Abstract
RNA polymerase II (pol II) transcribes genes encoding proteins and non-coding small nuclear (sn)RNAs. The carboxy-terminal domain (CTD) of the largest subunit of mammalian RNA polymerase II (pol II), comprising tandem repeats of the heptapeptide consensus tyr1ser2pro3thr4ser5pro6ser7, is required for expression of both gene types. Here, we show that mutation of ser7 to alanine causes a specific defect in snRNA gene expression. We also present evidence that phosphorylation of ser7 facilitates interaction with the snRNA gene-specific Integrator complex. These findings asign a biological function to this amino acid and highlight a gene type-specific requirement for a residue within the CTD heptapeptide, supporting the existence of a CTD code.
Human snRNA genes transcribed by pol II, including those encoding U1 and U2 spliceosomal RNAs, have specialized promoters comprising conserved proximal and distal sequence elements (PSE and DSE) (1). Rather than polyadenylation signals, 3′ box elements direct co-transcriptional formation of the primary 3′ end of transcripts (2, 3). The 3′ end of these pre-snRNAs is further processed in the cytoplasm to yields mature non-polyadenylated snRNAs (2). Removal of the CTD of the large subunit of mammalian pol II drastically affects expression of both snRNA and protein-coding genes (2-4). The CTD has a unique structure composed of multiple repeats containing residues that undergo reversible phosphorylation during transcription (5). For example, phosphorylation of ser5 by CDK7 facilitates promoter release and RNA capping, whereas ser2 phosphorylation by CDK9 is associated with processive elongation and 3′ processing (5, 6). No role has yet been ascribed to ser7.
The mammalian pol II CTD comprises 52 repeats, 25 of which deviate from the consensus at position 7. The mainly consensus repeats 1-25 activate snRNA 3′ processing more effectively than repeats 27-52, which have few serines at position 7 (2). In contrast, both halves of the CTD are equally effective in activating polyadenylation (7). We have tested the requirement for ser7 for expression of snRNA (U2G (2)) and mRNA (pCMV-hnRNPK (8)) templates in 293 cells by introducing mutations into consensus (Con) CTD repeats in an α-amanitin-resistant pol II large subunit (Rpb1) (9) (Figures 1A, S1A). The large subunit of endogenous pol II is very sensitive to inhibition by α-amanitin, facilitating complementation studies (9). A CTD with at least 25 consensus repeats ((Con)25) was used since this supports efficient production and co-transcriptional 3′ processing of transcripts from snRNA and protein-coding templates, while five CTD repeats (Δ5) do not (2, 4) (Figure S2A, B).
Figure 1. Ser7 is required for expression of snRNA but not protein-coding templates.
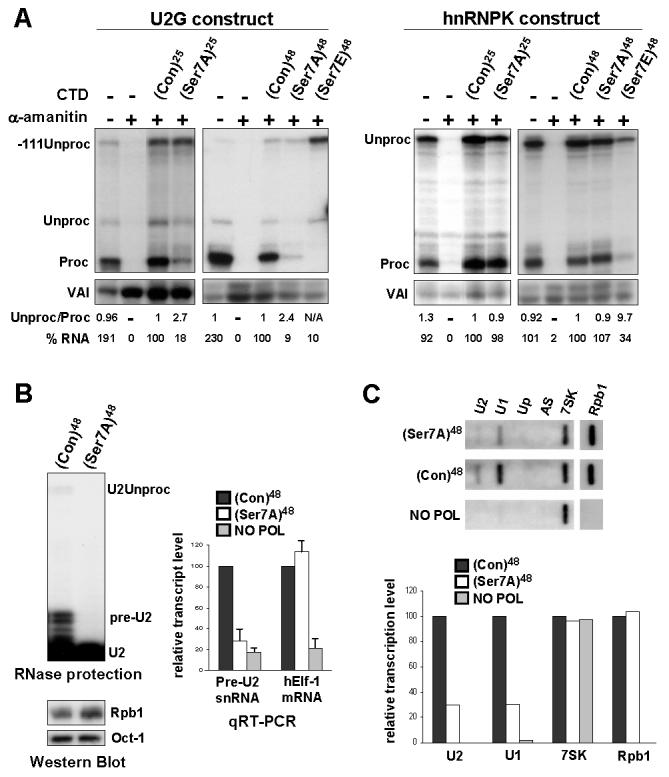
(A) RNase protection analysis of RNA transcribed from U2G or pCMV-hnRNPK constructs after ectopic expression of α-amanitin-resistant Rpb1 (see Figure S1A). (Con) designates consensus CTD heptapeptides. (B) RNase protection analysis of transcripts from endogenous U2 genes in cells stably expressing α-amanitin-resistant Rpb1 and Western blot analysis of Rpb1 expression. qRT-PCR analysis of U2 pre-snRNA and hElf-1 mRNA in total RNA normalized to 7SK RNA with α-amanitin-treated cells expressing no Rpb1 as negative control (NO POL). (C) Run-on analysis of endogenous U1 and U2 snRNA genes in cells transfected with α-amanitin-resistant Rpb1s. AS and Up are negative controls (see Materials and Methods). Quantitation of this data is shown in the bar graph below.
Mutation of ser7 to the non-phospho-acceptor alanine (Ser7A) in a background of 25 repeats reduces the level of properly processed U2G transcripts (Proc) and increases the ratio of unprocessed transcripts (Unproc). However, this mutation affects neither the level nor 3′ processing of hnRNPK transcripts (Figures 1A, S1B). Mutation of ser7 to alanine in a background of 48 consensus repeats (10) ((Con)48) has a similarly drastic effect on U2G transcripts, without affecting hnRNPK transcripts. This mutation does not affect expression of Rpb1 or ser2/ser5 phosphorylation (Figure S1C). Thus, ser7 is specifically required for efficient production of properly 3′ processed transcripts from an snRNA template.
Mutation of ser7 to the phospho-mimic glutamic acid (Ser7E) has a drastic effect on the level and 3′ processing of transcripts from both the U2G and hnRNPK templates (Figure 1A), possibly due to a defect in pol II recruitment/association caused by charged residues. The increase in -111Unproc may reflect a defect in termination of transcription of U2G resulting from the defect in 3′ processing (2, 11, Materials and Methods). The Ser7E mutation abolishes recognition by anti-Ser2-P (Figure S1C), reflecting a drop in ser2 phosphorylation and/or interference with antibody recognition. A drop in ser2 phosphorylation would account for the defect in 3′ processing and indicate that mutations can have secondary effects. Rpb2 is not affected by α-amanitin treatment in the same way as Rpb1 (Figure S1C), indicating that not all pol II-specific subunits are subject to α-amanitin-induced turnover in 293 cells.
Mutation of ser2 to alanine (Ser2A) affects 3′end formation of U2G and hnRNPK transcripts (Figure S2A, B, D) (6). Ser2A accumulates to a higher level than (Con)25 and Ser2E is undetectable (Figure S2C), suggesting that phosphorylation of ser2 is involved in Rpb1 turn-over. Introduction of alanine at position 5 (Ser5A) reduces steady state U2G and hnRNPK transcript levels and processing (Figure S2A, B, D). This likely reflects the requirement for ser5 phosphorylation for addition of the 5′ cap, which protects the RNA and activates 3′ processing (5, 12). Although introduction of glutamic acid at position 5 restores RNA levels, suggesting that capping now occurs, 3′ processing is still inefficient, demonstrating that a charged amino acid does not fully compensate for the lack of a serine. The increase in -111Unproc accompanies loss of processing of U2G transcripts in all cases (Figure S2A) likely reflects a termination defect. In contrast to ser7, mutations in ser2 and ser5 affect production of snRNAs and mRNAs in largely the same way. Mutation of ser2 to alanine does not reduce recognition by anti-Ser5-P and mutation of ser5 to alanine does not reduce recognition by anti-Ser2-P (Figure S2C), suggesting that both phosphorylation events can occur independently. Mutation of ser5 to glutamic acid reduces recognition by anti-Ser2-P antibody, reflecting a drop in ser2 phosphorylation and/or interference with antibody recognition. A drop in ser2 phosphorylation would again account for the defect in 3′ processing (Figure S2A, B).
To determine whether ser7 is required for expression of endogenous snRNA genes, we used cells with stably-integrated α-amanitin-resistant Rpb1 genes controlled by a tetracycline-regulated promoter (10) (Figure 1B). U2 pre-snRNA (pre-U2) and stable mature U2 snRNA (U2) are readily detected in RNA from cells expressing α-amanitin-resistant Rpb1 with 48 consensus repeats in the CTD (Con)48. A third minor protection product corresponds to transcripts that have escaped 3′ box-directed processing (2) (U2Unproc). Since fully processed snRNAs are very stable (13), there is little change in U2 levels when ser7 is mutated to alanine. However, accumulation of pre-U2 and U2Unproc is severely impaired, although expression of the α-amanitin-resistant Rpb1 is unaffected. qRT-PCR analysis of the RNA indicates that this mutation decreases the level of pre-U2 to less than 30%, whereas mRNA encoding the transcription factor hElf-1 is unaffected. Chapman et al (14) have independently determined that this mutation does not have a general effect on expression of protein-coding genes. Mutation of ser7 to alanine also reduces transcription of U1 and U2 genes to less than 30%, as measured by nuclear run-on analysis (Figure 1C), while transcription of the transfected CMV promoter-driven Rpb1 template and the pol III-dependent 7SK gene are unaffected. Taken together, these results indicate that ser7 is required for endogenous snRNA gene expression. Mutating ser7 to alanine does not affect the level of Rpb1 associated with γ-actin and GAPDH protein-coding genes, as measured by chromatin immunoprecipitation (ChIP) (Figure 2A), indicating that pol II is recruited efficiently. Unexpectedly, the mutant pol II is also recruited efficiently to U1 and U2 genes, indicating that transcription of snRNA genes is affected at a post-recruitment step.
Figure 2. Mutation of ser7 to alanine affects association of Integrator with snRNA genes.

(A) ChIP analysis of Rpb1 and TAP-Int9 associated with U1, U2, γ-actin and GAPDH promoters. (B) ChIP analysis of endogenous U2 genes using antibodies to Rpb1 (pol II) or phospho-serine7 (Ser7P). (C) Western blot analysis of GST-CTD “pull down” of Integrator using anti-Int11 antibodies (17). (D) Phosphorylation of ser7 is required for efficient interaction of Integrator with pol II. Disruption of this interaction may cause a defect in a post-recruitment step of transcription, in addition to affecting 3′ processing.
The transcription factor PTF/PBP/SNAPC, which recognizes the PSE (1, 15), and the Integrator complex, which plays a role in 3′ processing of snRNAs (16), are the only known factors specifically involved in expression of pol II-transcribed snRNA genes. Since Integrator interacts with the CTD (16), we analyzed the effect of ser7 mutation on recruitment of this complex. When the CTD contains 48 consensus repeats, TAP-tagged Integrator subunit 9/RC74 (16, 17) (TAP-Int9) is clearly detectable on snRNA genes but not on γ-actin and GAPDH genes (Figures 2A, S3A). Association with snRNA genes is lost when ser7 is mutated to alanine. In contrast, association of PTF with the promoters of snRNA genes is pol II-independent (Figure S3B).
When transcribing snRNA genes, pol II is phosphorylated on ser7 (Figure 2B), raising the possibility that CTD phosphorylation plays a role in Integrator recruitment. To investigate this, we performed “GST pull down” analysis using consensus or Ser7A repeats. Phosphorylation on ser2, ser5 and ser7 is detected after in vitro phosphorylation (Figure S3C). Int11/RC68 (16, 17), presumably as part of the Integrator complex, interacts strongly with the consensus repeats only after phosphorylation and the interaction increases with the number of repeats (Figure 2C). Mutation of ser7 to alanine has a drastic effect on Integrator binding, although ser2 and ser5 phosphorylation still occur (Figure S3D), strongly suggesting that ser7 phosphorylation participates in this interaction.
Taken together, these experiments suggest that phosphorylation of conserved ser7 residues within the CTD is critical for association with the Integrator complex in vivo. Disruption of this interaction would account for the defect in 3′ processing and may also be responsible for the defect in transcription (Figure 2D). It was proposed that different combinations of phosphorylation of ser2 and ser5 and proline isomerization could constitute a CTD “code” (18). Ser7 phosphorylation would provide an additional, important element of this code in mammals.
Supplementary Material
Materials and Methods, Figure S1-S3
Acknowledgments
We thank Z. Dominski for anti-RC68 antibody, J.B. Yoon and R.G. Roeder for anti-PTFγ antibody, B. Cullen for pCMV-hnRNPK, J. Corden for mutant CTD constructs, and P.R. Cook and N.J. Proudfoot for comments on the manuscript. This work was supported by MRC and Wellcome Trust Grants to S.M. and a Deutsche Forschungsgemeinschaft, SFB-Transregio 5 Grant to D.E..
References and Notes
- 1.Hernandez N. J Biol Chem. 2001;276:26733. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 2.Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. Embo J. 2003;22:925. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs EY, Ogiwara I, Weiner AM. Mol Cell Biol. 2004;24:846–55. doi: 10.1128/MCB.24.2.846-855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCracken S, et al. Nature. 1997;385:357. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 5.Phatnani HP, Greenleaf AL. Genes Dev. 2006;20:2922. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 6.Medlin J, et al. Embo J. 2005;24:4154. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong N, Bird G, Vigneron M, Bentley DL. Embo J. 2003;22:4274. doi: 10.1093/emboj/cdg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu S, Cullen BR. Rna. 2003;9:618. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber HP, et al. Nature. 1995;374:660. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 10.Chapman RD, Conrad M, Eick D. Mol Cell Biol. 2005;25:7665. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuello P, Boyd DC, Dye MJ, Proudfoot NJ, Murphy S. EMBO J. 1999;18:2867. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uguen P, Murphy S. Nucleic Acids Res. 2004;32:2987. doi: 10.1093/nar/gkh619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fury MG, Zieve GW. Exp Cell Res. 1996;228:160. doi: 10.1006/excr.1996.0311. [DOI] [PubMed] [Google Scholar]
- 14.Chapman RD, et al. (submitted to Science) 2007 [Google Scholar]
- 15.Yoon JB, Murphy S, Bai L, Wang Z, Roeder RG. Mol Cell Biol. 1995;15:2019. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baillat D, et al. Cell. 2005;123:265. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF. Mol Cell Biol. 2005;25:1489. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buratowski S. Nat Struct Biol. 2003;10:679. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods, Figure S1-S3


