Abstract
The phosphatidylinositol 3-kinase (PI3K)-signaling pathway has emerged as an important component of cytokine-mediated survival of hemopoietic cells. Recently, the protein kinase PKB/akt (referred to here as PKB) has been identified as a downstream target of PI3K necessary for survival. PKB has also been implicated in the phosphorylation of Bad, potentially linking the survival effects of cytokines with the Bcl-2 family. We have shown that granulocyte/macrophage colony-stimulating factor (GM-CSF) maintains survival in the absence of PI3K activity, and we now show that when PKB activation is also completely blocked, GM-CSF is still able to stimulate phosphorylation of Bad. Interleukin 3 (IL-3), on the other hand, requires PI3K for survival, and blocking PI3K partially inhibited Bad phosphorylation. IL-4, unique among the cytokines in that it lacks the ability to activate the p21ras–mitogen-activated protein kinase (MAPK) cascade, was found to activate PKB and promote cell survival, but it did not stimulate Bad phosphorylation. Finally, although our data suggest that the MAPK pathway is not required for inhibition of apoptosis, we provide evidence that phosphorylation of Bad may be occurring via a MAPK/ERK kinase (MEK)-dependent pathway. Together, these results demonstrate that although PI3K may contribute to phosphorylation of Bad in some instances, there is at least one other PI3K-independent pathway involved, possibly via activation of MEK. Our data also suggest that although phosphorylation of Bad may be one means by which cytokines can inhibit apoptosis, it may be neither sufficient nor necessary for the survival effect.
Survival and growth of hemopoietic cells exquisitely depends on the presence of appropriate cytokines. As such, they provide excellent model systems in which to study the signal transduction processes that are essential for inhibition of apoptosis. We and others (1–4) have provided evidence that PI3K can provide such an antiapoptosis signal in hemopoietic cells as well as several other cell types. Furthermore, there is now good evidence that PKB, a downstream target of PI3K (5, 6), is a key serine/threonine kinase required for inhibition of apoptosis (7–11), whereas p70S6 kinase activity is not required (12, 13).
The Bcl-2 family of related proteins contain protein–protein interaction domains that facilitate homo- and heterodimerization. Overexpression of some members (e.g., Bcl-2, Bcl-XL, A1, and Bag-1) in some systems promotes cell survival whereas others (e.g., Bax, Bad, and Bak) promote cell death. Together, a possible mechanism exists whereby the interactions resulting in homo- or heterodimerization of the various proteins define the fate of a cell (reviewed in refs. 14–17). While up- or down-regulation of these proteins may account for the survival of certain cell types in response to extracellular stimuli, it is also possible that survival factors use protein kinase pathways to alter the ability of these apoptotic proteins to promote survival or death. Bad (Bcl-2/Bcl-XL-associated death promoter; ref. 18) has emerged as a prime candidate for such a role. Bad dimerizes with Bcl-XL and Bcl-2 through interaction of the BH3 domain of Bad (19–21). Phosphorylation of Bad on two serine residues (112 and 136) in response to interleukin 3 (IL-3) promotes association with the 14-3-3 family of proteins, which may promote survival by allowing heterodimerization of Bcl-XL with Bax (22), thereby preventing the proapoptotic function of Bax. Alternatively, some other prosurvival function of Bcl-XL may be restored once Bad is sequestered to the cytoplasm. Consistent with this model, mutation of the phosphorylation sites on Bad that are phosphorylated in response to IL-3 treatment disrupts the ability of IL-3 to promote survival (22).
In more recent studies (23, 24), Bad has been identified as a potential target of PKB, linking the PI3K pathway directly to the apoptotic machinery. Bridging of the PI3K-signaling pathway to the Bcl-2 family of proteins through Bad represents an attractive model for the prosurvival function of PI3K. The evidence for a connection between PKB and Bad phosphorylation involved expression of a membrane-targeted PKB/Akt, or a kinase-deficient mutant. In some experiments, overexpression of a tagged version of Bad was used to monitor the phosphorylation. In none of the published reports has the endogenous level of PKB activity, as regulated by cytokines or growth factors, been used to stimulate the phosphorylation of endogenous levels of Bad. Therefore, the question of whether PKB-mediated phosphorylation of Bad is a key event in prevention of apoptosis downstream of PI3K activation has not been fully resolved. It is possible that phosphorylation of Bad is induced by activated forms of PKB, whereas the normal, regulated enzyme does not perform the same function.
We have shown previously that stimulation with IL-3, IL-4, or stem cell factor (SCF) requires PI3K activity for promoting survival (1). This was in contrast to the effects seen with granulocyte/macrophage colony-stimulating factor (GM-CSF), which maintains survival of MC/9 cells for prolonged periods of time in the absence of PI3K activity. We have investigated this difference seen between IL-3 and GM-CSF, which is of particular interest because the receptors for these cytokines share a common β subunit. In many cases, these receptors were shown to use identical signaling pathways. We show here that GM-CSF can promote survival in the absence of PKB activity. Furthermore, GM-CSF can induce PI3K- and PKB-independent phosphorylation of Bad, suggesting a possible explanation for its ability to block apoptosis independently of PI3K activity. Additionally, stimulation with IL-4 did not induce Bad phosphorylation, even though IL-4 activates both PI3K and PKB, indicating that at the very least the PI3K/PKB pathway requires cooperation from other signaling molecules to phosphorylate Bad. Surprisingly, inhibition of mitogen-activated protein kinase/ERK kinase (MEK) was found to inhibit both IL-3 and GM-CSF-dependent Bad phosphorylation, while having no effect on cell survival.
MATERIALS AND METHODS
Reagents.
Antibodies to Bad were from Transduction Laboratories (B36420) or Santa Cruz Biotechnology (SC-943). Anti-PKB antibodies were obtained from Upstate Biotechnology, Lake Placid, NY (06–558). Annexin-V-FITC was purchased from PharMingen.
Cell Culture.
MC/9 cells (American Type Culture Collection, Manassas, VA) were maintained at 37°C and 5% CO2 in a humidified incubator, in RPMI 1640 medium supplemented with 10% FCS and 10% WEHI-3 conditioned media as a source of IL-3.
PKB Assays.
Cells were washed free of cytokine with Hanks’ balanced salt solution and incubated in the above medium without cytokine for 3–5 hr before assay. Alternatively, cells were incubated overnight with 1% WEHI-3-conditioned medium before assay. At time of assay, cells were again washed and resuspended to 1.0 × 107 cells/ml in RPMI medium buffered with 20 mM Hepes, pH 7.4. Cells were incubated at 37°C for 15 min followed by addition of the PI3K inhibitors LY-294002 or wortmannin for 10 min. Cells were stimulated with either recombinant GM-CSF (100 ng/ml), synthetic IL-3 (10 μg/ml), synthetic IL-4 (10 μg/ml), or recombinant SCF (100 ng/ml) for various times. These concentrations of cytokines were shown previously to induce maximal increases in tyrosine phosphorylation. Reactions were stopped by rapid isolation of cells and lysis in ice-cold solubilization buffer (50 mM Tris⋅HCl, pH 7.7/0.5% Nonidet P-40/2.5 mM EDTA/10 mM NaF/0.2 mM Na3VO4/1 mM Na3MoO4/1 μg/ml microcystin-LR/0.25 mM phenylmethylsulfonyl fluoride/1 μM pepstatin/0.5 μg/ml leupeptin/10 μg/ml soybean trypsin inhibitor), immediately followed by removal of nuclei by centrifugation (20,000 × g, 1 min). Supernatants were incubated with 2 μg anti-PKB-α antibody (Upstate Biotechnology) at 4°C for 1 hr, with continuous mixing. This polyclonal antiserum immunoprecipitates both the α- and β-isoforms of PKB, as determined by immunoblotting (data not shown). Antibody was then captured with 20 μl of Protein-G Sepharose beads at 4°C for 1 hr. Beads were washed three times with fresh solubilization buffer containing 500 mM NaCl and once with kinase buffer (20 mM Hepes, pH 7.2/1 mM MgCl2/1 mM EGTA/1 mM DTT/0.25 mM phenylmethylsulfonyl fluoride/1 mM Na3VO4/0.5 μg/ml leupeptin). Beads were resuspended to 25 μl in kinase buffer containing 60 μM Crosstide (Upstate Biotechnology). ATP solution (5 μl) (200 μM ATP/10 μCi 32P-ATP in kinase buffer) was added followed by incubation for 15 min at 30°C. Reactions were stopped by spotting 20 μl of the reaction volume onto 2-cm2 sheets of P81 filter paper (Whatman), followed by extensive washing with 1% (vol/vol) phosphoric acid and measurement of associated radioactivity by liquid scintillation counting.
Apoptosis Measurements.
Cells undergoing apoptosis were quantitated by staining with Annexin-V-FITC, according to the manufacturer’s protocol (PharMingen) and measured by flow cytometry (Coulter EPICS V). Late apoptotic cells also were distinguished by their ability to take up propidium iodide. Cells staining with annexin-V alone and with both annexin-V and propidium iodide were combined, giving the total number of cells at both early and late stages of apoptosis.
Immunoprecipitation and Blotting of Bad.
For determining the electrophoretic mobility shift of Bad, MC/9 cells stimulated under various conditions were lysed with ice-cold solubilization buffer (20 mM Tris⋅HCl, pH 7.4/137 mM NaCl/0.25% Nonidet P-40/1.5 mM MgCl2/1 mM EDTA/10 mM NaF/0.2 mM Na3VO4/1 mM Na3MoO4/1 μg/ml microcystin-LR/0.25 mM phenylmethylsulfonyl fluoride/1 μM pepstatin/0.5 μg/ml leupeptin/10 μg/ml soybean trypsin inhibitor) and incubated on ice for 10 min. Samples were centrifuged (20,000 × g, 1 min) and supernatants were transferred to clean tubes. Five micrograms of anti-Bad mAb (B36420; Transduction Laboratories, Lexington, KY) was added and the samples were rotated overnight at 4°C. Bad-immunocomplexes were captured with 20 μl of Protein-G Sepharose beads at 4°C for 1 hr. Beads were washed three times with fresh solubilization buffer and resuspended in 1× reducing sample buffer followed by boiling for 5 min. Samples were fractionated in a 12.5% polyacrylamide gel with a 118:1 acrylamide/bisacrylamide ratio and transferred to nitrocellulose. Blots were blocked with 3% skim milk solution for 1 hr and then incubated with 1 μg/ml anti-Bad antibody (either SC-943 from Santa Cruz or B36420 from Transduction Laboratories) overnight at room temperature. Primary antibody was detected with horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence (ECL; Amersham) according to the manufacturer’s protocol.
Metabolic Labeling.
MC/9 cells were starved of cytokine as described above, washed in phosphate-free medium, and then placed in phosphate-free RPMI 1640 medium buffered with 10 mM Hepes, pH 7.4, with 1 mCi/ml 32P-labeled orthophosphate at 37°C for 2 hr. Bad was immunoprecipitated from detergent-solubilized lysates as described above. Immunoprecipitates were fractionated on a 12.5% gel with an acrylamide/bisacrylamide ratio of 118:1 and dried under heat and vacuum. 32P-labeled Bad was detected by autoradiography and quantified by either excision from the gel followed by liquid scintillation counting or by using a PhosphorImager (Molecular Dynamics).
Inhibition of MEK and MAPK.
MEK inhibition was achieved by preincubating MC/9 cells with various concentrations of PD98059 (Biomol, Plymouth Meeting, PA) for 30 min followed by stimulation with cytokines. MEK inhibition was confirmed by probing detergent-solubilized cell lysates with a phosphospecific Erk antibody (New England Biolabs), which only has affinity for active, tyrosine-phosphorylated forms of p42erk2 and p44erk1. Alternatively, p44erk1 was immunoprecipitated and its activation by cytokines was determined in an in vitro kinase assay as described previously (25).
RESULTS
Dependence of Some But Not All Cytokines on PI3K Activity for Inhibition of Apoptosis.
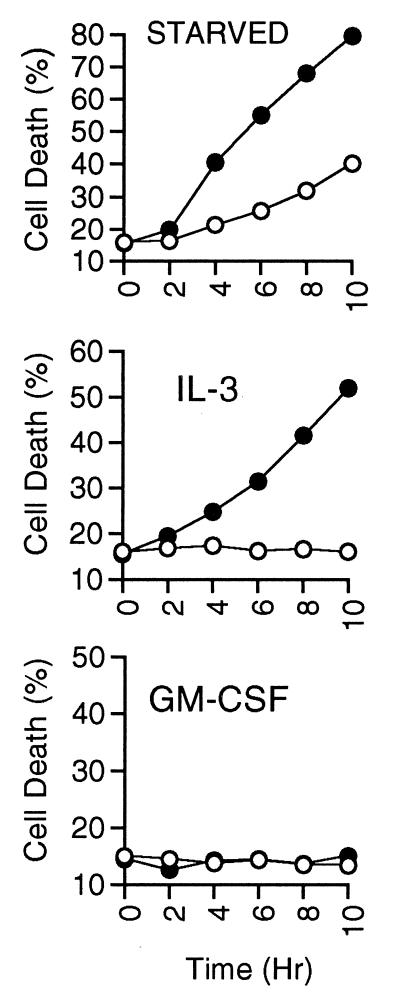
In previous studies, we have shown that in the presence of some cytokines, including IL-3, IL-4, and SCF, inhibition of PI3K led to death of hemopoietic cells by apoptosis. However, in the presence of GM-CSF, complete inhibition of PI3K was unable to induce apoptosis. We have extended our original findings that used DNA fragmentation analysis by using a more quantitative measure of apoptosis, binding of annexin V at the cell surface. We measured the extent of apoptosis induced over a time course in cells treated with LY-294002 in the presence of IL-3 or GM-CSF, or deprived of cytokine, as measured by annexin-V and propidium iodide staining, which represent a combination of early and late stages of apoptosis (Fig. 1). LY-294002 accelerated the rate of cell death induced by withdrawal of cytokine. Similarly, addition of LY-294002 to IL-3-stimulated cells induced cell death to >50% by 8 hr. In contrast, GM-CSF-stimulated cells were unaffected by LY-294002 throughout the time course, and cells examined after 24 hr in LY-294002 were less than 15% apoptotic as determined by annexin V and propidium iodide staining (data not shown). Use of another PI3K inhibitor, wortmannin, yielded similar results.
Figure 1.
Cell death resulting from PI3K inhibition. MC/9 cells were washed three times and resuspended in complete medium containing the indicated cytokine, or medium without cytokine (starved). LY-294002 (25 μM; closed circles) or vehicle (open circles) were added at time 0, and at the indicated hours duplicate aliquots of cells were removed, washed, stained with annexin-V-FITC and propidium iodide, and analyzed by using flow cytometry. We found that cells undergoing apoptosis quickly showed an increase in annexin-V binding but excluded propidium iodide (early apoptosis). At later time points the percentage of propidium iodide staining cells gradually increased (late apoptosis). We are reporting here the total amount of annexin-V-FITC and propidium iodide staining, which is representative of populations containing cells at both stages of apoptosis. Results are representative of at least four independent experiments.
Cytokine-Mediated Activation of PKB.
MC/9 cells were starved in the absence of cytokine, followed by stimulation with either IL-3, IL-4, GM-CSF, or SCF. Activity of PKB in these cells was assessed by immunoprecipitation of endogenous enzyme and in vitro kinase assay using a peptide substrate. As can be seen in Fig. 2, each of the cytokines was able to activate PKB, with IL-3, GM-CSF, and IL-4 giving similar potencies, and SCF giving the greatest activation. In general, these results are consistent with the ability of the cytokines to cause elevation of PI(3,4)P2 and PI(3,4,5)P3 in the same cell type, as we have shown previously (26). However, IL-4 was shown previously to be less effective than IL-3 or GM-CSF in stimulating formation of 3-phosphoinositides, but it is almost as potent as IL-3 or GM-CSF in stimulating PKB activity.
Figure 2.
Cytokine activation of PKB and requirement for PI3K. (A) MC/9 cells starved of cytokine for 3–5 hr were stimulated with the indicated cytokines at concentrations that have been shown previously to induce maximal tyrosine phosphorylation and PI3K activity, for the indicated times. Also, some cells were pretreated with LY-294002 (25 μM) for 10 min and stimulated for 5 min with cytokine. Cells were pelleted and lysed in ice-cold solubilization buffer (see Materials and Methods). PKB was immunoprecipitated and its activity was measured in an in vitro kinase assay by using Crosstide as substrate. Individual experiments have been normalized to the percentage of stimulation induced by a 5-min treatment with IL-3, which was always performed concurrently. Typical maximum stimulations for IL-3, IL-4, and GM-CSF ranged between 8- and 12-fold above unstimulated samples, which generally were in the 2,000–3,000 cpm range. (B) Cells were prepared as above and pretreated with LY-294002 (25 μM) or vehicle alone for 10 min followed by stimulation with GM-CSF for the indicated times. PKB immunoprecipitation and kinase assay was performed as above. The results presented are from three experiments using duplicate samples, with error bars representing SEM.
We next confirmed that activation of PKB depended on the activity of PI3K by inhibition with LY-294002 (Fig. 2) or wortmannin (results not shown). These inhibitors were able to completely block PKB activation in response to each of the cytokines, at concentrations where they completely blocked PI3K activity in these cells (25). There was no indication that GM-CSF was able to activate PKB in the absence of PI3K activity, so the ability of GM-CSF to promote cell survival was independent of both PI3K and PKB activity.
Phosphorylation of Bad Is Induced by Several Cytokines But Not IL-4.
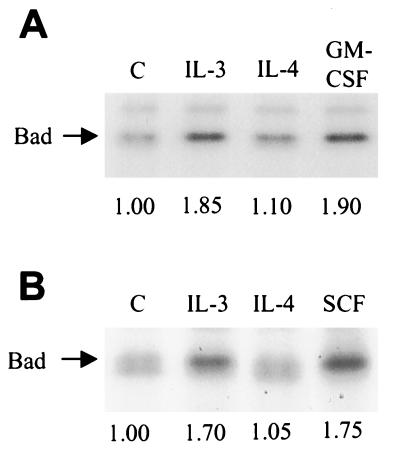
Bad, a member of the Bcl-2 family of proteins, has been shown previously to undergo phosphorylation in response to IL-3, which may account partially for IL-3-dependent survival (22). Bad phosphorylation on two sites (Ser-112 and Ser-136) alters its mobility during migration through polyacrylamide (22). Thus, we used this indicator to examine whether the cytokines we were testing induced a similar mobility shift. Consistent with the original report (22) and more recent work (24), IL-3-stimulation of MC/9 cells for various times caused a modification of Bad such that it migrated slower in polyacrylamide than from unstimulated cells, indicative of IL-3-mediated phosphorylation (Fig. 3A). GM-CSF was able to induce a complete shift to the slower migrating form after 10 min of stimulation (Fig. 3B). We also examined IL-4, which, surprisingly, was not able to induce a mobility shift of the lower band (Fig. 3B). These preparations were from immunoprecipitated Bad, which were detected with the B36420 anti-Bad antibody. Detergent-solubilized whole-cell lysates from another experiment were separated on SDS-polyacrylamide gels and probed with a different antibody (SC-943 from Santa Cruz Biotechnology), which produced identical results (Fig. 3C). These results were consistent in four independent experiments. It should be noted that in immunoblots of whole-cell lysates, the B36420 antibody detected a prominent band at approximately 23 kDa, which was just below the doublet of 24 and 25 kDa attributed to Bad. The 23-kDa polypeptide was not immunoprecipitated from cell extracts and was not detected by the SC-943 antibody, suggesting that it contained a cross-reacting epitope detected only by the B36420 antibody when the protein was denatured.
Figure 3.
Bad mobility shift is induced by treatment with IL-3, GM-CSF, or SCF but not IL-4. (A) Cells were starved of cytokine for 3–5 hr and stimulated with IL-3 for the indicated times, or were left unstimulated. Cells were isolated and solubilized as described in Materials and Methods. Bad was immunoprecipitated (5 μg of B36420, Transduction Laboratories) and separated by SDS/PAGE followed by transfer to nitrocellulose and immunoblotting for Bad. (B) Immunoprecipitated Bad from cells stimulated with IL-3, IL-4, or GM-CSF for 10 min and immunoblotted with B36420. (C) Whole-cell lysates from a similar experiment immunoblotted with anti-Bad antibody SC-943 (Santa Cruz Biotechnology). Results shown are representative of at least four independent experiments.
To assess whether IL-4 may be stimulating an increase in phosphorylation of Bad at one site, which would not alter its mobility on gels, we performed in vivo 32P-phosphate labeling. Bad immunoprecipitated from cells stimulated with IL-3, GM-CSF, or SCF had an increase of greater than 70% in radioactivity compared with unstimulated cells (Fig. 4). Strikingly, IL-4 stimulation did not induce any significant increase in Bad phosphorylation (Fig. 4). Because our results in Fig. 2 confirmed that PKB was activated by these cytokines, including IL-4, these findings suggest that activation of endogenous PKB alone is not able to induce phosphorylation of Bad.
Figure 4.
IL-4 does not stimulate Bad phosphorylation. (A) Cells were starved of cytokine for 3–5 hr and then metabolically labeled with 32P-labeled orthophosphate for 2 hr, followed by stimulation with the indicated cytokine for 10 min. Bad was immunoprecipitated as described in Fig. 3 and fractionated by SDS/PAGE. 32P-labeled Bad was detected by autoradiography and quantitated by either a PhosphorImager or by excising the bands and counting by liquid scintillation. This experiment was performed in duplicate, with one set of samples shown. (B) Identical experiment as A, but SCF was tested instead of GM-CSF. The numbers beneath each lane correspond to the average stimulation above untreated for each set of duplicates.
Bad Phosphorylation by IL-3 But Not GM-CSF Depends on PI3K Activity.
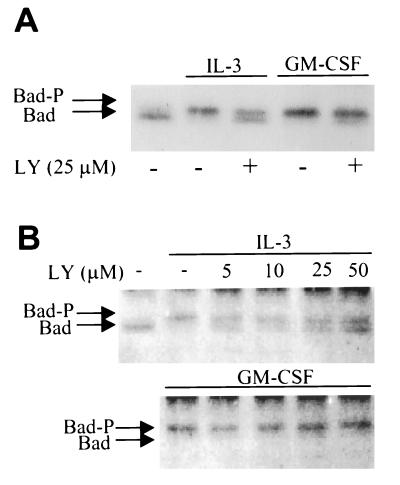
We next evaluated the phosphorylation of Bad induced by IL-3 and GM-CSF after inhibition of PI3K (Fig. 5). PI3K inhibitors partially inhibited, but did not completely block, the mobility shift of Bad induced by IL-3. In contrast, GM-CSF-induced phosphorylation of Bad did not depend on PI3K, because a complete bandshift occurred at concentrations of either LY-294002 or wortmannin that completely blocked PI3K activity. The effects of LY-294002 on IL-3-induced mobility shift of Bad appeared to be maximal at concentrations between 5 and 10 μM, consistent with the ability of this compound to inhibit PI3K. Higher doses (up to 50 μM) did not further affect the ratio between upper and lower bands, indicating that at least partial phosphorylation of Bad is LY-294002-insensitive.
Figure 5.
PI3K inhibition partially blocks IL-3- but not GM-CSF-induced Bad phosphorylation. (A) Cells were preincubated with LY-294002 (25 μM) or vehicle alone for 10 min, followed by stimulation with IL-3 or GM-CSF for 10 min. Bad was immunoprecipitated as described in Fig. 3 and mobility shift was examined by immunoblotting with a polyclonal anti-Bad antibody (SC-943). (B) Cells were pretreated with the concentrations of LY-294002 indicated above the lanes for 10 min and stimulated with IL-3 or GM-CSF for 10 min. Bad was immunoprecipitated and immunoblotted with B36420 (Transduction Laboratories) to determine mobility shift. These results are representative of three independent experiments.
MEK Inhibition Blocks Bad Phosphorylation.
Some results have suggested that a MAPK-dependent pathway is involved in inhibition of apoptosis by cytokines (27), although p21ras-dependent survival was shown more recently to be independent of MAPK (28). Our results with IL-4 suggest that MAPK is not necessary for cell survival (1), and furthermore, in the absence of PI3K activity, MAPK activation cannot rescue cells from apoptosis (25). We examined a potential role for MEK in Bad phosphorylation by treating cells with PD98059, a potent and selective inhibitor of both MEK1 and MEK2 (29). Preincubation of cells with PD98059 blocked p44erk1 and p42erk2 tyrosine phosphorylation after stimulation with IL-3 or GM-CSF (Fig. 6A), and at these concentrations the bandshift of Bad induced by cytokines was completely blocked in the case of IL-3, indicating that at least one of the phosphorylation events on Bad occurs as a result of MEK activation. The mobility shift of Bad in response to GM-CSF was partially reduced under the same conditions. Also, activity of PKB was not blocked by incubation with PD98059 (data not shown). A question that arises from these studies is whether inhibition of MEK blocks IL-3- or GM-CSF-mediated survival. PD98059 had no effect on survival supported by IL-3 or GM-CSF (Fig. 6C), indicating that other signaling pathways (both PI3K-dependent and -independent) can provide survival signals independently of PD98059-sensitive Bad phosphorylation.
Figure 6.
MEK inhibition blocks Bad phosphorylation without effect on survival. (A) Cells were preincubated with PD98059 (PD) at the indicated concentrations for 30 min followed by stimulation with IL-3 or GM-CSF for 5 min. Detergent solubilized cell lysates were fractionated by SDS/PAGE and immunoblotted with anti-phospho-MAPK. (B) Bad was immunoprecipitated from the same cells lysates and immunoblotted to determine mobility shift. These results are representative of three independent experiments. (C) MC/9 cells were washed and resuspended in IL-3, GM-CSF, or media alone (Unstim) and incubated with 50 μM PD98059 (solid bars) or vehicle (shaded bars) for 11 hr. Apoptosis was determined by annexin V binding by using flow cytometry. Results shown are averages of duplicate samples and are representative of two independent experiments.
DISCUSSION
The requirement for PI3K signaling in the survival of many cell types has been well documented in recent years. However, it has also become clear that some stimuli appear to bypass the need for PI3K activity. Molecular biology approaches using dominant, negative and membrane-targeted PKB have suggested that this serine/threonine kinase provides the next level of death suppression downstream of PI3K. Because there have been no reports of a naturally occurring PKB kinase that operates independently of PI3K, the ability of certain survival factors to act independently of PI3K may be mediated by signaling pathways that do not involve PKB. For example, Philpott and coworkers (30) have reported recently that primary sympathetic neurons are able to maintain survival in the presence of either PI3K inhibitors or a dominant, negative PI3K construct only if they are also stimulated with nerve growth factor (NGF), suggesting that NGF contributes to survival through alternate pathways. Our results have demonstrated that GM-CSF also can promote cell survival independently of PI3K (1) and PKB (this study). Therefore, further characterization of these signaling pathways is crucial to aid in our understanding of how survival factors inhibit apoptosis.
Recently, Bad has been shown to undergo phosphorylation in response to IL-3 and platelet-derived growth factor (PDGF), and this phosphorylation may depend on PI3K/PKB activity (23, 24). It should be pointed out that in most of these studies, overexpression of Bad and expression of activated PKB were used to provide evidence for the importance of PKB and phosphorylation of Bad in blocking apoptosis. Inhibition of Bad phosphorylation by LY-294002 and wortmannin also is documented, although again Bad was overexpressed in these studies. Because an anti-AU1 antibody was used to immunoprecipitate AU1-tagged Bad, only this expressed form was examined. It is not clear whether overexpression of Bad alters its susceptibility to kinases that would not normally come in spatial proximity to endogenous Bad.
In our studies, we have monitored the phosphorylation of endogenous Bad and we have examined the activity of the endogenous PKB enzyme. We have used the MC/9 cell line that is absolutely dependent on any one of several cytokines for survival. If one begins with the hypothesis that activation of endogenous PKB leads directly to phosphorylation of Bad, then all cytokines that activate PKB should cause an increased phosphorylation of Bad. When cells were stimulated with IL-3, blocking the activation of PI3K and PKB using PI3K inhibitors partially reduced the phosphorylation of Bad. This would be consistent with evidence that only one of the two key sites on Bad (Ser-136) depends on PKB for its phosphorylation (23). However, when GM-CSF-treated cells were incubated with PI3K inhibitors, to completely block PI3K and PKB activation, there was no change in the ability of GM-CSF to stimulate phosphorylation of Bad. Clearly, this result provides a possible explanation of why GM-CSF is able to maintain cell survival in the absence of PI3K and PKB activity, whereas IL-3 is not. Further studies will be required to determine whether association of Bad with 14-3-3, for example, depends on PI3K activity in the case of IL-3 stimulation, but not GM-CSF.
Another important observation is that IL-4 was able to stimulate PKB activity to a level similar to that of IL-3 or GM-CSF and, yet, it was unable to significantly stimulate phosphorylation of Bad. The lack of Bad phosphorylation after treatment with IL-4 provides strong evidence that PKB alone, when it is activated by cytokines, is not able to induce phosphorylation of Bad, thereby contradicting the above hypothesis. These results also suggest that IL-4 utilizes other mechanisms for maintaining survival, independent of phosphorylation of Bad, although we have shown previously that PI3K is necessary for this property (1).
An alternate kinase stimulated by cytokines that may contribute to Bad phosphorylation is Raf-1 (31). This would be consistent with IL-4 being unable to induce phosphorylation of Bad, because IL-4 does not activate Raf-1. Nevertheless, because IL-3 and GM-CSF both stimulate the p21ras pathway in our system (32), it remains unlikely that GM-CSF is utilizing Raf-1 differently compared with IL-3. Another possibility is that there is another Raf-like kinase that may be activated by GM-CSF, but not by IL-3.
Conflicting evidence regarding a potential role for the MEK-ERK pathway in preventing apoptosis has emerged in recent years (27, 28). Our studies have argued against such a cytoprotective role, because IL-4 can promote survival but is unable to stimulate MAPK (33), and other groups also have provided evidence that blocking MAPK does not lead to apoptosis (34, 35). Furthermore, our finding that the MEK inhibitor PD98059 did not prevent IL-3- or GM-CSF-mediated survival also supports this conclusion. However, a connection beween MAPK activation and Bad phosphorylation appears likely, because PD98059 blocked the IL-3-stimulated mobility shift of Bad. GM-CSF-induced Bad phosphorylation was less sensitive to PD98059, with approximately 50% of dual-phosphorylated Bad reduced to a faster migrating form. This result is in contrast to the findings of Datta et al. (23), who reported that MEK inhibition had no effect on PDGF-induced Bad phosphorylation.
In summary, the multiple events involved in regulation of apoptosis continue to increase in complexity. Although PI3K activity seems to be important for survival of hemopoietic cells in response to many cytokines, there are exceptions to this rule, as exemplified by the effect of GM-CSF. Furthermore, although phosphorylation of Bad in response to some cytokines may contribute to cell survival, it is not absolutely required. Even in the case of cytokines that stimulate phosphorylation of Bad, when the phosphorylation is blocked by inhibition of MEK, there is no increase in apoptosis. Although it is possible that PKB activity may contribute to phosphorylation of Bad as one means of promoting cell survival, our evidence suggests that activation of PKB alone is insufficient to promote phosphorylation of Bad. Furthermore, phosphorylation of Bad may be unaffected by the complete inhibition of PI3K and PKB, as in the case of GM-CSF. The identity of other Bad kinases will be an important avenue to explore. Finally, the ability of IL-4 to stimulate PKB and to mediate PI3K-dependent cell survival does not involve phosphorylation of Bad. Therefore, other phosphorylation events (e.g., phosphorylation of other Bcl-2 family members) may be able to fulfill the same function that has been attributed to phosphorylation of Bad.
Acknowledgments
This work was supported by the Medical Research Council of Canada. M.P.S. was supported by a Cancer Research Society Studentship, and V.D. was supported by a joint Medical Research Council/BC Lung Association Scholarship.
ABBREVIATIONS
- GM-CSF
granulocyte/macrophage colony-stimulating factor
- IL
interleukin
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- SCF
stem cell factor
References
- 1.Scheid M P, Lauener R W, Duronio V. Biochem J. 1995;312:159–162. doi: 10.1042/bj3120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao R, Cooper G M. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 3.Minshall C, Arkins S, Freund G G, Kelley K W. J Immunol. 1996;156:939–947. [PubMed] [Google Scholar]
- 4.Vemuri G S, McMorris F A. Development. 1996;122:2529–2537. doi: 10.1242/dev.122.8.2529. [DOI] [PubMed] [Google Scholar]
- 5.Burgering B M, Coffer P J. Nature (London) 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 6.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed N N, Grimes H L, Bellacosa A, Chan T O, Tsichlis P N. Proc Natl Acad Sci USA. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R S, Kaplan D R, Greenberg M E. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 9.Kauffman-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Nature (London) 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 10.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheid M P, Charlton L, Pelech S L, Duronio V. Biochem Cell Biol. 1996;74:595–600. doi: 10.1139/o96-064. [DOI] [PubMed] [Google Scholar]
- 13.Yao R, Cooper G M. Oncogene. 1996;11:1604–1614. [Google Scholar]
- 14.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 15.Chinnaiyan A M, Dixit V M. Curr Biol. 1996;6:555–562. doi: 10.1016/s0960-9822(02)00541-9. [DOI] [PubMed] [Google Scholar]
- 16.Jacobson M D. Curr Biol. 1997;7:277–281. doi: 10.1016/s0960-9822(06)00125-4. [DOI] [PubMed] [Google Scholar]
- 17.Reed J C. Nature (London) 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 18.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 19.Zha J, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer S J. J Biol Chem. 1997;272:24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 20.Ottilie S, Diaz J L, Horne W, Chang J, Wang Y, Wilson G, Chang S, Weeks S, Fritz L C, Oltersdorf T. J Biol Chem. 1997;272:30866–30872. doi: 10.1074/jbc.272.49.30866. [DOI] [PubMed] [Google Scholar]
- 21.Kelekar A, Chang B S, Harlan J E, Fesik S W, Thompson C B. Mol Cell Biol. 1997;17:7040–7046. doi: 10.1128/mcb.17.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 23.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 24.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 25.Scheid M P, Duronio V. J Biol Chem. 1996;271:18134–18139. doi: 10.1074/jbc.271.30.18134. [DOI] [PubMed] [Google Scholar]
- 26.Gold M R, Duronio V, Saxena S P, Schrader J W, Aebersold R. J Biol Chem. 1994;269:5403–5412. [PubMed] [Google Scholar]
- 27.Kinoshita T, Yokota T, Arai K, Miyajima A. EMBO J. 1995;14:266–275. doi: 10.1002/j.1460-2075.1995.tb07000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita T, Shirouzu M, Kamiya A, Hashimoto K, Yokoyama S, Miyajima A. Oncogene. 1997;15:619–627. doi: 10.1038/sj.onc.1201234. [DOI] [PubMed] [Google Scholar]
- 29.Pang L, Sawada T, Decker S J, Saltiel A R. J Biol Chem. 1995;270:13585–13588. doi: 10.1074/jbc.270.23.13585. [DOI] [PubMed] [Google Scholar]
- 30.Philpott K L, McCarthy M J, Klippel A, Rubin L L. J Cell Biol. 1997;139:809–815. doi: 10.1083/jcb.139.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H G, Rapp U R, Reed J C. Cell. 1996;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 32.Duronio V, Welham M J, Abraham S, Dryden P, Schrader J W. Proc Natl Acad Sci USA. 1992;89:1587–1591. doi: 10.1073/pnas.89.5.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welham M J, Duronio V, Sanghera J S, Pelech S L, Schrader J W. J Immunol. 1992;149:1683–1693. [PubMed] [Google Scholar]
- 34.Virdee K, Tolkovsky A M. J Neurochem. 1996;67:1801–1805. doi: 10.1046/j.1471-4159.1996.67051801.x. [DOI] [PubMed] [Google Scholar]
- 35.Kummer J L, Rao P K, Heidenreich K A. J Biol Chem. 1997;272:20490–20494. doi: 10.1074/jbc.272.33.20490. [DOI] [PubMed] [Google Scholar]