Abstract
The structural and DNA binding behavior is described for an analog of the vnd/NK-2 homeodomain, which contains a single amino acid residue alanine to threonine replacement in position 35 of the homeodomain. Multidimensional nuclear magnetic resonance, circular dichroism, and electrophoretic gel retardation assays were carried out on recombinant 80-aa residue proteins that encompass the wild-type and mutant homeodomains. The mutant A35T vnd/NK-2 homeodomain is unable to adopt a folded conformation free in solution at temperatures down to −5°C in contrast to the behavior of the corresponding wild-type vnd/NK-2 homeodomain, which is folded into a functional three-dimensional structure below 25°C. The A35T vnd/NK-2 binds specifically to the vnd/NK-2 target DNA sequence, but with an affinity that is 50-fold lower than that of the wild-type homeodomain. Although the three-dimensional structure of the mutant A35T vnd/NK-2 in the DNA bound state shows characteristic helix–turn–helix behavior similar to that of the wild-type homeodomain, a notable structural deviation in the mutant A35T analog is observed for the amide proton of leucine-40. The wild-type homeodomain forms an unusual i,i-5 hydrogen bond with the backbone amide oxygen of residue 35. In the A35T mutant this amide proton resonance is shifted upfield by 1.27 ppm relative to the resonance frequency for the wild-type analog, thereby indicating a significant alteration of this i,i-5 hydrogen bond.
Keywords: nuclear magnetic resonance, structure, folding, DNA binding
The homeodomain is the 60-aa residue DNA binding domain of the protein product from a homeobox containing gene (1). The homeodomain containing protein acts as a transcription regulator and some are involved in different aspects of embryonic development such as segmental patterning and limb formation (1). Mutations in the homeodomain often are associated with abnormal development and disease (2–13). Furthermore, changes as subtle as single site amino acid residue replacements in a given homeodomain have been shown to result in significant developmental abnormalities in species ranging from the fruit fly to human (2–9). Knowledge of the structural changes in the homeodomain or modifications of the interactions with DNA induced by such site mutations should provide important clues toward a better understanding of the biological basis of the corresponding developmental abnormalities. This ability to relate anomalies in function to single amino acid residue mutations in a transcriptional protein provides one of the simplest and most direct opportunities to investigate the structure–function relationship.
The homeodomain of interest in this study comes from the vnd (i.e., ventral nervous system defective) gene (9, 14) from Drosophila melanogaster, which was first cloned and characterized as the NK-2 gene (15). The vnd/NK-2 gene is a neural gene regulator that initiates the neural program of development in part of the central nervous system of Drosophila embryos (16–19). Recently, a number of vnd mutant alleles were reported that produce transcriptional defects in early embryogenesis and are embryonically lethal (9). One of the lethal vnd mutants was shown to encode the vnd/NK-2 homeodomain protein with a single amino acid replacement in the homeodomain, where alanine in position 35 of the homeodomain is replaced by threonine (9). Alanine usually is found in this position, with serine occurring in this position in about 20% of the homeodomains characterized thus far (1). Threonine has not been found at position 35 in any of the many homeodomains that have been described.
The vnd/NK-2 homeodomain binds (Kd ≈ 1 nM) to the unusual DNA consensus sequence 5′-CAAGTG-3′ (L.-H. Wang, R. Chemelik, and M.N., unpublished data). The three-dimensional (3D) solution structures of the wild-type vnd/NK-2 homeodomain in the free and DNA bound states determined by using NMR have been described recently (20–22). This homeodomain contains three helical segments in analogy with other known homeodomains, where helix II and helix III make up the well-known helix–turn–helix DNA binding motif (23–27). The alanine residue at position 35 is in helix II and its side-chain methyl group makes extensive contacts across the turn with residues at the N-terminal region of helix III. In this study we demonstrate that the presence of threonine in position 35 (i.e., the mutated A35T vnd/NK-2 homeodomain) is sufficient to completely prevent vnd/NK-2 from adopting the native 3D structure. In addition, we show that the affinity of the A35T mutant homeodomain for its cognate DNA is reduced by a factor of 50 compared with that of the wild-type homeodomain.
MATERIALS AND METHODS
Construction of the A35T vnd/NK-2 Expression System.
The site-directed A35T mutation in the vnd/NK-2 homeodomain was performed by PCR to convert the relevant alanine GCC codon to ACC for threonine in the vnd/NK-2 homeobox DNA by using the wild-type vnd/NK-2 homeobox DNA as the template (28). Two restriction sites, NdeI and BamHI, were introduced at the 5′ and 3′ ends of the DNA fragment by appropriately designed primers. An additional mutation was made to eliminate an undesirable NdeI site within the vnd/NK-2 homeobox DNA by converting CATATG to CATACG. This second mutation in the DNA, part of which codes for Y14 in the homeodomain, does not change the amino acid sequence, because both TAT and TAC represent codons for tyrosine. The PCR products were purified by using a 2% agarose gel and Mermaid kit (Bio 101). The final DNA fragment of 263 bp was digested with NdeI and BamHI and ligated into the NdeI–BamHI sites of the pET-15b plasmid vector (Novagen) to express an 80 amino acid residue protein that encompasses the A35T vnd/NK-2 homeodomain (20) with a His-Tag fused to the N terminus (29).
The pET-15b plasmid with the A35T vnd/NK-2 homeodomain insert was transformed into the cloning host NovaBlue (Novagen) and plated on Luria–Bertani (LB)–ampicillin agar. Eighteen clones grown overnight were screened by PCR and 9 positive clones, which produced DNA of ≈260 bp in PCR, were identified. Plasmids isolated from cells grown from three positive clones were sequenced by Loftstrand Laboratories (Gaithersburg, MD) and two plasmids were found to have the correct sequence.
The plasmid with the A35T vnd/NK-2 insert was then transformed into the expression host BL21(DE3)plysS (Novagen) and plated on LB–ampicillin/chloramphenicol agar. Clones were screened further for A35T vnd/NK-2 expression and a clone called BL21(DE3)plysS/A35T vnd/NK-2, which has high A35T vnd/NK-2 expression, was used for large scale expression and purification of the protein.
Purification of A35T vnd/NK-2.
The BL21(DE3)plysS/A35T vnd/NK-2 cells were grown in LB–ampicillin/chloramphenicol media, and isopropyl β-d-thiogalactoside was used to induce the expression of A35T vnd/NK-2 protein (29). Cells were harvested and resuspended in a binding buffer containing 6 M guanidine, 5 mM imidazole, 500 mM NaCl, and 20 mM Tris⋅HCl at pH 7.9. A French press was used to break cells and cell debris was removed by centrifugation. The supernatant fraction containing A35T vnd/NK-2 protein was loaded onto a Ni2+ affinity column (Novagen) that was equilibrated with the binding buffer. After extensive washing with the binding buffer, the A35T vnd/NK-2 was eluted with buffer containing 6 M guanidine, 200 mM imidazole, 500 mM NaCl, and 20 mM Tris⋅HCl at pH 7.9. The eluted protein was diluted 50-fold with a thrombin buffer containing 2.5 mM CaCl2, 150 mM NaCl, and 20 mM Tris⋅HCl at pH 7.5. To cut the His-Tag, immobilized thrombin (Calbiochem) was added to the solution and after incubation removed by centrifugation. The reaction mixture then was concentrated, dialyzed against the binding buffer for buffer exchange, and loaded onto another Ni2+ column. The solution passing through the second column was collected and dialyzed against water extensively to refold the protein. The solution also was dialyzed against water at pH 3.0 to remove Ni2+ bound to the protein (found by mass spectroscopy to be bound to the protein) and further purified by reverse-phase HPLC (30). The purity of the protein was demonstrated by gel electrophoresis, NMR, and mass spectroscopy. The amino acid sequence of the 80 amino acid residue protein containing the mutant A35T vnd/NK-2 homeodomain is given in Fig. 1.
Figure 1.
Sequence of the 80 amino acid residue protein that encompasses the mutant A35T vnd/NK-2 homeodomain. The T in position 35 of the homeodomain is underlined.
To prepare 15N single-labeled and 15N/13C double-labeled A35T vnd/NK-2 protein, Martek 9-N and Martek 9-CN (Martek Biosciences, Columbia, MD) were used instead of LB in the above procedure. The extent of 15N and 13C incorporation was 97% as determined by mass spectroscopy (30). The yield of pure-labeled and unlabeled mutant A35T vnd/NK-2 protein from either 1,000 ml LB, Martek 9-N, or Martek 9-CN media was approximately 13 mg.
Electrophoretic Gel Retardation Assays.
Gel retardation assays were used to determine the dissociation constant of the A35T vnd/NK-2 bound to the vnd/NK-2 target DNA (31). The wild-type vnd/NK-2 homeodomain protein used in the gel retardation experiment was purified under native conditions (29, 32). Both of the 18-mer single strands, 5′-TGTGTCAAGTGGCTGTAG-3′ and 5′-CTACAGCCACTTGACACA-3′, of the vnd/NK-2 target DNA were synthesized and purified by Midland Certified Reagent Company. Their purity was confirmed by the absence of impurity peaks in their MS spectra. Each DNA single-stranded 18-base fragment was labeled with [γ-32P]ATP by using T4 polynucleotide kinase and then annealed to form double-stranded DNA. The double-stranded DNA was purified using 6% agarose gel and Mermaid kit. Both wild-type and mutant A35T vnd/NK-2 proteins at concentrations ranging from 1 pM to 1 μM individually were mixed with 10 pM 32P-labeled double-stranded DNA in a buffer containing 20 mM Hepes (pH 7.9), 90 mM NaCl, 0.4 mM EDTA, 0.3 mg/ml BSA, and 10% glycerol. The solutions were incubated on ice for 1 hr before being loaded onto a 10% native polyacrylamide gel. Two gels loaded with the wild-type and mutant homeodomain/DNA solutions, respectively, were run at 300 V and 4°C in one electrophoresis unit. After the gels were dried, free and protein bound DNA bands were visualized by autoradiography. The relative affinities of the homeodomain for the DNA were determined from the protein concentrations of the lanes containing equal amounts of free and bound DNA.
Preparation of the A35T vnd/NK-2–DNA Complex.
The complex of the singly 15N-labeled or doubly 15N- and 13C-labeled A35T vnd/NK-2 homeodomain bound to a 16-mer double-strand vnd/NK-2 target DNA was prepared as an aqueous, i.e., H2O or D2O solution. Both 16-mer single strands, 5′-ACAGCCACTTGACACA-3′ and 5′-TGTGTCAAGTGGCTGT-3′, of the vnd/NK-2 target DNA were synthesized and purified by Midland Certified Reagent Company. Their purity was demonstrated by the absence of impurity peaks in their MS and NMR spectra. The two strands then were mixed in an equal molar amount and annealed to form the double-stranded DNA. To form the A35T vnd/NK-2–DNA complex, an aqueous solution of the protein (50 μM, pH 8.0) was titrated into the solution (50 μM, pH 8.0) containing the DNA, which was stirred continuously until a DNA-to-protein ratio of 1:0.9 was reached. The solution then was concentrated to about 1.0 mM and the pH was adjusted to a value of 6.8. The samples were sealed in Shigemi NMR tubes (Shigemi, Allison Park, PA) for NMR measurements. The analogous preparation of the wild-type vnd/NK-2 homeodomain protein and its DNA complex used in an NMR study was described previously (20).
CD Spectropolarimetry.
CD spectra were recorded with unbuffered aqueous solutions of 50 μM free protein and 50 μM of the protein/DNA complex on a Jasco J-600 spectrometer. The sample temperature was controlled by circulating water/ethanol mixture from a water bath.
NMR Spectroscopy.
Sample concentrations were all approximately 1 mM. NMR measurements were performed on a Bruker AMX600 spectrometer equipped with a triple resonance probe and a single z-axis gradient coil. Gradient pulses were supplied by a home built gradient amplifier. Spectra were processed by using nmrpipe and nmrdraw programs on a Silicon Graphics Indigo 2 workstation (33). HSQC, CBCANH, CBCA(CO)NH, HCCH–TOCSY, HBHA(CBCACO)NH, NOESY–HSQC, and HMQC–NOESY–HSQC pulse sequences were implemented with pulsed-field gradients for coherence selection and water suppression (34–38). For the NOESY experiments, τmix = 100 msec. Linear prediction, zero filling, window function, and polynomial baseline correction were employed in the data processing.
RESULTS
DNA Binding Assay.
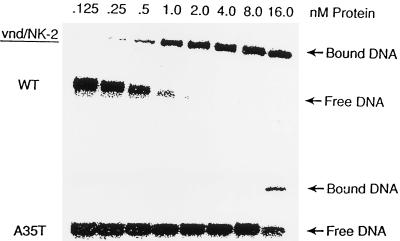
The concentration dependence of the affinity of wild-type and mutant A35T vnd/NK-2 homeodomain proteins for the 18 bp DNA duplex (containing 5′-CAAGTG-3′ as a central segment of the DNA) electrophoresed on a native polyacrylamide gel was measured. The gel retardation data show high-affinity binding of the wild-type vnd/NK-2 homeodomain to DNA in the nanomolar range in accord with our previous results (32). In contrast, the mutant A35T vnd/NK-2 binds to this same 18 bp target DNA sequence with an affinity that is 50-fold lower than that of the wild-type vnd/NK-2 homeodomain (Fig. 2). The 18 bp DNA fragment contains the same 16 bp segment used for the NMR studies. The affinity of the 16 bp segment for the wild-type vnd/NK-2 is only a factor of two less than that of the 18 bp segment.
Figure 2.
Gel mobility assay of wild-type and mutant A35T vnd/NK-2 homeodomains binding to the DNA containing the vnd/NK-2 consensus sequence.
CD.
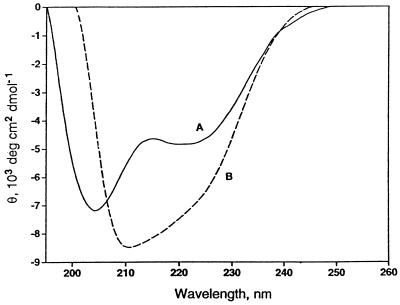
Fig. 3 shows the CD spectra of the A35T vnd/NK-2 homeodomain in both the free state (curve A) and bound to the vnd/NK-2 target DNA (curve B). The increase in the magnitude of the mean residue ellipticity at 222 nm, θ222, upon binding to the DNA shows that the helix content of the DNA bound A35T vnd/NK-2 is higher than that of the free mutant homeodomain.
Figure 3.
CD spectra of curve A, a 50 μM solution of free A35T vnd/NK-2 unbuffered at pH 6.0 and 0°C, and curve B, the complex formed with 50 μM 16-mer target DNA (double-stranded) at pH 6.0 and 0°C.
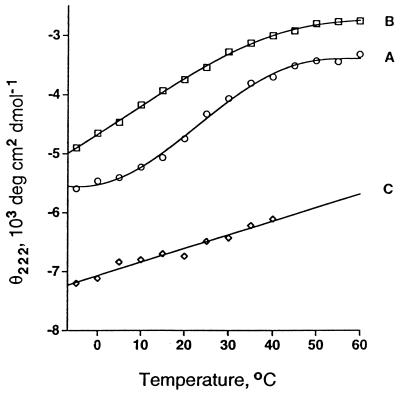
CD spectra of the wild-type vnd/NK-2 free in solution together with spectra of the mutant A35T vnd/NK-2, both in the free and DNA bound states, were recorded at various temperatures from −5°C to 60°C. The respective temperature dependencies of θ222 are depicted in curves A, B, and C in Fig. 4. The wild-type vnd/NK-2 homeodomain free in solution shows a stable θ222 below 10°C and above 50°C and a conformation transition temperature, Tm ≈ 25°C. In contrast, θ222 for the free mutant A35T vnd/NK-2 decreases monotonically down to −5°C. The implication of these results is that the free mutant A35T vnd/NK-2 does not undergo a transition to a thermally stable folded conformation at low temperatures analogous to that found for the wild-type homeodomain. The values of θ222 for the DNA bound mutant A35T vnd/NK-2 up to 40°C (curve C in Fig. 4) are quite similar to those found for the wild-type vnd/NK-2 bound to the DNA. These observations suggest that the DNA bound mutant A35T vnd/NK-2 adopts a folded structure with helix content similar to that found for the wild-type homeodomain bound to DNA (21).
Figure 4.
Temperature dependencies of the mean residue ellipticities (θ222) at 222 nm of 50 μM solutions of free wild-type vnd/NK-2 homeodomain at pH 4.5 (curve A), free mutant A35T vnd/NK-2 at pH 6.0 (curve B), and A35T vnd/NK-2 bound to DNA at pH 6.8 determined by subtracting the ellipticity of unbound target 16-mer DNA from that of the A35T vnd/NK-2–DNA complex (curve C).
NMR.
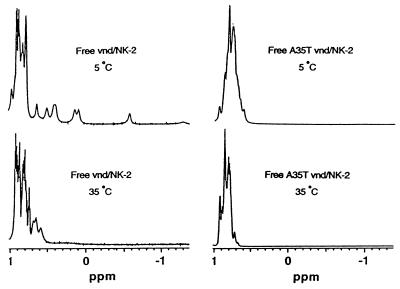
Fig. 5 shows the high field portions of the one-dimensional (1D) 1H NMR spectra of free wild-type and mutant A35T vnd/NK-2 homeodomain at 5°C and 35°C. A number of peaks from 0.7 to −1.2 ppm can be seen in the NMR spectrum of free vnd/NK-2 homeodomain at 5°C. These resonances have been assigned to protons that make up the hydrophobic core (L16, L26, and L40) of the folded conformation of the wild-type homeodomain (22). The wild-type homeodomain in the free state is folded at low temperatures (Tm ≈ 25°C). With increasing temperature, these high field peaks shift to a lower field, broaden initially, and then sharpen and become part of the resonance envelope around 0.8 ppm. The appearance of these high field peaks is thus characteristic of the folding of the vnd/NK-2 homeodomain. In contrast to the behavior of the wild-type homeodomain free in solution, no high field peaks above 0.7 ppm are seen in the spectra of the free mutant homeodomain at temperatures down to −5°C. Furthermore, a two-dimensional 1H NOESY spectrum of this mutant homeodomain does not show any cross peaks characteristic of the folded conformation. These results demonstrate that the mutant A35T vnd/NK-2 free in solution does not adopt a folded conformation.
Figure 5.
Upfield region of the 1D 1H NMR spectra of free wild-type (pH 4.4) and mutant A35T vnd/NK-2 (pH 5.0) recorded at 5°C and 35°C.
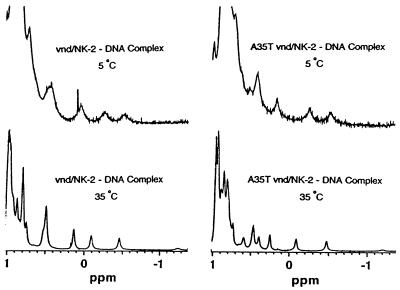
The 1D 1H NMR spectra at 5°C and 35°C of the wild-type and mutant A35T vnd/NK-2 homeodomain bound to DNA are presented in Fig. 6. For both wild-type and mutant DNA complexes, high field peaks are observed above 0.7 ppm that are characteristic of the formation of the hydrophobic core of the protein. In addition, for both wild-type and mutant homeodomains the positions of these high field resonances are quite similar (21). Both the wild-type and mutant homeodomains are structurally stable at temperatures up to at least 42°C when they are bound to the vnd/NK-2 cognate DNA.
Figure 6.
Upfield region of the 1D 1H NMR spectra of the wild-type and mutant A35T vnd/NK-2–DNA complexes (pH 6.8) recorded at 5°C and 35°C.
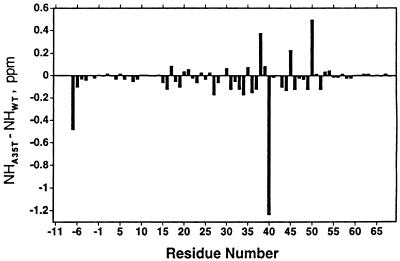
The backbone resonances of the DNA bound mutant A35T vnd/NK-2 were assigned using 3D CBCANH and CBCA(CO)NH pulse sequences. Side chain resonance assignments were made using 3D HCCH TOCSY and HBHA(CBCACO)NH pulse experiments. Most of the resonances were found at positions that are, in general, analogous to those found for the wild-type analog. The primary exception (Fig. 7) is that of the amide proton of L40, which is shifted upfield by 1.27 ppm relative to the chemical shift of the wild-type analog. This upfield shift would represent an increase of 0.6 Å in a hydrogen bond distance (39). Because the i,i-5 hydrogen bond distance is 2.4 Å in the wild-type vnd/NK-2 (PDB structure 1vnd), the implication is that there is no analogous hydrogen bond in the A35T mutant. Smaller amide proton shift differences associated with I38, K45, and Q50 may be related to slight distortions of the C-terminal region of helix II and the N-terminal portion of helix III. Various 3D 15N and 13C edited NOESY and NOESY–HSQC spectra, as well as a fourth-dimensional 13C edited HMQC–NOESY–HSQC spectrum of the A35T vnd/NK-2–DNA complex were recorded and analyzed. Many long-range NOESY cross peaks that are characteristic of spectra of a stable folded protein were observed in the spectra. These observations confirm that the mutant A35T vnd/NK-2 adopts a stable folded conformation with the usual helix–turn–helix motif when bound to the DNA. Furthermore, the conformation of the mutant A35T vnd/NK-2 bound to the target DNA appears to be homologous to that determined previously for the wild-type homeodomain.
Figure 7.
Histogram showing the difference in chemical shift between the individual amide proton resonances of the wild-type and mutant A35T vnd/NK-2.
DISCUSSION
The decision to investigate the mutant homeodomain was based on the determination that the origin of early embryonic lethality mapped to a single base change in a codon of the vnd/NK-2 homeobox-containing gene that corresponds to the replacement of alanine by threonine in position 35 of the homeodomain. We were quite surprised to find that the single alanine to threonine substitution in position 35 of the vnd/NK-2 homeodomain completely eliminated the capability of the protein to adopt its 3D structure free in solution, especially given that this position is surface exposed (22). To the best of our knowledge, a conservative substitution of the type described here that produces such a catastrophic structural result has not been reported previously. Homology modeling using the coordinates of the wild-type homeodomain provided no significant insight into the nature of the interactions associated with the threonine side chain, which might explain the inability of the mutant protein to fold. The side chain methyl protons of A35 in the wild-type homeodomain make extensive contacts with (i.e., come within 5 Å of) the amide oxygen atoms of E32, L40, and T41 as, well as with the side chains of P42 and V45. Furthermore, serine in position 35 does not appear to give rise to any unfavorable interactions, although the side chain hydroxyl group of S35 in the engrailed homeodomain (22) forms a hydrogen bond with the amide oxygen of L40. Presumably, replacement of the A35 methyl group with the side chain of T in this position causes sufficient distortion to the helix–turn–helix scaffold to obviate formation of the hydrophobic core that is required to maintain the tertiary structure of the protein.
The binding of the mutant A35T vnd/NK-2 homeodomain to the target DNA was demonstrated by electrophoretic gel retardation and CD, as well as from the NMR results. The gel mobility studies have demonstrated that the affinity of the mutant homeodomain is 50-fold lower than the affinity of the wild-type homeodomain for the same target DNA sequence. The lower binding affinity shown by the mutant homeodomain presumably is largely because of the entropic penalty required to fold the protein upon binding to the DNA. The CD results also show the increase of helix content of A35T vnd/NK-2 upon binding to DNA. The values of the mean residue ellipticities at 222 nm of the A35T vnd/NK-2 bound to DNA over the temperature range of −5 to 40°C indicate that the thermal stability of the mutant homeodomain bound to the DNA is analogous to that of the wild-type vnd/NK-2. This observation is further supported, with the exception of L40, by the similarity of the amide proton chemical shifts of the wild-type and the mutant homeodomains. The significant change in the chemical shift of the amide proton of L40 in the A35T mutant strongly suggests an alteration in the nature of the i,i-5 hydrogen bond between the amide proton of L40 and the amide oxygen atom of T35 relative to that of the corresponding hydrogen bond in the wild-type homeodomain. The full determination of the 3D structure of the A35T vnd/NK-2–DNA complex is in progress.
The structural basis of the early embryonic lethality in the mutant allele may be divided into three possible distinct but related events. The first event, which would be the inability of the homeodomain region of the full-length protein to fold after translation, could result in decreased cellular levels of the protein because of degradation by endogenous proteases. The second event could directly involve transcription regulation. Here, cellular concentrations of the homeodomain containing protein could be too low to allow for binding to the target DNA sequence (or sequences) given the reduced binding affinity of the mutant homeodomain. The third event could involve a subtle alteration of the 3D structure of the mutant homeodomain-containing protein relative to that of the wild-type analog in the DNA bound state. Such a structural perturbation might alter specificity of protein–protein interactions involved in the cascade of events leading to embryonic development. Although the data presented here do not bridge the gap between structural modifications and alterations in function, they do provide interesting correlations and permit us to design more specific experiments.
Acknowledgments
We thank Dr. James M. Gruschus and Dr. Nico Tjandra for helpful discussions and assistance in NMR data acquisition and processing.
ABBREVIATIONS
- 1D
one-dimensional
- 3D
three-dimensional
- LB
Luria–Bertani
References
- 1.Duboule D. Guidebook to the Homeobox Genes. New York: Oxford Univ. Press; 1994. [Google Scholar]
- 2.de Kok Y J, Cremers C W, Ropers H-H, Cremers F P. Hum Mutat. 1997;10:207–211. doi: 10.1002/(SICI)1098-1004(1997)10:3<207::AID-HUMU5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Benassayag C, Seroude L, Boube M, Erard M, Cribbs D L. Mech Dev. 1997;63:187–198. doi: 10.1016/s0925-4773(97)00040-3. [DOI] [PubMed] [Google Scholar]
- 4.Benassayag C, Boube M, Seroude L, Cribbs D L. Genetics. 1997;146:939–949. doi: 10.1093/genetics/146.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 6.Vastardis H, Karimbux N, Guthua S W, Seidman J G, Seidman C E. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Golden S, Wu L, Maxson R. Hum Mol Genet. 1996;5:1915–1920. doi: 10.1093/hmg/5.12.1915. [DOI] [PubMed] [Google Scholar]
- 8.Sornson M W, Wu W, Dasen J S, Flynn S E, Norman D N, O’Connell S M, Gukovsky I, Carriere C, Ryan A K, Miller A P, et al. Nature (London) 1996;384:327–333. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez F, Martin-Morris L E, Velasco L, Chu H, Sierra J, Rosen D R, White K. EMBO J. 1995;14:3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson S A, Qiu M, Bulfone A, Eisenstat D D, Meneses J, Pedersen R, Rubenstein J L. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 11.Mortlock D P, Innis J W. Nat Genet. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- 12.Stoffers D A, Zinkin N T, Stanojevic V, Clarke W L, Habener J F. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 13.Morell R, Carey M L, Lalwani A K, Friedman T B, Asher J H. Hum Hered. 1997;47:38–41. doi: 10.1159/000154387. [DOI] [PubMed] [Google Scholar]
- 14.White K, DeCelles N L, Enlow T C. Genetics. 1983;104:433–448. doi: 10.1093/genetics/104.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, Nirenberg M. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skeath J B, Pangoniban G F, Carroll S B. Development (Cambridge, UK) 1994;120:1517–1524. doi: 10.1242/dev.120.6.1517. [DOI] [PubMed] [Google Scholar]
- 17.Mellerick D, Nirenberg M. Dev Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez F, Campos-Ortega J A. Neuron. 1990;5:81–89. doi: 10.1016/0896-6273(90)90036-f. [DOI] [PubMed] [Google Scholar]
- 19.Nirenberg M, Nakayama K, Nakayama N, Kim Y S, Mellerick D, Wang L-H, Webber K, Lad R. Ann NY Acad Sci. 1995;758:224–242. doi: 10.1111/j.1749-6632.1995.tb24830.x. [DOI] [PubMed] [Google Scholar]
- 20.Desiree H H, Tsao D H H, Gruschus J M, Wang L-H, Nirenberg M, Ferretti J A. Biochemistry. 1994;33:15053–15060. doi: 10.1021/bi00254a014. [DOI] [PubMed] [Google Scholar]
- 21.Gruschus J M, Tsao D H H, Wang L-H, Nirenberg M, Ferretti J A. Biochemistry. 1997;36:5372–5380. doi: 10.1021/bi9620060. [DOI] [PubMed] [Google Scholar]
- 22.Tsao D H H, Gruschus J M, Wang L-H, Nirenberg M, Ferretti J A. J Mol Biol. 1995;251:297–307. doi: 10.1006/jmbi.1995.0435. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Stark M R, Johnson A D, Wolberger C. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 24.Wolberger C, Vershon A K, Liu B, Johnson A D, Pabo C O. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 25.Kissinger C R, Liu B, Martin-Bianco E, Kornberg T B, Pabo C O. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 26.Otting G, Qian Y Q, Billeter M, Müller M, Affolter M, Gehring W J, Wüthrich K. EMBO J. 1990;9:3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian Y Q, Billeter M, Otting G, Müller M, Gehring W J, Wüthrich K. Cell. 1989;59:573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 28.Horton R M, Pease L R. In: Directed Mutagenesis: A Practical Approach. McPherson M J, editor. Oxford, U.K.: Information Press; 1991. pp. 217–247. [Google Scholar]
- 29.Novagen. pET System Manual. 6th Ed. Madison, WI: Novagen; 1995. [Google Scholar]
- 30.Xiang B, Ferretti J A, Fales H M. Anal Chem. 1998;70:2188–2190. [Google Scholar]
- 31.Carey J. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 32.Weiler S, Gruschus J M, Tsao D H H, Yu L, Wang L-H, Nirenberg M, Ferretti J A. J Biol Chem. 1998;273:10994–11000. doi: 10.1074/jbc.273.18.10994. [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Cavanagh J, Fairbrother W J, Palmer A G, III, Skelton N J. Protein NMR Spectroscopy. New York: Academic; 1996. [Google Scholar]
- 35.Grzesiek S, Bax A. J Magn Reson. 1992;99:201–207. [Google Scholar]
- 36.Grzesiek S, Bax A. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 37.Grzesiek S, Bax A. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 38.Vuister G W, Clore G M, Gronenborn A M, Powers R, Garrett D S, Tschudin R, Bax A. J Magn Reson B. 1993;101:210–213. [Google Scholar]
- 39.Tjandra N, Bax A. J Am Chem Soc. 1997;119:8076–8082. [Google Scholar]