected except for the uppermost injected cell in the Right column (by Texas red localization, Middle). Cells were incubated for 12 hr at 37°C before in situ hybridization with a digoxigenin-labeled oligo(dT30) probe and fluorescein isothiocyanate antidigoxigenin Fab. Arrowhead highlights a typical injected cell, and arrow indicates an uninjected cell (Top). Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (Bottom). (Bar = 20 μm.)
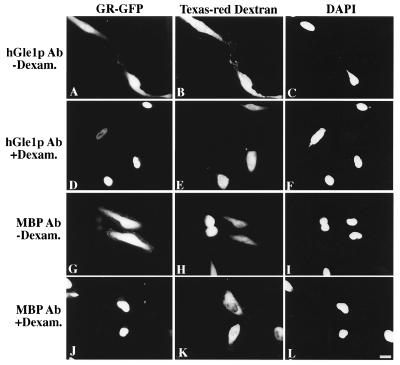
Because hGle1p appeared localized at the NPCs (Fig. 2), the inhibition of export may result from indirect physical obstruction of the NPC. To address this issue, nuclear import activity was measured in HeLa cells expressing a GR-GFP fusion (38). Transfected cells were cytoplasmically injected with Texas red–dextran and either anti-hGle1p or anti-MBP antibodies, and incubated for 12 hr at 37°C. In a given field only a subset of the cells were injected as reflected by the Texas red staining (Fig. 5 Center). In the absence of the agonist dexamethasone, the GR-GFP was predominantly cytosolic (Fig. 5 A and G). When dexamethasone was added to induce nuclear translocation, nuclear accumulation of GR-GFP was observed (Fig. 5 D and J). Injection of anti-hGle1p antibodies had no detectable effect on nuclear import of GR-GFP. In nuclear coinjection experiments, the export of a coinjected Rev-NES substrate also was not blocked by the anti-hGle1p antibodies (data not shown). Thus, NPCs are not sterically blocked by anti-hGle1p antibodies, and hGle1p is not an essential element of the GR-GFP import pathway. Thus, anti-hGle1p antibodies specifically inhibit poly(A)+ RNA export, implying that hGle1p functions in export.
Figure 5.
Nuclear import is not affected by microinjection of anti-hGle1p antibody. HeLa cells transiently expressing GR-GFP (A, D, G, and J) were co-microinjected with Texas red–dextran and either affinity-purified rabbit antibody: anti-hGle1p (2 mg/ml, A–F) or anti-MBP (2 mg/ml, G–L). The anti-MBP antibody served as a control for nonspecific effects of antibody injection. Texas red–dextran (Center, B, E, H, and K) differentiates between injected and uninjected cells. In a given field, only a subset of the cells were cytoplasmically injected. Cells were incubated for 12 hr at 37°C, and those in D–F and J–L were treated with medium containing 10 μg/ml dexamethasone for 30 min before fixation and processing for fluorescence microscopy. Cells in A–C and G–I were untreated. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (C, F, I, and L). (Bar = 20 μm.)
DISCUSSION
Here we report the identification of a human homologue of the yeast RNA export factor Gle1p, and further demonstrate that hGle1p is required for the export of poly(A)+ RNA in human cells. Our results suggest hGle1p is a component of the vertebrate mRNA export pathway. Interestingly, hGle1p does not harbor a recognizable LR-NES. Because the yeast Gle1p requires a LR-NES to function (33), there may be mechanistic differences for poly(A)+ RNA export between species.
Evidence for participation of hGle1p in nuclear poly(A)+ RNA export is based not only on its structural and functional homology to a yeast poly(A)+ RNA export factor, but also on the block in poly(A)+ RNA export observed with coincident inhibition of hGle1p activity. How is Gle1p functioning in the mRNA export pathway? A cascade of processing events must occur for maturation of pre-mRNA to an export-competent substrate (10). In particular, pre-mRNAs are retained in the nucleus by interaction with the splicing machinery (4–6). Thus, export is prevented effectively until proper splicing is completed. hGle1p is clearly distinct from hnRNP proteins in that it has no apparent RNA-binding sites and is localized predominantly at the NPCs. This suggests hGle1p is confined both spatially and temporally at the end of this processing pathway and may be positioned to act at a terminal step that commits mature RNA messages for export.
That hGle1p does not contain a LR-NES suggests several models for its mechanism of action. hGle1p may not require NES activity for function, Region 3 in hGle1p may bind to another exported protein with an actual NES, or hGle1p may possess a novel NES that is not functionally recognized by yeast. If Gle1p shuttles between the nucleus/NPC and the cytoplasm, it may serve as a linker between the shuttling hnRNP proteins, an mRNA-specific β transport factor, and the NPC. Alternatively, Gle1p may act to remove an inhibitory block to export, possibly by triggering removal of nonshuttling hnRNP proteins. For example, the nuclear retention signals in the nonshuttling hnRNP proteins prevent the transport of hnRNPs regardless of the presence of the NES in hnRNP A1 protein (45, 46). It is also possible that Gle1p is required for the import of an essential export factor. In any of these scenarios, the NES in Gle1p may serve to dictate the directionality of transport and/or carefully restrict Gle1p access to the nucleoplasm and unspliced pre-mRNAs.
In contrast, Gle1p may be a stable/structural component of the NPC. In terms of the yeast protein, such a NPC-bound, NES-containing nucleoporin may function to recycle the β NES receptor. A connection between Gle1p and the NPC is supported by genetic interactions between yeast Gle1p and several yeast nucleoporins with primary roles in nuclear export (33, 34, 44). However, functional links between yeast Gle1p and the LR-NES β receptor Crm1p/Xpo1p have not been established (22), and yeast Gle1p and Crm1p/Xpo1p do not interact in the yeast two-hybrid assay (J.L.W., R.M., and S.R.W., unpublished results). Work with the human β Crm1p/exportin also has suggested that it does not have a role in mRNA export (16) and therefore may not be linked to hGle1p function. Testing the dynamics of hGle1p and revealing the network of structural interactions during the mRNA export mechanism will be necessary to differentiate between these models.
Although human and yeast cells facilitate export of poly(A)+ RNA export with a similar factor, the absence of a recognizable LR-NES in hGle1p also provides some insight into the evolution of mRNA export pathways. Yeast cells may utilize a more restricted range of NES export pathways or NES-containing factors for the transport of all RNA types, reflected by complementation only when Gle1p (yeast or human) has a LR-NES (Fig. 3). This simpler system may reflect the comparatively small fraction of yeast genes with introns, less than 5% of all S. cerevisiae messages. In light of recent transport competition studies in Xenopus oocytes showing inhibition of mRNA export by LR-NES conjugates (21), the lack of a LR-NES in hGle1p suggests the LR-NES factors that mediate human mRNA export remain to be identified.
The differences between yeast and vertebrate mRNA export may be restricted to just one factor (e.g., adapters for Region 3). Here we have shown for yeast Gle1p that both the coiled-coil N-terminal region and the region C-terminal to the NES also are essential for function. The structural and functional conservation between these other essential regions in yeast and human Gle1p (Regions 2 and 4, Fig. 3) suggests there will be shared elements for mRNA export across species. The precise role for Gle1p in mRNA export may be revealed by analyzing its unique regions. The combined actions of several different proteins may determine the rate of export, including the hnRNP and cap-binding proteins (19) as well as a series of Gle1p-like factors. The recent characterization of Mex67p (20), a second LR-NES mRNA export factor in yeast, highlights this possibility. Interestingly, the putative human Mex67p (TAP) also lacks a recognizable LR-NES (20). Finally, all types of RNA export pathways inherently will intersect at the point of translocation through the NPC. This is evidenced, for example, by inhibiting multiple classes of RNA export with blocked Nup98 function (47). We predict that the study of both yeast and human systems will prove a powerful approach toward delineating the mRNA export machinery in all species. Our future analysis will focus on discovering the links between Gle1p and the transported RNPs.
Acknowledgments
We thank D. Schafer and N. Watson for tissue culture and microinjection technology; P. Thomas for DNA sequencing; I. Macara for the GR-GFP plasmid; B. Burke for sharing dexamethasone; N. Chaudhary for anti-lamin B antibodies; and M. K. Iovine and M. Bucci for discussion. J.L.T.E. performed this work as a Fellow of the American Heart Association. This work was supported by a grant from the National Institutes of Health, GM51219, to S.R.W.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: GFP, green fluorescent protein; GR, glucocorticoid receptor; GST, glutathione S-transferase; hn, heterogeneous nuclear; LR, leucine-rich; MBP, maltose-binding protein; NES, nuclear export sequence; NPC, nuclear pore complex; RNP, ribonucleoprotein. Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF058922).
References
- 1.Lee M S, Silver P A. Curr Opin Genet Dev. 1997;7:212–219. doi: 10.1016/s0959-437x(97)80131-1. [DOI] [PubMed] [Google Scholar]
- 2.Nakielny S, Dreyfuss G. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 3.Ullman K S, Powers M A, Forbes D J. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 4.Legrain P, Rosbash M. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 5.Chang D D, Sharp P A. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 6.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 7.Eckner R, Ellmeier W, Birnstiel M L. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I W. Nature (London) 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Carmichael G G. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daneholt B. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 11.Zapp M L, Green M R. Nature (London) 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 12.Daly T J, Cook K S, Gray G S, Maione T E, Rusche J R. Nature (London) 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 13.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 14.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 15.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 17.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Gopal M, Fransen J, Grosfeld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izaurralde E, Mattaj I W. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 20.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasquinelli A E, Powers M, Lund E, Forbes D, Dahlberg J E. Proc Natl Acad Sci USA. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 23.Seedorf M, Silver P A. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 25.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 26.Michael W M, Eder P S, Dreyfuss G. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M S, Henry M, Silver P A. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 28.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 29.Aitchison J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 30.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doye V, Hurt E. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 32.Corbett A H, Silver P A. Microbiol Mol Bio Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy R, Wente S. Nature (London) 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 34.Del Priore V, Snay C A, Bahr A, Cole C N. Mol Biol Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Iovine M K, Wente S R. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iovine M K, Watkins J L, Wente S R. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey K L, Richards S A, Lounsbury K M, Macara I G. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shafer K, Hug C, Cooper J A. J Cell Biol. 1995;128:61–70. doi: 10.1083/jcb.128.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis L I, Blobel G. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhary N, Courvalin J C. J Cell Biol. 1993;122:295–306. doi: 10.1083/jcb.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wente S R, Blobel G. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dayhoff M O, Barker W C, Hunt L T. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 44.Murphy R, Watkins J L, Wente S R. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakielny S, Dreyfuss G. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimm C, Lund E, Dahlberg J E. Proc Natl Acad Sci USA. 1997;94:10122–10127. doi: 10.1073/pnas.94.19.10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powers M A, Forbes D J, Dahlberg J E, Lund E. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]