Abstract
Yeast cells mutated in YRB2, which encodes a nuclear protein with similarity to other Ran-binding proteins, fail to export nuclear export signal (NES)-containing proteins including HIV Rev out of the nucleus. Unlike Xpo1p/Crm1p/exportin, an NES receptor, Yrb2p does not shuttle between the nucleus and the cytoplasm but instead remains inside the nucleus. However, by both biochemical and genetic criteria, Yrb2p interacts with Xpo1p and not with other members of the importin/karyopherin β superfamily. Moreover, the Yrb2p region containing nucleoporin-like FG repeats is important for NES-mediated protein export. Taken together, these data suggest that Yrb2p acts inside the nucleus to mediate the action of Xpo1p in at least one of several nuclear export pathways.
Macromolecular movement across the nuclear envelope is a highly orchestrated process (reviewed in ref. 1). It is now well established that proteins destined for the nuclear interior contain NLSs (termed nuclear localization sequences) that target the protein to the nucleus. A general picture has emerged whereby an NLS-bearing protein is recognized by cytoplasmic transporters termed importin (or karyopherin) and transportin. The resulting complex docks at (2, 3) and moves through the nuclear pore complex (NPC) by a still poorly understood mechanism (see reviews in ref. 4). Once in the nuclear interior, the cargo dissociates from the transport machinery and the importins then are recycled back to the cytoplasm.
At least some proteins move out of the nucleus in a manner that is, in principle, the reciprocal of nuclear protein entry. In particular, certain proteins, such as HIV Rev, contain short stretches of amino acids that target them for export out of the nucleus (5, 6). These so-called nuclear export signals (NESs) are recognized by a highly conserved export receptor, termed exportin/Xpo1p or Crm1p, which is related to the importins (7–10). The general model is that proteins such as exportin recognize NES-containing proteins in the nucleus and deliver them to and through the NPC. Once in the cytoplasm, release of the exported protein occurs and exportin reenters the nucleus for another round of transport. The observation that exportin shuttles between the nucleus and the cytoplasm (7) lends support to this model.
The biological significance of protein export is intimately connected to the mechanism for RNA export. Rev is specialized for the export of viral RNAs, whereas export of cellular mRNA also depends on a set of hnRNP proteins (reviewed in ref. 11). It is presumably the RNA–protein complex that is recognized by the export machinery. In fact, mutation of several importin-like proteins in yeast results in blocks in mRNA export (7, 12). Taken together, it is probable that there will be a number of distinct “export pathways” defined by the type of cargo being transported.
The GTPase, Ran (13, 14) (Gsp1p in yeast), critically defines the movement of macromolecules in both directions across the nuclear envelope. The asymmetric distribution of the major Ran regulators may provide an important key to how Ran mediates both import and export. Ran in the GDP bound state is probably high in the cytoplasm because of the cytoplasmic location of the RanGAP (Rna1p in yeast) (15). Conversely, Ran-GTP would be the preferred state in the nucleus where the Ran nucleotide exchanger, Rcc1 (Prp20p in yeast), is located (16). One current view is that Ran-GDP is required for importin–substrate complexes to form and enter the nucleus (17). Conversely, Ran-GTP is thought to be required for NES-containing proteins to associate with exportin and move out of the nucleus (8, 18). Hydrolysis of GTP by Ran, which has been shown to be required for nuclear transport (13, 14, 19), simply could be needed to generate Ran-GDP for multiple rounds of transport.
A second class of proteins that affect the activity of Ran, termed Ran-binding proteins (RanBPs), has been identified. These RanBPs have in common a stretch of about 100 aa that compose their Ran-binding domains. Mammalian RanBP1 and its yeast counterpart, Yrb1p, were identified by their preferred affinity for the GTP-bound form of Ran (20–22). Further, these proteins stimulate the RanGAP-dependent GTPase activity of Ran (22, 23) and the disassembly of RanGTP–importin complex (24). Yeast Yrb1p is located in the cytoplasm and at the NPCs (22). It is essential for growth, and cells bearing temperature-sensitive mutations in YRB1 fail to properly transport proteins into the nucleus (22), thus providing the first biological function for RanBPs. Evidence from experiments with mammalian cells indicates that RanBP1 may gain access to the nuclear interior, suggesting an intranuclear role as well (25). Interestingly, some yeast YRB1 mutants also show defects in the export of mRNA out of the nucleus (22). In addition, mammalian RanBP1 binds to importin-β and, in doing so, may affect its ability to interact with importin-α and other nuclear transport factors (24, 26).
The recent completion of the DNA sequence of the yeast Saccharomyces cerevisiae genome has provided a complete picture of the genetic make-up of this simple eukaryote. Analysis of the genome indicates that there are only three genes encoding proteins that contain the Ran-binding region, YRB1 being the first to be described (22, 27). The others are NUP2, encoding a nonessential nucleoporin (28), and YRB2 (29, 30), which, in addition to its Ran-binding domain, also contains two FXFG and three FG repeats characteristic of some nucleoporins. However, unlike Yrb1p, Yrb2p is located inside the nucleus and is not essential for growth at normal temperatures, but cells deleted for YRB2 fail to grow at 15°C (e.g., they are cold-sensitive) (29, 30). In addition, unlike the situation with YRB1, YRB2 mutants show no defects in nuclear protein import or mRNA export (29, 30).
The nuclear localization of Yrb2p led us to investigate whether or not it might be involved in export of macromolecules out of the nucleus. We now show that yeast cells missing Yrb2p fail to properly export NES-bearing proteins out of the nucleus. Consistent with its role in nuclear export, we also demonstrate an interaction between Yrb2p and Xpo1p, the NES receptor.
MATERIALS AND METHODS
Yeast Strains.
PSY1134 (MATα his3Δ200 leu2Δ1 trp1 ura3–52), PSY1135 (MATα Δyrb2∷HIS3 his3Δ200 leu2Δ1 trp1 ura3–52), PSY1002 [MATα Δyrb2∷HIS3 (29)], and PSY1003 [MATa Δyrb2∷HIS3 (29)] were used for the analysis of YRB2 function. A nup49–313 mutant strain (31) was used for the protein-shuttling assay. For the interaction trap assay, EGY48 carrying a lacZ reporter plasmid pSH18–34 (32) and EGY42 (gift of R. Brent, Massachusetts General Hospital) was used. XPO1 wild-type and xpo1–1 temperature-sensitive strains (7) were provided by K. Weis (University of California, San Francisco). pPS1312, a xpo1–1 TRP1 plasmid (described below), was introduced into the xpo1–1 strain, and the preexisting xpo1–1 HIS3 plasmid (pKW457) was eliminated, yielding PSY1109. Strains FY23 (MATa leu2Δ1 trp1Δ63 ura3–52, gift of F. Winston, Harvard Medical School), JLY506 [MATa nup2–4 (28)], JLY543 (MATα srp1–31 ade2 his3 leu2 trp1 ura3, gift of G. Fink, Massachusetts Institute of Technology), PSY713 [MATα prp20–1 (33)], PSY714 [MATa rna1–1 (34)], PSY716 [MATα yrb1∷HIS3/pyrb1–1 (22)], and PSY717 [MATα yrb1∷HIS3/pyrb1–2 (22)], PSY967 [MATα kap123∷HIS3 (12)], T255 [MATa mtr10–1 (35)], ESY42 (MATa Δkap104, gift of E. Shen), and Y1705 [MATα cse1–1 (36)] were used for export assays.
Plasmids.
Plasmids encoding the NLS-NES-GFP fusion protein were constructed as follows. Primers were designed to generate PCR fragments encoding the ADH promoter and the simian virus 40 (SV40) NLS, the protein kinase inhibitor (PKI) NES, and green fluorescent protein (GFP). These fragments were inserted into a GFP plasmid pKG50 (gift of J. Kahana, Harvard Medical School, Boston) to yield pPS1372, respectively, so that the resulting NLS-NES-GFP encodes a 63-kDa fusion protein. pPS1494, encoding Rev-GFP, was constructed similarly (M. Lee, unpublished data).
A 1.1-kb BamHI-XbaI fragment containing the YRB2 ORF prepared from pPS848 (29) was inserted into a galactose-inducible GFP plasmid to generate pPS1369. The GFP-YRB2 ORF was placed downstream of the YRB2 promoter to generate pPS1523. YRB2∷HA and its deletion derivatives (29) were cloned into pRS314 (37).
pPS1312, a xpo1–1 TRP1 plasmid, was constructed by cloning a SalI-BamHI fragment carrying xpo1–1, which was prepared from pKW457 (7), into pRS314 by using the same restriction sites. CSE1 gene, including its promoter region, was fused to the GFP to generate pPS1536.
Protein Export Assay.
Cells containing pPS1372 (NLS-NES-GFP) or pPS1494 (Rev-GFP) were exposed to the restrictive temperature for 1–2 hr for temperature-sensitive mutants or for 12–15 hr for cold-sensitive mutants. For pPS1494, expression of the fusion gene was carried out for 2 hr before the temperature shift. Cells were then viewed either directly by fluorescence microscopy or after fixing with formaldehyde for 30 min, which did not alter the GFP signal.
Detection of Protein Interactions.
For interaction trap assay, pJG4–5 (32) with or without genes involved in nuclear transport was cointroduced into EGY48/pSH18–34 with pEG202 (32) or its derivatives. Meanwhile, pJG4–5 and pEG202 derivatives were introduced separately into EGY42 and EGY48/pSH18–34, respectively. These strains were crossed to each other to generate diploid strains containing both pJG4–5 and pEG202 derivatives. The expression of the fusion genes was checked by immunoblotting with 12CA5 anti-HA mouse mAb or anti-LexA serum. To test for interactions, cells were streaked onto selective media without leucine, and β-galactosidase activity of whole-cell lysates was assayed (38) after cultivating in liquid media. The mean values of four independent experiments were plotted as a bar graph.
Protein interaction assays with GST-Yrb2 fusion proteins from yeast lysates were carried out as described previously (29). Proteins bound to the GST fusion proteins were detected by immunoblotting.
RESULTS
YRB2 Is Required for NES-Dependent Nuclear Export.
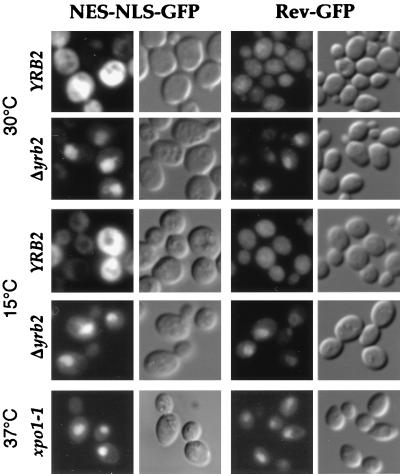
To investigate nuclear protein export, we analyzed the distribution of a reporter protein containing the SV40 NLS and the NES from the protein kinase inhibitor fused to the GFP. As has been reported previously (7), when expressed in wild-type cells at either 30°C or 15°C, the NLS-NES-GFP accumulates in the cytoplasm (Fig. 1), consistent with the action of the NES dominating over that of the NLS. In contrast, in cells deleted for YRB2 (Δyrb2), the NLS-NES-GFP accumulated in the nucleus at both 30°C and 15°C (Fig. 1), consistent with its inability to exit the nucleus once inside. This behavior is similar to that observed with cells mutated for XPO1 (7), which encodes exportin and is required for NES-dependent protein export (Fig. 1 and ref. 7).
Figure 1.
NES-mediated protein export in Δyrb2 cells. A NLS-NES-GFP- or Rev-GFP-encoding plasmid was introduced into wild-type, Δyrb2, or xpo1–1 cells. The localization of the GFP fusion proteins was examined at 30°C, after a 15-hr shift to 15°C or after a 1-hr shift to 37°C by fluorescence microscopy.
We also examined a fusion of the HIV Rev to GFP. When expressed in wild-type cells at either 30°C or 15°C, like the case with NLS-NES-GFP, Rev-GFP accumulates in the cytoplasm. This is consistent with the Rev NES predominating over the activity of the endogenous Rev NLS. However, as in xpo1–1 cells, Rev-GFP accumulated in the nuclei of Δyrb2 cells at both 30°C and 15°C (Fig. 1). In contrast to the case of xpo1–1 and Δyrb2 mutants, we did not observe any significant export defect of the NLS-NES-GFP reporter in other nuclear transport mutants surveyed (Table 1). This includes cells bearing mutations in CSE1, KAP104, KAP123, and MTR10, which all encode members of the importin-β family. In some cases, we instead observed a stronger fluorescence signal in the cytoplasm. This was the case for cells mutated in RNA1, SRP1, and, in particular, YRB1 (Table 1). This is consistent with a block in nuclear import rather than export of the reporter fusion protein in these cells. Taken together, from all analyzed mutants, only xpo1–1 and Δyrb2 strains show nuclear accumulation of NES-carrying fusion proteins, suggesting that YRB2 is necessary for proper NES-dependent nuclear protein export.
Table 1.
Distribution of NLS-NES-GFP in nuclear transport mutants
| Strains containing the NLS-NES-GFP construct | Localization of GFP signal*
|
||
|---|---|---|---|
| Cytoplasmic | Both | Nuclear | |
| Wild type | + | ||
| Δxpo1::LEU2/pXPO1 | + | ||
| Δxpo1::LEU2/pxpo1-1 | + | ||
| Δyrb2::HIS3/pYRB2 | + | ||
| Δyrb2::HIS3 | + | ||
| cse1-1 | + | ||
| Δkap104 | + | ||
| Δkap123 | + | ||
| mtr10-1 | + | ||
| prp20-1 | + | ||
| nup2-4 | + | ||
| rna1-1 | + | ||
| srp1-31 | + | ||
| Δyrb1::HIS3/pyrb1-1 | + | ||
| Δyrb1::HIS3/pyrb1-2 | + | ||
Wild-type and mutant cells bearing the NLS-NES-GFP reporter were examined by fluorescence microscopy after shifting to the nonpermissive temperatures. The level of defect was scored as “Cytoplasmic” when no nuclear signal was observed, “Both” when similar to “Cytoplasmic” with additional nuclear signal in 20–40% of total cells, and “Nuclear” for >90% of total cells with strong nuclear fluorescence.
Yrb2p Is Not a Shuttling Protein.
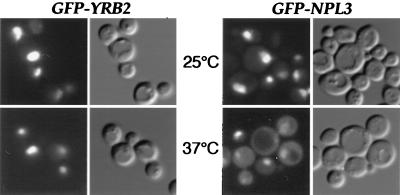
We previously reported that Yrb2p is exclusively located inside the nucleus (29). However, as with other proteins involved in nuclear transport including Xpo1p (7), it is possible that Yrb2p shuttles in and out of the nucleus and that its rate of reimport is high compared with export. To test the possibility that Yrb2p can exit the nucleus, we constructed a gene fusion between Yrb2p and GFP. The resulting protein fusion, GFP-Yrb2p, is functional in that it restores normal growth to Δyrb2 cells at 15°C (data not shown). To assay Yrb2p nuclear export, we placed GFP-Yrb2p under control of the repressible GAL1 promoter and expressed the fusion protein in nup49–313 cells. Cells bearing the temperature-sensitive nup49–313 allele are defective in nuclear import of proteins such as Yrb2p at 37°C but not in export, making it is possible to uncouple the two processes as we have demonstrated previously (31). Therefore, a shuttling protein such as the hnRNP Npl3p is nuclear at the permissive temperature but accumulates in the cytoplasm at the nonpermissive temperature because of failure to reenter the nucleus (Fig. 2 and ref. 31). GFP-Yrb2p, on the other hand, remains entirely nuclear at both 25°C and 37°C, indicating that under these conditions it does not leave the nucleus (Fig. 2). Thus, we conclude that Yrb2p does not cycle between the nucleus and the cytoplasm.
Figure 2.
Yrb2p does not shuttle between the nucleus and the cytoplasm. The nup49–313 strain expressing GFP-Yrb2p or GFP-Npl3p was used for the protein-shuttling assay. After expression/repression at 25°C, cells were shifted to 37°C and incubated for 5.5 hr and the fusion proteins were localized by fluorescence microscopy.
Yrb2p Interacts with Xpo1p, the NES Receptor.
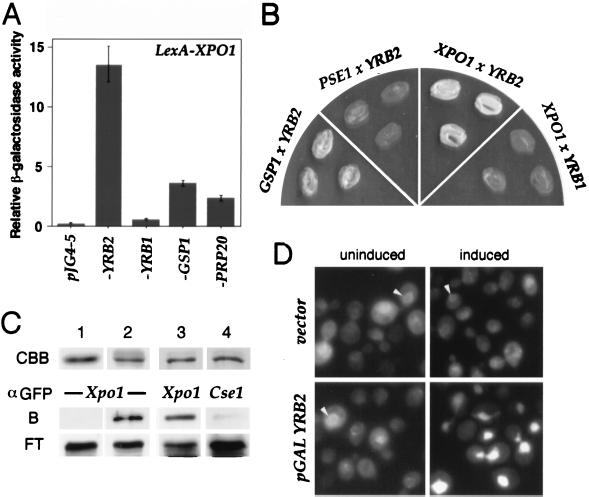
Because the action of Yrb2p may parallel that of other export factors, we tested for interactions of Yrb2p with Xpo1p as well as other nuclear transport factors. To test for interactions by two-hybrid analysis (32), we constructed a fusion between lexA and XPO1 that expressed a protein of the appropriate size in yeast but did not activate expression of either the lacZ reporter as evidenced by the lack of β-galactosidase activity or LEU2 as evidenced by the cells’ inability to grow in the absence of leucine. However, when coexpressed with Yrb2p fused to an activator domain, the lacZ reporter was expressed as evidenced by the ability of the double transformants to turn blue on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal)-containing plates and by direct measure of the resulting β-galactosidase activity (Fig. 3A). Similarly, the LEU2 reporter was also expressed as evidenced by the ability of the double transformants to grow in the absence of leucine (Fig. 3B). No such strong interactions were observed when LexA-Xpo1p was combined with either an empty vector or with a Yrb1p-activator-containing plasmid (Fig. 3 A and B). We also detect some weaker interactions between LexA-Xpo1p and Gsp1p (39), which encodes the yeast Ran, as well as with Prp20p (40), which encodes the Gsp1p nucleotide exchanger (Fig. 3A). The former interaction is consistent with the prediction that, as with all other importin-like proteins examined thus far (41–43), exportin interacts with Ran (7, 8) and the latter is consistent with our previous report that Yrb2p binds to Prp20p (29). Finally, when we tested for interaction between Yrb2p and Pse1p, a different importin-like protein (12, 44), we observed no interaction by two-hybrid analysis (Fig. 3B).
Figure 3.
Interactions between YRB2 and XPO1. (A) EGY42 × EGY48 diploid strains containing plexA-XPO1 (bait) and the indicated pJG4–5 derivatives (prey) were grown in selective media. After 2 hr of induction with galactose, β-galactosidase activity was measured as described in Materials and Methods. (B) EGY48 containing the indicated tester genes were streaked onto leucine drop-out media and grown at 30°C. (C) Cells expressing the GST-YRB2 fusion as well as XPO1-GFP or CSE1-GFP were lysed, and the resulting lysate was mixed with glutathione-Sepharose beads. After washing, proteins bound to the beads were detected by α-GFP immunoblotting. CBB, GST (lane 1) or GST-Yrb2 fusion protein (lanes 2–4) stained with Coomassie brilliant blue; B, bound; FT, flow through. (D) Cells containing a GAL1-YRB2 or a vector as well as a plasmid encoding XPO1-GFP were grown in raffinose media followed by 2 hr in galactose (induced) or glucose (uninduced) and were viewed by fluorescence microscopy. The nuclear rim is indicated with arrowheads.
To further test the predicted interaction between Yrb2p and Xpo1p, the following biochemical experiments were carried out. A gene fusion encoding a fully functional GST-Yrb2 fusion protein was expressed in yeast that also expressed the functional Xpo1-GFP protein fusion (7). The GST-Yrb2p was purified and copurifying proteins were analyzed by immunoblotting with anti-GFP antibodies. We found by this analysis that a significant fraction of the Xpo1-GFP remained associated with Yrb2p (Fig. 3C, lanes 2 and 3). On the other hand, no Xpo1-GFP was found complexed with GST alone (Fig. 3C, lane 1). Moreover, when the same experiment was carried out in cells expressing a functional fusion of GFP to Cse1p, another member of the importin-β family, essentially no binding to Yrb2p was detected (Fig. 3C, lane 4).
The intracellular distribution of Xpo1p is also affected by the level of Yrb2p. A functional Xpo1-GFP fusion protein localizes in the nucleus, the cytoplasm, and predominantly at the nuclear envelope in wild-type cells (Fig. 3D and ref. 7). However, overexpression of Yrb2p from the strong GAL1 promoter caused accumulation of Xpo1-GFP inside the nucleus (Fig. 3D). Because Xpo1-GFPp normally shuttles between the nucleus and the cytoplasm, this result suggests that overexpressed nuclear Yrb2p traps Xpo1p in the nucleus and may, in turn, block the exit of NES-containing proteins out of the nucleus. This appears to be the case because overexpression of Yrb2p also resulted in accumulation of the NLS-NES-GFP inside the nucleus (data not shown).
Genetic Interactions Between YRB2 and XPO1.
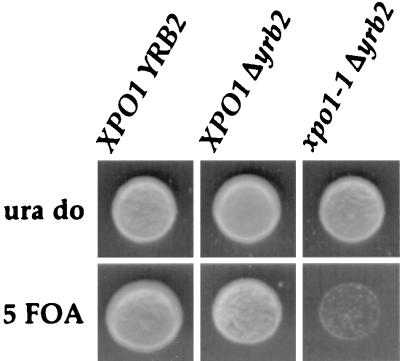
To seek independent confirmation that Yrb2p acts in concert with Xpo1p, we turned to genetic tests of synthetic lethality. Haploid Δyrb2 cells covered by YRB2 on a URA3-containing plasmid were crossed to cells bearing the temperature-sensitive xpo1–1 allele on a TRP1-containing plasmid. The resulting diploid was sporulated and subjected to tetrad analysis. The resulting spores were allowed to germinate and their genotypes were determined. Those bearing both the YRB2 gene knockout and the xpo1–1 mutation then were tested for their ability to lose the wild-type YRB2-containing plasmid on 5-fluoroorotic acid plates. As shown in Fig. 4, the growth of the double mutant was severely inhibited unless it maintained wild-type YRB2, thus indicating a synthetic lethal relationship between these two genes. This result in combination with the above interaction data indicates that Yrb2p acts in the same “pathway” as Xpo1p. However, it remains a formal possibility that Yrb2p also acts in a parallel transport pathway as well.
Figure 4.
Synthetic relationship of YRB2 and XPO1. Δyrb2 cells containing a YRB2 CEN URA3 plasmid were crossed to xpo1–1 cells. After sporulation and dissection of the resulting tetrads, haploid progeny with the indicated genotypes were spotted onto a uracil drop-out plate (ura do) or a plate with 1 mg/ml 5-fluoroorotic acid (5 FOA) and grown at 25°C.
The Nucleoporin-Like Repeats of Yrb2p Are Important for Export Function.
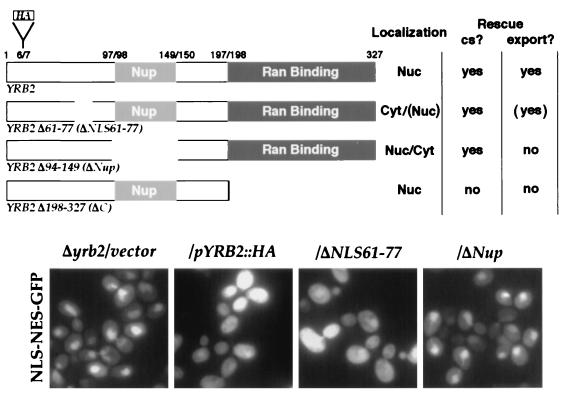
The ability of several previously characterized YRB2 deletion mutants to support NLS-NES-GFP protein export was tested. We found that a mutant Yrb2p lacking the C terminus did not rescue the NES-mediated export defect (Fig. 5). In addition, a chimeric protein with the Yrb2p Ran-binding region replaced with that from Yrb1p did not rescue the export defect (data not shown). Neither of these mutated proteins restores growth at low temperatures (29). Surprisingly, Yrb2p lacking the region that contains the five FG repeats (ΔNup) did not support nuclear export (Fig. 5) even though the mutant can rescue the Δyrb2 cold-sensitive growth defect (29).
Figure 5.
The FG repeats are important for Yrb2p activity. Δyrb2 cells expressing NLS-NES-GFP and the indicated Yrb2p derivatives were grown at 30°C and shifted to 15°C for 13 hr, and the localization of the NLS-NES-GFP fusion protein was determined by fluorescence microscopy. All mutant Yrb2 proteins are expressed at levels similar to the intact protein. Parentheses indicate that a fraction of the protein still appears in either the nucleus or cytoplasm.
One possible explanation for the these results is that the decreased amount of the nuclear localized Yrb2pΔNup (29) could result in its inability to sustain protein export. To test this possibility, we constructed an additional YRB2 mutant that is mislocalized to the cytoplasm. There are two sequences similar to the canonical NLS at positions 33–38 and 61–77 (29) in Yrb2p. When hemagglutinin (HA)-tagged versions of Yrb2p lacking these sequences (ΔNLS33–38 and ΔNLS61–77) were localized by immunofluorescence, ΔNLS33–38 was still nuclear but significant amounts of ΔNLS61–77 accumulated in the cytoplasm (data not shown), suggesting that amino acids 61–77 function as a bipartite NLS. Importantly, the ΔNLS61–77 mutant is capable of rescuing the cold sensitivity of the Δyrb2 mutant as well as the nuclear export defect (Fig. 5), indicating the lower nuclear level of Yrb2p does not necessarily result in the loss of export activity, but rather that the FG-containing region of Yrb2p is important for the proper function of Yrb2p in nuclear protein export.
DISCUSSION
In this report, we show that yeast cells deleted for YRB2, which encodes one of only three Ran-binding protein family members in yeast, fail to export two NES-containing proteins from the nucleus. Consistent with its role in protein export, we also find by three independent criteria that Yrb2p interacts with Xpo1p, a receptor that recognizes NES-containing proteins for export. Taken together, we propose that Yrb2p is acting to promote nuclear protein export together with Xpo1p.
Mutations in other transport factors examined in this report show no protein export defects (see Table 1). However, unlike the case for cells deleted for YRB2 (29, 30), all of these mutants, with the exception of cse1–1, have been reported to show defects in nuclear protein import (1). For example, a temperature-sensitive mutant in SRP1 (45), which encodes the importin-α subunit, shows only protein import defects and not defects in export of mRNAs (46) or, as we show here, NES-dependent protein export. Similarly, KAP104, KAP123, and MTR10 encode members of the importin-β family and have been implicated in import of specific RNA-binding proteins into the nucleus (42, 44, 47). In contrast, a human homologue of CSE1, CAS, which also encodes an importin-β-like protein, has been implicated in recycling of importin-α from the nucleus to the cytoplasm (24). Our results indicate that, thus far, XPO1 encodes the only importin-β-like protein important for NES-dependent export. Moreover, temperature-sensitive mutations in essential genes encoding three Ran/Gsp1p regulators, Rna1p (the Ran/Gsp1p GAP), Prp20p (the Ran/Gsp1p nucleotide exchange factor), and Yrb1p (a GAP coactivator), all show no defect in NES-dependent protein export. In fact, for SRP1, RNA1, and YRB1, we observe an increase in cytoplasmic accumulation of the NLS-NES reporter, suggesting that the corresponding proteins may function primarily in protein import. The fact that the same mutants show defects of both protein import and mRNA export could, in principle, be a result of failure to import an essential mRNA export factor, such as the mRNA-binding protein Npl3p, which is important for mRNA export (31).
That cells deleted for YRB2 still grow at 30°C (29, 30), albeit somewhat less well than wild-type cells, suggests that Yrb2p facilitates but is dispensable for protein export. Consistent with this view, the failure to export NES-containing proteins is apparent, but not complete, at 30°C but worsens after cells are shifted to the nonpermissive temperature. Moreover, when combined with the xpo1–1 temperature-sensitive mutation, the growth of the resulting double mutant is severely impaired, indicating that both genes participate in the same pathway. XPO1 is, on the other hand, essential for growth at all temperatures (7); however, nonconditional alleles exist that still show defects in protein export (48). These observations taken together indicate that Yrb2p and Xpo1p mediate at least one of several export pathways. Although there still remains a debate about the requirement of XPO1 for mRNA export (7, 48), for which Yrb2p is not required (29, 30), the role played by Yrb2p may be more specific for NES-mediated protein export and not for mRNA export.
We propose the following model for the action of Yrb2p in nuclear protein export. Yrb2p may associate with Xpo1p and with Ran via its Ran-binding domain in nucleoplasm. Yrb2p also forms a complex with Prp20p (29), perhaps assisting Yrb2p to properly interact with Gsp1p in its GTP-bound state (30). Xpo1p has been shown to bind Ran-GTP in the presence of NES-containing proteins (8). Yrb2p may further stabilize this complex and/or target it to the NPC, where Yrb2p could be replaced by interaction of Xpo1p with FG-containing nucleoporins (48, 49). Both of these steps could occur without Yrb2p, but at lower efficiency, deduced from the nonessentiality of YRB2. However, the presence of Yrb2p may significantly increase the rate or efficiency of these reactions. Further studies undoubtedly will reveal yet more factors affecting this complex process.
Acknowledgments
We thank K. Weis, E. Shen, M. Lee, J. Kahana, and M. Seedorf for reagents and C. Cole for comments on the manuscript. T.T. and H.K. were supported by fellowships from the Japanese Society for the Promotion of Science and the Deutsche Forschungsgemeinschaft, respectively. This work was supported by grants from the National Institutes of Health and by the Novartis/DFCI Drug Discovery Program.
ABBREVIATIONS
- NLS
nuclear localization sequence
- NES
nuclear export signal
- NPC
nuclear pore complex
- GFP
green fluorescent protein
References
- 1.Corbett A H, Silver P A. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 3.Moroianu J, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 5.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 6.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 7.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 9.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 11.Lee M S, Silver P A. Curr Opin Genet Dev. 1997;7:212–219. doi: 10.1016/s0959-437x(97)80131-1. [DOI] [PubMed] [Google Scholar]
- 12.Seedorf M, Silver P A. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melchior F, Paschal B, Evans J, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 15.Hopper A K, Traglia H M, Dunst R W. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtsubo M, Okazaki H, Nishimoto T. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Görlich D, Panté N, Kutay U, Aebi U, Bischoff F R. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S A, Carey K L, Macara I G. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 19.Schlenstedt G, Saavedra C, Loeb J D J, Cole C N, Silver P A. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coutavas E, Ren M, Oppenheim J D, D’Eustachio P, Rush M G. Nature (London) 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 21.Lounsbury K M, Beddow A L, Macara I G. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- 22.Schlenstedt G, Wong D H, Koepp D M, Silver P A. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bischoff F R, Krebber H, Smirnova E, Dong W, Ponstingl H. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutay U, Bischoff F R, Kostka S, Kraft R, Görlich D. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 25.Richards S A, Lounsbury K M, Carey K L, Macara I G. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi N C, Adam E J H, Visser G D, Adam S A. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouspenski I I, Mueller U W, Matynia A, Sazer S, Elledge S J, Brinkley B R. J Biol Chem. 1995;270:1975–1978. doi: 10.1074/jbc.270.5.1975. [DOI] [PubMed] [Google Scholar]
- 28.Loeb J D J, Davis L I, Fink G R. Mol Biol Cell. 1993;4:209–222. doi: 10.1091/mbc.4.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taura T, Schlenstedt G, Silver P A. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 30.Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M S, Henry M, Silver P A. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 32.Gyuris J, Golemis E A, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 33.Koepp D M, Wong D H, Corbett A H, Silver P A. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corbett A H, Koepp D M, Lee M S, Schlenstedt G, Hopper A K, Silver P A. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff A M. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 39.Belhumeur P, Lee A, Tam R, DiPaolo T, Fortin N, Clark M W. Mol Cell Biol. 1993;13:2152–2161. doi: 10.1128/mcb.13.4.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aebi M, Clark M W, Vijayraghavan U, Abelson J. Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- 41.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pemberton L F, Rosenblum J S, Blobel G. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff F R. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rout M P, Blobel G, Aitchison J D. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 45.Yano R, Oakes M, Tabb M M, Nomura M. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loeb J D J, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G R. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aitchison J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 48.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. Current Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 49.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]