Abstract
Interleukin 3-dependent murine 32D cells do not detectably express members of the ErbB receptor family and do not proliferate in response to known ligands for these receptors. 32D transfectants were generated expressing human ErbB4 alone (32D.E4) or with ErbB2 (32D.E2/E4). Epidermal growth factor (EGF), neuregulin 1-β (NRG1-β), betacellulin (BTC), transforming growth factor-α (TGF-α), heparin binding-EGF (HB-EGF), and amphiregulin were analyzed for their ability to mediate mitogenesis in these transfectants. 32D.E4 responded mitogenically to NRG1-β and BTC. Surprisingly, EGF also induced significant DNA synthesis and TGF-α was negligibly mitogenic on 32D.E4 cells, whereas HB-EGF and amphiregulin were inactive. Although coexpression of ErbB2 with ErbB4 in 32D.E2/E4 cells did not significantly alter DNA synthesis in response to NRG1-β or BTC, it greatly enhanced mitogenesis elicited by EGF and TGF-α and unmasked the ability of HB-EGF to induce proliferation. EGF-related ligands that exhibited potent mitogenic activity on 32D.E2/E4 cells at low concentrations induced adherence, morphological alterations, and up-regulation of the Mac-1 integrin and FcγRII/III at higher concentrations. While 125I-EGF could be specifically crosslinked to both 32D.E4 and 32D.E2/E4 cells, its crosslinking capacity was greatly enhanced in the cotransfected cells. The ability of the various ligands to mediate proliferation and/or adhesion in the two transfectants correlated with their capacity to induce substrate tyrosine phosphorylation and to initiate and sustain activation of mitogen-activated protein kinase. We conclude that the ability of ErbB4 to mediate signal transduction through EGF-like ligands is broader than previously assumed and can be profoundly altered by the concomitant expression of ErbB2.
The ErbB receptor family is comprised of four members termed epidermal growth factor (EGF) receptor (ErbB1), ErbB2, ErbB3, and ErbB4. All ErbB receptors possess similar structural features including a highly glycosylated extracellular ligand binding domain with two cysteine-rich clusters, a single transmembrane domain, and a large cytoplasmic region containing a tyrosine kinase motif flanked by juxtamembrane and carboxy-terminal residues. These receptors are broadly expressed in mammalian tissues of epithelial, mesenchymal, and neuronal origin and are known to play fundamental roles during development and lineage determination (1). For example, targeted gene disruption of ErbB2 and ErbB4 in mice causes abnormal neural and cardiac development resulting in death during mid-embryogenesis (2, 3). The ability of EGF and EGF-related ligands to bind to the various members of the ErbB receptor family has been the subject of intense investigation. While the number of ErbB receptors has not expanded since the identification of ErbB4, almost 30 EGF-related ligands have been isolated (1, 4). ErbB2 has been found to be frequently amplified and/or overexpressed in a variety of human carcinomas (1, 4–6). Its overexpression has been correlated with poor prognosis and reduced survival of patients with breast and ovarian carcinomas (1, 4, 7, 8). Overexpression of ErbB1 and ErbB3 has also been observed in several types of cancer (9–11). However, the clinical relevance of their overexpression remains unclear. While there have been no reports documenting ErbB4 overexpression in human tumor tissues or cell lines, a recent clinical study revealed that coexpression of ErbB2 and ErbB4 is frequent and has prognostic significance in childhood medulloblastoma (12). Thus, the relevance of overexpression of one ErbB receptor in determining the clinical outcome of a specific neoplasm may depend on the simultaneous expression of another ErbB member.
EGF contains a prototypical 50 amino acid domain defined by six cysteine residues that form three disulfide bonds and a core arginine that stabilizes protein orientation (1). This EGF-like domain is present in all ErbB ligands and has been demonstrated to be sufficient for receptor activation. ErbB1 has been demonstrated to directly bind to EGF, transforming growth factor-α (TGF-α), betacellulin (BTC), heparin binding-EGF (HB-EGF), amphiregulin (AR), and epiregulin (13–18). ErbB ligands known to bind ErbB3 and ErbB4 include neuregulin 1 (NRG1), NRG2, NRG3, acetylcholine receptor-inducing activity, and glial-derived growth factor (19–26). Recently, BTC has also been demonstrated to bind to ErbB2/ErbB3 complexes and to ErbB4 (27, 28). Interestingly, it has been demonstrated that substitution of amino acids 177–181 of NRG1-β for those present in the amino terminus of EGF produces a bifunctional agonist that interacts with both ErbB1 and ErbB4 with similar high affinities (29).
Several lines of evidence indicate that receptor specificity for the various ligands may be broader than previously assumed (1, 4). This increased diversity may result from the propensity of ErbB receptors to form heterodimeric complexes upon ligand occupancy (30–32). ErbB2 has been demonstrated to be the preferred partner for all ErbB receptors (6, 31, 33). While ErbB2 binds no known ligand with high affinity, it exhibits high basal tyrosine kinase activity that can be further enhanced by heterodimerization in response to ligand binding in the presence of coexpressed ErbB partners. For example, it has been shown that NRG1-β binds to ErbB3 with moderate affinity and to ErbB2/ErbB3 heterodimers with high affinity (20, 22, 23). Furthermore, coexpression of ErbB2 with ErbB1 has been demonstrated to enhance EGF-mediated transformation of rodent fibroblasts and EGF binding affinity (34). ErbB2 and ErbB3 have also been found to cooperate in mediating in vitro transformation, and NRG-induced transformation of NIH 3T3 cells by ErbB4 has been shown to require the presence of ErbB2 or ErbB1 (11, 35, 36).
Interleukin 3 (IL-3)-dependent murine hematopoietic cells, which are thought to be naive for ErbB receptor expression, have been used to elucidate which EGF-like ligands can bind to and signal through which combinations of ErbB receptors (27, 28, 30, 37–39). In a recent study, it was unexpectedly found that EGF and BTC can bind to and signal through ErbB2/ErbB3 complexes by a mechanism that requires coexpression of both receptors (28). In the present study, we have utilized 32D cells to provide evidence that EGF and TGF-α can serve as ligands for ErbB4 and that ErbB2 and ErbB4 coexpression modifies and strengthens signal transduction induced by several ErbB ligands.
METHODS
Growth Factors, Antibodies, and Cell Lines.
Recombinant human EGF was purchased from Upstate Biotechnology (Lake Placid, NY), TGF-α was from Intergen (Purchase, NY); NRG1-β, BTC, HB-EGF, and AR were from R & D Systems. Rabbit antisera utilized for detection of human ErbB receptors by immunoblot analysis were generated against synthetic peptides derived from human ErbB2 (residues 1,218–1,232) or ErbB4 (residues 1,285–1,308; Ab-2; Neomarkers, Fremont, CA). mAbs utilized for flow cytometry include anti-ErbB2 (Ab-2; Oncogene Science) and anti-ErbB4 (Ab-1; Neomarkers), fluorescein isothiocyanate (FITC)-conjugated anti-Mac-1 (Caltag, South San Francisco, CA), FITC-labeled anti-FcγRII/III (PharMingen), and FITC-conjugated goat anti-mouse (Caltag). Antisera used for immunoprecipitation include rabbit anti-ErbB2 peptide serum and anti-ErbB4 mAb (Ab-1, Neomarker). The 4G10 anti-phosphotyrosine (anti-pTyr) mAb was utilized in immunoprecipitation and immunoblot analysis (Upstate Biotechnology). The rabbit antiserum utilized for detecting activated mitogen-activated protein kinase (MAPK) was from New England Biolabs. The murine IL-3-dependent 32D myeloid progenitor cell, EGF-dependent Balb/MK epithelial, and NR6 fibroblast lines have been described (34, 40, 41). 32D cells were transfected with LTR2-erbB2neo and/or LTR2-erbB4 gpt expression vectors by electroporation (11, 28, 41, 42). Human ErbB4 cDNA was cloned by reverse transcription–PCR (RT-PCR) based on the published sequence (43). Briefly, single stranded cDNA was synthesized utilizing total RNA isolated from the MDA-MB-453 cell line. ErbB4 cDNA was then amplified by PCR reaction. Sense and anti-sense strand primers used were 5′-GTCGACACCATGAAGCCGGCGAACGGACTTT-3′ and 5′-CTAGTCGACTTACACCACAGTATTCCGGTGTC-3′. The amplified ErbB4 cDNA was digested with SalI and cloned into Bluescript SKII (Stratagene). LTR2-erbB4 gpt was generated by inserting the SalI fragment into the XhoI site of the LTR2-gpt expression vector (41). Stable transfectants were generated by their ability to grow in medium containing 750 μg/ml geneticin to confirm neo expression and/or 80 μM mycophenolic acid and hypoxanthine, aminopterin, and thymidine to confirm gpt expression.
Characterization of ErbB Receptor Expression by RT-PCR, Immunoblot, and Flow Cytometric Analysis.
Amplification of gene products corresponding to murine ErbB2, ErbB3, or ErbB4 transcripts was carried out by RT-PCR utilizing total RNA isolated from murine cell lines, whole embryo (day 10 postconception) or adult brain as previously described (28). The sense strand and anti-sense strand primers used to detect murine transcripts were 5′-GAAGTTCAGGGATACATGAT-3′ and 5′-TGTCTTTGCAGGTTGGGGCACAA-3′ for ErbB2; 5′-ACCTAACCTCCGAGTGGTCCGG-3′ and 5′-ACTGGTTAGGATTGGGCCCGAA-3′ for ErbB3; 5′-GTTCGCCCCAGAACGGAATCCTCG-3′ and 5′-TCACACCACAGTATTCCGGTGTCT-3′ for ErbB4; and 5′-CAGGAAGGAAGGCTGGAAGA-3′ and 5′TTCTACAATGAGCTGCGTGTG-3′ for β-actin. Detection of ErbB receptors by flow cytometry and immunoblot analysis was performed as described (28, 42, 44, 45).
Binding and Covalent Affinity Crosslinking Analysis.
Crosslinking was performed essentially as described (22, 28). Cells (107) were suspended in RPMI 1640 medium containing 0.1% BSA and 10 mM Hepes with 125I-EGF (50 nM) in the absence or presence of a 100-fold excess of unlabeled EGF and incubated for 3 h at 16°C. The crosslinking agent, BS3 (2.0 mM), was then added, and samples were incubated for 30 min at room temperature. Lysates were immunoprecipitated with anti-ErbB2 or anti-ErbB4, and immunoprecipitated proteins were resolved on an SDS/5% polyacrylamide gel. Displacement binding of 125I-EGF was assayed as described (28). 32D.E2/E4 cells (5 × 104 cells per 100 μl) were incubated for 3 h at 16°C with 125I-EGF (50 nM) in the absence or presence of various concentrations of unlabeled EGF, NRG1-β, BTC, TGF-α, or HB-EGF. Free 125I-EGF was removed by pelleting the cells through phthalate oil, and 125I-EGF bound to the cell surface was measured by γ counting.
Proliferation and Differentiation Assays.
Ligand-induced DNA synthesis in the 32D transfectants was performed as described (28, 45). Briefly, 32D.E4 or 32D.E2/E4 cells were washed twice and resuspended in RPMI 1640 medium with 15% fetal bovine serum (2 × 105 cells/ml), and exposed to various concentrations of the different ErbB ligands. DNA synthesis was measured after 36 h, and [3H]thymidine (0.5 μCi/ml; 1 Ci = 37 GBq) was added for the last 4 h of the incubation period. Cells were harvested on an automated cell harvester and triplicate samples were counted in a β counter. For detection of monocytic cell surface antigens, cells were untreated or treated with ErbB ligands (100 ng/ml) for 24 h. Live cells were incubated with FITC-conjugated anti-Mac-1 or anti-FcγRII/III and analyzed by flow cytometry as described (41, 44). Adherence was determined in ligand-treated or untreated cultures after 24 h by counting unattached and attached cells with a hemocytometer. Attached cells were removed from the tissue culture substrate by trypsin digestion. Two samples were analyzed for each treatment.
Analysis of EGF-Like Ligand-Induced Tyrosine Phosphorylation and MAPK Activation in the 32D Transfectants.
For detection of tyrosine-phosphorylated proteins, cells were serum starved for 4 h and either untreated or treated with various ligands (100 ng/ml) for 10 min at 37°C. Lysates were prepared in buffer containing 1% Nonidet P-40 and immunoprecipitated with anti-pTyr and subsequently immunoblotted with anti-pTyr as described (11, 28, 46). For detection of activated MAPK, protein (100 μg) from untreated cells or cells treated for 5, 10, 30, or 60 min with the various ligands were resolved on an SDS/12% polyacrylamide gel and subsequently immunoblotted with the anti-phosphospecific MAPK serum.
RESULTS
Analysis of Endogenous ErbB Receptor Expression in 32D Cells and Exogenous Human ErbB2 and ErbB4 Expression in 32D Transfectants.
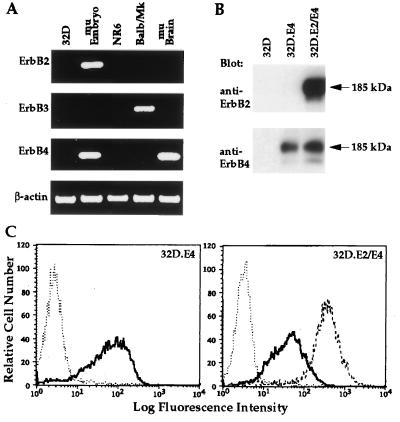
To confirm that the IL-3-dependent murine myeloid progenitor 32D cell line utilized in this study does not endogenously express any ErbB receptor, RT-PCR analysis was performed utilizing oligonucleotide primers pairs designed to differentially detect murine ErbB2, ErbB3, and ErbB4. Alimandi et al. (28) have recently demonstrated that this line does not detectably express ErbB1. As shown in Fig. 1A, no amplified cDNA products were detected from 32D or NR6 murine fibroblast RNA preparation as determined by RT-PCR analysis utilizing the three oligonucleotide primer pairs. Amplified DNA products of the expected size were detected from RNA isolated from whole murine embryo (day 10 postconception) for ErbB2, Balb/MK epithelial cells for ErbB3 and whole murine embryo and adult murine brain for ErbB4 (Fig. 1A). These results provide definitive evidence that 32D cells do not detectably express any known ErbB receptor.
Figure 1.
Characterization of endogenous and exogenous ErbB receptor expression in 32D cells and transfectants. (A) Analysis of endogenous ErbB2, ErbB3, and ErbB4 expression in 32D cells. The amplified cDNA products generated by RT-PCR analysis of total RNA from 32D cells, day 10 murine embryo (mu Embryo), NR6 cells, Balb/MK cells, and adult murine brain (mu Brain) by oligonucleotide primers that recognize murine ErbB2, ErbB3, ErbB4, or β-actin are shown. (B) Immunoblot analysis to examine expression of ErbB2 and ErbB4 in the two 32D transfectants. Anti-ErbB2 or anti-ErbB4 serum was utilized for immunoblot (Blot) analysis of proteins from lysates of 32D, 32D.E4, or 32D.E2/E4 cells as designated. The 185-kDa mature forms of ErbB2 and ErbB4 are marked by arrows. (C) Flow cytometric analysis to determine the relative levels of cell surface expression of ErbB2 and ErbB4 on the two 32D transfectants. The histograms for untransfected 32D cells (⋅⋅) or 32D.E4 cells (—) incubated with anti-ErbB4 serum are shown (Left). The histograms for 32D (⋅⋅⋅) or 32D.E2/E4 cells incubated with anti-ErbB4 (—) or anti-ErbB2 (– – –) (Right).
To determine the spectrum of EGF-like ligands capable of interacting with ErbB4 or coexpressed ErbB2 and ErbB4, 32D cells were electroporated with expression vectors containing human ErbB4 cDNA alone or together with human ErbB2, and transfectants were termed 32D.E4 and 32D.E2/E4, respectively. ErbB receptor expression in the two transfectants was assessed by immunoblot and flow cytometric analysis (Fig. 1 B and C). Each transfectant expressed readily detectable levels of the appropriate receptor(s) as determined by both methods. Furthermore, similarities in immunoblot protein levels and flow cytometric mean fluorescence intensities provided evidence that the amounts of ErbB4 expressed on the individual transfectant were roughly equivalent to those expressed on the cotransfectant.
ErbB2 Coexpression Increases the Range and Potency of ErbB Ligands Capable of Eliciting Mitogenesis in 32D Transfectants Expressing ErbB4.
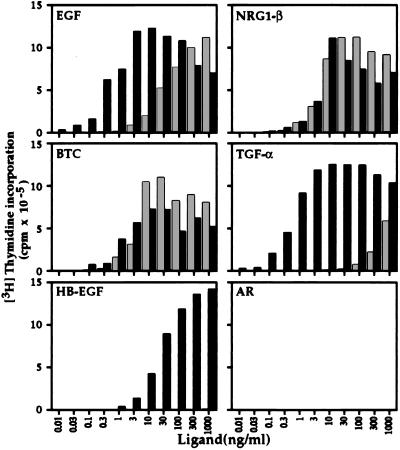
Several ligands, including EGF, NRG1-β, BTC, TGF-α, HB-EGF, and AR, were tested for their ability to mediate DNA synthesis in the 32D transfectants (Fig. 2). It has been previously demonstrated that none of these ligands are mitogenically active on 32D cells or 32D transfectants expressing human ErbB2 alone (28, 30, 39). NRG1-β and BTC induced readily detectable mitogenesis in 32D.E4 cells. Surprisingly, 32D.E4 cells also underwent significant mitogenesis in response to EGF treatment. TGF-α induced weak DNA synthesis at high concentrations in 32D.E4 cells, while HB-EGF and AR were not active at any concentration analyzed. Dose-response analysis revealed that the concentrations of EGF and TGF-α required to initiate detectable DNA synthesis were 10- and 100-fold greater, respectively, than those for NRG1-β and BTC.
Figure 2.
Analysis of the ability of various EGF-like ligands to mediate mitogenesis in 32D.E4 and 32D.E2/E4 cells. Dose-response analysis of [3H]thymidine incorporation induced by EGF, NRG1-β, BTC, TGF-α, HB-EGF, or AR in 32D.E4 cells (░⃞) or 32D.E2/E4 cells (▪) are shown. Data points are the mean of triplicate samples and are expressed as amount [3H]thymidine incorporation in cpm.
NRG1-β, BTC, EGF, TGF-α, and HB-EGF were all able to evoke mitogenesis in 32D.E2/E4 cells, while AR remained inactive (Fig. 2). Expression of ErbB2 did not significantly alter the ability of NRG1-β or BTC to induce DNA synthesis in the 32D transfectants. However, EGF and TGF-α elicited detectable mitogenesis at significantly lower concentrations in 32D.E2/E4 cells when compared with 32D.E4 cells. Although HB-EGF was slightly less active than the other ligands in terms of the concentration required to detect mitogenesis, it was able to induce a robust mitogenic response in 32D.E2/E4 cells and maximal [3H]thymidine incorporation achieved by HB-EGF was similar to the other ligands. Interestingly, the maximal levels of [3H]thymidine incorporation reached in response to EGF, NRG1-β, TGF-α, and BTC treatment of 32D.E2/E4 cells at higher concentrations were less than those observed at lower concentrations.
Certain ErbB Ligands Mediate Adherence, Morphological Alterations, and Up-Regulation of Monocytic Differentiation Markers in 32D.E2/E4 But Not in 32D.E4 Cells.
Alterations in the adherence properties of the two transfectants were investigated utilizing a single high dose of each ligand (Table 1). Both transfectants grew in suspension in the presence of IL-3. Some 32D.E4 cells weakly attached to the tissue culture substrate within 24 h after treatment with NRG1-β or BTC, while none of the other ligands analyzed induced this effect. NRG1-β, BTC, and EGF induced pronounced adherence of 32D.E2/E4 cells, and TGF-α did so to a lesser extent. The majority of cells that did not attach to the substrate attached to each other, forming large clumps. HB-EGF and AR did not affect adherence of the cotransfectant. Morphological changes in cell shape and size were also observed in 32D.E2/E4 cells treated with EGF, NRG1-β, BTC, and TGF-α (data not shown). Many attached cell formed elongated filopodia, and cell size increased even when they remained in suspension.
Table 1.
Effects of EGF-like ligands on adherence and monocytic differentiation markers in the 32D transfectants
| Ligand | Adherence*
|
Mac1†
|
FcγRII/III†
|
|||
|---|---|---|---|---|---|---|
| E4‡ | E2/E4‡ | E4‡ | E2/E4‡ | E4‡ | E2/E4‡ | |
| 0 | – | – | 10.6 | 12.9 | 35.0 | 33.3 |
| EGF | – | +++ | 12.1 | 49.2 | 37.0 | 107.0 |
| NRG1-β | +/− | +++ | 16.0 | 39.1 | 47.6 | 124.6 |
| BTC | +/− | +++ | 17.8 | 40.2 | 52.5 | 144.0 |
| TGF-α | – | ++ | 11.2 | 35.1 | 31.7 | 84.4 |
| HB-EGF | – | – | 11.1 | 12.4 | 33.1 | 39.0 |
Adherence was based on the percent of cells that attached to the tissue culture substrate before or after stimulation with ligand (100 ng/ml) for 24 h. −, No adherence; +/−, <10%; +, 10–25%; ++, 25–50%; +++, >50%.
The numbers shown represent the mean fluorescence intensity of staining by FITC-conjugated anti-Mac1 and anti-FcγRII/III antibodies as determined by flow cytometry before or after stimulation with ligand (100 ng/ml) for 24 h.
E4 and E2/E4 are the abbreviation of 32D.E4 and 32D.E2/E4, respectively.
The murine Mac-1 integrin and FCγRII/III antigens are known to be up-regulated during monocytic differentiation of 32D cells (41, 44). Therefore, expression of these cell surface antigens before and after treatment of 32D.E4 and 32D.E2/E4 cells with the various ligands was analyzed by flow cytometry (Table 1). Treatment of 32D.E4 cells with NRG1-β and BTC slightly enhanced the mean fluorescence intensity of Mac-1 and FcγRII/III staining, while EGF, TGF-α, and HB-EGF did not alter expression of either antigen. Exposure of 32D.E2/E4 cells to EGF, NRG1-β, BTC, and TGF-α, but not to HB-EGF, led to striking increases in the mean fluorescence intensity for both cell surface markers. The degree of Mac1 and FcγRII/III up-regulation paralleled the level of attachment observed when 32D.E2/E4 cells were exposed to the different ligands. These results provide evidence that several ErbB ligands are able to induce significant morphological alterations, adherence, and up-regulation of differentiation antigens only when ErbB2 is coexpressed with ErbB4. Furthermore, it is likely that decreases in DNA synthesis observed in 32D.E2/E4 cells at higher concentrations of EGF, NRG1-β, BTC, and TGF-α can be attributed to the differentiation effects induced by these factors.
Characterization of EGF Binding to 32D.E4 and 32D.E2/E4 Transfectants.
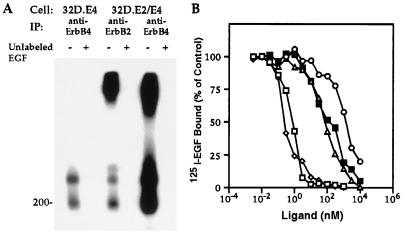
The capacity of EGF to elicit mitogenesis in cells expressing only ErbB4 or coexpressing ErbB2 and ErbB4 has not been previously reported. To assess the capacity of EGF to interact with these receptors in the 32D.E4 or 32D.E2/E4 transfectants, covalent affinity crosslinking analysis was performed. It has been shown that no detectable 125I-EGF-crosslinked proteins could be observed in anti-ErbB2 immunoprecipitates from 32D.E2 cells (28). As shown in Fig. 3A, 125I-EGF-crosslinked proteins with molecular sizes of ≈190 and 350 kDa were weakly observed in anti-ErbB4 immunoprecipitates from 32D.E4 cells. In agreement with the cooperative effect of ErbB2 and ErbB4 in mediating EGF-induced biological responses, 125I-EGF-crosslinked species were much more readily detected in lysates from 32D.E2/E4 cells immunoprecipitated with anti-ErbB4. Similar crosslinked species were also visualized in anti-ErbB2 immunoprecipitates from the same transfectant. Higher molecular sized crosslinked species of >500 kDa, indicative of multi-oligomerization, were only observed in immunoprecipitates from 32D.E2/E4 cells. Addition of a 100-fold excess of unlabeled EGF before crosslinking abolished the detection of all crosslinked species from both transfectants.
Figure 3.
Characterization of EGF binding to 32D.E4 and 32D.E2/E4 cells. (A) Covalent affinity crosslinking of 125I-EGF to the two 32D transfectants. 32D.E4 or 32D.E2/E4 cells were exposed to 125I-EGF in the absence (−) or presence (+) of a 100-fold excess unlabeled EGF. After crosslinking, cell lysates from the designated transfectants were immunoprecipitated (IP) with anti-ErbB2 or anti-ErbB4 serum, and immunoprecipitated proteins were resolved by SDS/PAGE. The molecular mass marker is shown in kDa. (B) Displacement analysis of 125I-EGF binding to 32D.E2/E4 cells in the presence of increasing concentrations of unlabeled EGF (▪), NRG1-β (◊), BTC (□), TGF-α (▵), and HB-EGF (○). Each data point represents the mean of two determinations and are expressed as percent of 125I-EGF binding in the absence of unlabeled ligand.
The relatively low affinity of radiolabeled EGF for 32D.E4 cells precluded accurate displacement analysis with unlabeled ligands (data not shown). However, binding of 125I-EGF to 32D.E2/E4 cells was much more efficient, and displacement with unlabeled EGF, NRG1-β, BTC, TGF-α, or HB-EGF are shown in Fig. 3B. The efficiency of the different EGF-like ligands to displace radiolabeled EGF was compatible with their capacity to induce biological effects in this transfectant. Taken together, these data provide evidence that EGF can weakly bind to ErbB4 and that binding is greatly enhanced by ErbB2 expression.
Analysis of the Tyrosine Phosphorylation Pattern and Activation of MAPK Induced by the Different EGF-Like Ligands in 32D.E4 and 32D.E2/E4 Cells.
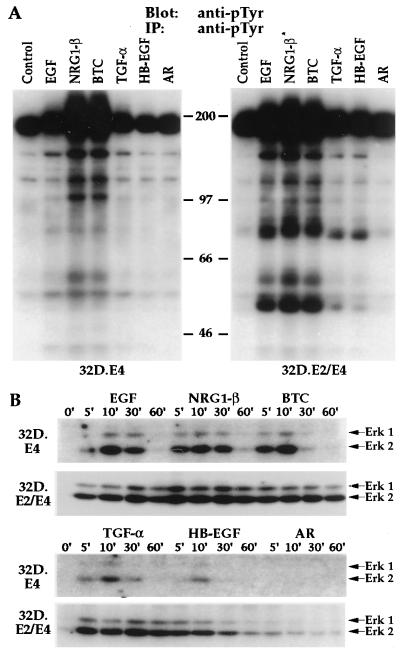
The ability of the various ligands to mediate tyrosine phosphorylation of cellular proteins was evaluated in 32D.E4 and 32D.E2/E4 cells. As shown in Fig. 4A, relatively high basal phosphorylation of a protein corresponding in molecular size to ErbB4 was detected after immunoprecipitation of lysates from 32D.E4 cells and subsequent immunoblot analysis with anti-pTyr. Enhanced phosphorylation of this molecule was observed in response to stimulation with EGF, NRG1-β, BTC, and TGF-α, but not HB-EGF or AR. The increase in phosphotyrosine content and the more diffuse nature of the high molecular size protein were very apparent in response to NRG1-β or BTC treatment. These differences were also observed, albeit to a lesser extent, after EGF or TGF-α stimulation. Furthermore, enhanced or de novo tyrosine phosphorylation of multiple protein species ranging from ≈40 to 160 kDa were detected in lysates from 32D.E4 cells stimulated with EGF, NRG1-β, BTC, and TGF-α, while HB-EGF and AR did not alter substrate phosphorylation.
Figure 4.
The various EGF-like ligands induce differential tyrosine phosphorylation and MAPK activation in the 32D.E4 and 32D.E2/D4 transfectants. (A) Detection of EGF-like ligand-induced tyrosine phosphorylation of cellular proteins in 32D.E4 and 32D.E2/E4 cells. Lysates from untreated cells (Control) or cells treated with EGF, NRG1-β, BTC, TGF-α, HB-EGF, or AR were immunoprecipitated (IP) and subsequently immunoblotted (Blot) with anti-pTyr as designated. Molecular mass markers are given in kDa. (B) The effects of EGF-like ligand treatment on the potency and duration of MAPK activation in 32D.E4 and 32D.E2/E4 cells. Lysates from untreated cells (O) or cells treated with the various EGF-like ligands for 5, 10, 30, or 60 min were subjected to immunoblot analysis with an antibody that specifically recognizes the activated forms of Erk1 and Erk2 marked by arrows.
An identical analysis of 32D.E2/E4 cells revealed similar high level basal tyrosine phosphorylation of proteins in the size range of ErbB2 and ErbB4. EGF, NRG1-β, and BTC induced equivalent increases in the phosphotyrosine content and the diffuse nature of proteins in this molecular size range. Substrate phosphorylation was also dramatically enhanced in response to treatment of 32D.E2/E4 cells with these three factors in comparison to that observed in 32D.E4 cells. TGF-α and HB-EGF also elicited de novo or increased tyrosine phosphorylation of several proteins but to a lesser degree than EGF, NRG1-β, or BTC, while AR remained inactive.
It has been reported that ligand occupancy of various ErbB receptor combinations results in activation of MAPK (47). Because activation of MAPK results from integration of multiple upstream signaling events, we analyzed the duration and potency of MAPK activation induced by the various ligands in the 32D.E4 and 32D.E2/E4 transfectants. As shown in Fig. 4B, no activated MAPK proteins were detected before ligand stimulation. Treatment of 32D.E4 cells with EGF resulted in detectable activation of both Erk1 and Erk2 at 5 min that was increased and sustained through 30 min. NRG1-β and BTC treatment resulted sustained activation for 60 min with slightly different potencies. TGF-α induced similar kinetics but with much weaker potency when compared with EGF treatment. Although HB-EGF was biologically inert on this transfectant, weak MAPK activation was discernible at 10 min after treatment. Analysis of the 32D.E2/E4 transfectant revealed that EGF, NRG1-β, BTC, and TGF-α all mediated sustained and potent Erk1 and Erk2 activation through 60 min. HB-EGF treatment resulted in weaker activation that was sustained for 30 min and decreased at 60 min. Immunoblot analysis with a combination of anti-Erk1 and anti-Erk2 sera that recognize both the inactive and active forms revealed that there were equivalent levels of these proteins in each lane (data not shown). Thus, the potency and kinetics of MAPK activation by the various ligands was remarkably similar to the biological activity evoked by these factors in the two transfectants.
DISCUSSION
The results of our present study provide the first documentation that EGF is capable of acting as a mitogen for ErbB4. Because EGF was shown to bind ErbB4 with weak affinity, it is likely that efficient EGF-mediated mitogenic signaling requires high-density receptor expression such that achieved in the 32D.E4 transfectant. In fact, 32D.E4 transfectants expressing lower levels of cell surface ErbB4 only negligibly responded to EGF (unpublished observations). Furthermore, EGF was unable to mediate tyrosine phosphorylation or mitogenesis in IL-3-dependent Ba/F3 cells transfected with ErbB4 (27, 38). However, in contrast to the results obtained in our present study, neither NRG1-β or BTC induced mitogenesis of Ba/F3 cell expressing ErbB4 (27, 38). Thus, it is likely that the level of ErbB4 expression achieved in Ba/F3 was lower than that attained in 32D transfectants. Recent studies have provided conflicting evidence regarding the ability of EGF to stimulate proliferation in NIH 3T3 cells expressing exogenous ErbB4. In two studies, EGF treatment did not induce proliferation of NIH 3T3 cells transfected with ErbB4 in serum-free monolayer assays (36, 48). In contrast, ErbB4-transfected NIH 3T3 cells formed large colonies in soft agar in the presence of EGF in a separate study (49). Whether differences in level of ErbB4 expression or variation in the expression of other ErbB receptors in the NIH 3T3 lines utilized in the different studies influenced the results obtained remains to be determined. We have also utilized murine NR6 transfectants to analyze ErbB ligand responses (unpublished observations). NR6 cells do not express any ErbB receptor as determined by RT-PCR (Fig. 1A and ref. 42). While EGF did induce a weak mitogenic response in NR6 cells transfected with ErbB4, NRG1-β, and BTC were far superior to EGF in eliciting DNA synthesis. Taken together, the results of these different studies indicate that EGF is a poor ligand for ErbB4 when it is not coexpressed with other ErbB receptors.
Several studies have revealed that ErbB2 is the preferred and common auxiliary subunit for the other three ErbB family members (6, 31, 33). A recent study utilizing titration calorimetry and small-angle x-ray scattering analysis demonstrated a requirement for two EGF molecules to induce dimerization of ErbB1. However, these data could not distinguish whether each EGF molecule exclusively binds to one receptor resulting in subsequent receptor dimerization or whether bivalent binding of both EGF molecules to two receptors is required to mediate homodimeric receptor interactions (50). A second study has provided strong evidence that ErbB ligands are bivalent in their binding capacity (51). This model predicts that one site on EGF or other ErbB ligands would bind to ErbB4 and a second asymmetric site would bind to ErbB2, creating a stable heterodimer. Although direct binding of NRG1-β or EGF to ErbB2 had not been reported previously, results from the above mentioned study demonstrated that these ligands are able to bind to soluble ErbB2 with an extremely weak affinity as determined by Biacore and affinity labeling analysis (51). The ability of ErbB2 to weakly interact with ErbB ligands would predict that ErbB2 could cooperate in mediating bivalent binding of any ligand that also possesses the capacity to bind ErbB4. The results of our present study demonstrating enhanced signaling by EGF and TGF-α in 32D.E2/E4 cells strongly support this concept. Similar findings were observed in NR6 cells coexpressing these receptors (unpublished observations).
High concentrations of EGF, NRGβ-1, BTC, and TGF-α mediated significant adherence, morphological alterations, and cell surface up-regulation of the Mac-1 integrin and FcγRII/III antigen only in 32D cells coexpressing ErbB2 and ErbB4. It has been well documented that several NRG isoforms promote differentiation of certain breast carcinoma cell lines, resulting in the production of milk proteins and the up-regulation of ICAM-1 and CD44 adhesion molecules (52, 53). While our results were not obtained in the context of a mammary epithelial cell background, they do suggest that the levels of coexpressed receptors as well as the concentration of their cognate ligands can influence whether proliferation or differentiation predominates. Thus, one could envision that the prognostic importance of ErbB2 overexpression would depend on the level of expression of other ErbB receptors as well as the availability of ligands that could bivalently interact with the coexpressed receptors.
It is difficult to explain why HB-EGF was unable to mediate mitogenesis of 32D.E4 cells, while it readily did so in the cotransfectant. A recent study demonstrated that HB-EGF can bind to NIH 3T3 cells expressing exogenous ErbB4 and mediate chemotaxis but not mitogenesis (48). The inability of HB-EGF to induce DNA synthesis in the 32D.E4 cells is probably not due to the absence of proteoglycans on these cells, because addition of heparin did not affect HB-EGF action in this transfectant. Furthermore, HB-EGF was demonstrated to be mitogenic in 32D cells expressing ErbB1 without the addition of heparin (28). NR6 cells cotransfected with ErbB2 and ErbB4 were also shown to readily respond mitogenically to HB-EGF, while NR6 transfectants expressing only ErbB4 were not responsive to this ligand (unpublished observations). Thus, HB-EGF appears to require the coexpression of both receptors to allow efficient mitogenic signaling. It remains to be determined whether HB-EGF is capable of binding and mediating chemotaxis in 32D or NR6 cells expressing only ErbB4.
It has recently been demonstrated that ErbB3, unlike other ErbB receptors, is kinase defective (54). Thus, heterodimerization with a second ErbB receptor is obligatory for mediating signal transduction through ErbB3. It has been demonstrated that EGF and BTC, like NRG1-β, can elicit mitogenesis in 32D cells coexpressing ErbB2 and ErbB3 (28). Interestingly, TGF-α and HB-EGF were not able to mediate signal transduction in the 32D cotransfectant expressing ErbB2 and ErbB3, while both ligands were active on 32D.E2/E4 and 32D.E1 cells. These results suggest that EGF and BTC contain multiple binding sites can differentially interact with ErbB receptors or that the binding sites present on TGF-α and HB-EGF are distinct from those that exist on EGF and BTC.
While NRG1-β has been demonstrated to bind to ErbB3 when it is expressed alone (20), EGF and BTC did not detectably bind to and could not be crosslinked to either ErbB2 or ErbB3 unless these receptors were coexpressed in 32D cells (28). These results indicate that EGF and BTC bind to ErbB2 and ErbB3 by a mechanism requiring the simultaneous expression of two distinct receptors. In contrast, the results obtained in the present study suggest that NRG1-β, BTC, EGF, and TGF-α bind directly to ErbB4 with varying degrees of affinity, resulting in graded proliferative responses. However, coexpression of ErbB2 with ErbB4 dramatically increased the binding affinity and reduced the amounts of EGF or TGF-α required to initiate DNA synthesis in the 32D cotransfectant, suggesting that ErbB2 may cooperate with ErbB4 by mediating bivalent binding of certain ligands or by impairing down-regulation of ErbB2/ErbB4 complexes. Furthermore, tyrosine phosphorylation of intracellular substrates was expanded, the potency and duration of MAPK activation was increased and differentiation antigens were dramatically up-regulated in the cotransfectant in response to NRG1-β, BTC, EGF, and TGF-α. Therefore, rather than being essential for activation of ErbB4 by these four ligands, ErbB2 expression appears to modify and strengthen signal transduction, potentially altering gene transcription and diversifying biological responses.
In summary, the results of the present study provide evidence that coexpression of ErbB2 with ErbB4 has profound effects on the ability of ligands to mediate proliferation and/or differentiation in a model system. The recent evaluation of a large series of patients with childhood medulloblastoma revealed that ErbB2 and ErbB4 were coexpressed in >50% of the tumor tissues along with concurrent NRG1 expression in many of these samples (12). Primitive cells of the cerebellum are thought to be the targets for development of medulloblastoma. ErbB2 is thought to be expressed in the developing murine brain and absent in the adult brain, while ErbB4 expression is retained in the adult brain (see Fig. 1A and refs. 1, 32, 34, 55). Thus, it can be postulated that aberrant deregulation of ErbB2 in tissues that normally express ErbB4 and are exposed to certain ErbB ligands by autocrine, paracrine or juxtacrine means could have profound effects on the progression of certain human neoplasms.
Acknowledgments
We thank Dr. Cary Lai for providing us the unpublished sequence for murine ErbB4, Dr. Stuart S. Aaronson for helpful discussion during the course of this work, and Helen Goode and Nancy Cruz for superb secretarial support.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: EGF, epidermal growth factor; NRG1-β, neuregulin 1-β; BTC, betacellulin; HB-EGF, heparin binding-EGF; AR, amphiregulin; IL-3, interleukin 3; MAPK, mitogen-activated protein kinase; FITC, fluorescein isothiocyanate; RT-PCR, reverse transcription–PCR.
References
- 1.Pinkas-Kramarski R, Alroy I, Yarden Y. J Mammary Gland Biol Neoplasia. 1997;2:97–107. doi: 10.1023/a:1026343528967. [DOI] [PubMed] [Google Scholar]
- 2.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Nature (London) 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 3.Lee K-F, Simon H, Chen H, Bates B, Hung M-C, Hauser C. Nature (London) 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 4.Salomon D S, Brandt R, Ciardiello F, Normanno N. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 5.Hynes N E, Stern D F. Biochim Biophys Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 6.Karunagaran D, Tazhar E, Beerli R R, Chen X, Graus-Porta D, Wen D, Seger R, Hynes N E, Yarden Y. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 7.Slamon D J, Clark G M, Wong S G, Levin W J, Ullrich A, McGuire W L. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 8.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A, Press M F. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 9.Kraus M H, Fedi P, Starks V, Muraro R, Aaronson S A. Proc Natl Acad Sci USA. 1989;86:9193–9197. [Google Scholar]
- 10.Gullick W J. Br Med Bull. 1991;47:87–98. doi: 10.1093/oxfordjournals.bmb.a072464. [DOI] [PubMed] [Google Scholar]
- 11.Alimandi M, Romano A, Curia M C, Muraro R, Fedi P, Aaronson S A, Di Fiore P P, Kraus M H. Oncogene. 1995;10:1813–1821. [PubMed] [Google Scholar]
- 12.Gilbertson R J, Perry R H, Kelly P J, Person A D J, Lunec J. Cancer Res. 1997;57:3272–3280. [PubMed] [Google Scholar]
- 13.Savage C R, Jr, Inagami T, Cohen S. J Biol Chem. 1972;247:7612–7621. [PubMed] [Google Scholar]
- 14.Marquart H M, Hunkapiller W, Hood L E, Todaro G J. Science. 1984;223:1079–1082. doi: 10.1126/science.6320373. [DOI] [PubMed] [Google Scholar]
- 15.Shoyab M G, Plowman G, McDonald V L, Bradley J G, Todaro G J. Science. 1989;243:1074–1076. doi: 10.1126/science.2466334. [DOI] [PubMed] [Google Scholar]
- 16.Higashiyama S, Abraham J A, Miller J, Fiddes J C, Klagsbrun M. Science. 1991;251:936–939. doi: 10.1126/science.1840698. [DOI] [PubMed] [Google Scholar]
- 17.Shing Y, Christofori G, Hanahan D, Ono Y, Sasada R, Igarashi K, Folkman J. Science. 1993;259:1604–1607. doi: 10.1126/science.8456283. [DOI] [PubMed] [Google Scholar]
- 18.Toyada H, Komurasaki T, Uchida D, Takayama Y, Isobe T, Okuyama T, Hanada K. J Biol Chem. 1995;270:7495–7500. doi: 10.1074/jbc.270.13.7495. [DOI] [PubMed] [Google Scholar]
- 19.Plowman G D, Green J M, Culouscou J-M, Carlton G W, Rothwell V M, Buckley S. Nature (London) 1993;366:473–475. doi: 10.1038/366473a0. [DOI] [PubMed] [Google Scholar]
- 20.Carraway K L, III, Sliwkovski M X, Akita R, Platko J V, Guy P M, Nuijens A, Diamonti A J, Vandlen R L, Cantley L C, Cerione R A. J Biol Chem. 1994;269:14303–14306. [PubMed] [Google Scholar]
- 21.Kita Y A, Barff J, Luo Y, Wen D, Brankow D, Hu S, Liu N, Prigent S A, Gullick W J, Nicolson M. FEBS Lett. 1994;349:139–143. doi: 10.1016/0014-5793(94)00644-x. [DOI] [PubMed] [Google Scholar]
- 22.Sliwkowski M X, Sehaefer G, Akita R, Lofgren J A, Fitzpatrick V D, Nuijens A, Frendly B M, Cerione R A, Vandlen R L, Carraway K L., III J Biol Chem. 1994;269:14661–14665. [PubMed] [Google Scholar]
- 23.Tazhar E, Levkowitz G, Karunagaran D, Yi L, Peles E, Lavi S, Chang D, Liu N, Yayon A, Wen D, Yarden Y. J Biol Chem. 1994;269:25226–25233. [PubMed] [Google Scholar]
- 24.Carraway K L, III, Weber J L, Unger M J, Ledesma J, Yu N, Gassman M. Nature (London) 1997;387:512–516. doi: 10.1038/387512a0. [DOI] [PubMed] [Google Scholar]
- 25.Chang H, Riese D, Gilbert W, Stern D F, Mamahan U J. Nature (London) 1997;387:509–512. doi: 10.1038/387509a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, Sliwkowski M X, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, Godowski P J. Proc Natl Acad Sci USA. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riese D J, II, Bermingham Y, van Raaij T M, Buckley S, Plowman G D, Stern D F. Oncogene. 1996;12:345–353. [PubMed] [Google Scholar]
- 28.Alimandi M, Wang L-M, Bottaro D, Lee C-C, Kuo A, Frankel M, Fedi P, Tang C, Lippman M, Pierce J H. EMBO J. 1997;16:5608–5617. doi: 10.1093/emboj/16.18.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbacci E G, Guarino B C, Stroh J G, Singleton D H, Rosnack K J, Moyer J D, Anrews G C. J Biol Chem. 1995;270:9585–9589. doi: 10.1074/jbc.270.16.9585. [DOI] [PubMed] [Google Scholar]
- 30.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin B J, Sela M, Yarden Y. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 31.Tazhar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin B J, Yarden Y. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alroy I, Yarden Y. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 33.Graus-Porta D, Beerli R R, Daly J M, Hynes N E. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kokai Y, Myers J N, Wada T, Brown V I, LeVea C M, Davis J G, Dobashi K, Greene M. Cell. 1989;58:287–292. doi: 10.1016/0092-8674(89)90843-x. [DOI] [PubMed] [Google Scholar]
- 35.Wallasch C, Weiss F U, Niederfellner G, Jallal B, Issing W, Ullrich A. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K, Sun J, Liu N, Wen D, Chan D, Thomason A, Yoshinaga S K. J Biol Chem. 1996;271:3884–3890. [PubMed] [Google Scholar]
- 37.Riese D J, II, van Raaij T M, Plowman G D, Andrews G C, Stern D F. Mol Cell Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riese D J, Kim E D, Elenius K, Buckley S, Klagsbrun M, Plowman G D, Stern D. J Biol Chem. 1996;271:20047–20052. doi: 10.1074/jbc.271.33.20047. [DOI] [PubMed] [Google Scholar]
- 39.Pinkas-Kramarski R, Shelly M, Glathe S, Ratzkin B J, Yarden Y. J Biol Chem. 1996;271:19029–19032. doi: 10.1074/jbc.271.32.19029. [DOI] [PubMed] [Google Scholar]
- 40.Weissman B, Aaronson S A. Cell. 1983;32:599–606. doi: 10.1016/0092-8674(83)90479-8. [DOI] [PubMed] [Google Scholar]
- 41.Pierce J H, Ruggiero M, Fleming T P, Di Fiore P P, Greenberger J S, Varticovski L, Schlessinger J, Rovera G, Aaronson S A. Science. 1988;239:628–631. doi: 10.1126/science.3257584. [DOI] [PubMed] [Google Scholar]
- 42.Di Fiore P P, Segatto O, Taylor W G, Aaronson S A, Pierce J H. Science. 1990;248:79–83. doi: 10.1126/science.2181668. [DOI] [PubMed] [Google Scholar]
- 43.Plowman G D, Culouscou J-M, Whitney G S, Green J M, Carlton G W, Foy L, Neubauer M G, Shoyab M. Proc Natl Acad Sci USA. 1993;90:1746–1750. doi: 10.1073/pnas.90.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce J H, Dimareo E, Cox G W, Lombardi D, Ruggiero M, Varesio L, Wang L-M, Ghosh-Choudhury G, Sakaguchi A Y, Di Fiore P P, Aaronson S A. Proc Natl Acad Sci USA. 1990;87:5613–5617. doi: 10.1073/pnas.87.15.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L-M, Myers M G, Sun X-J, Aaronson S A, White M, Pierce J H. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 46.Fedi P, Pierce J H, Di Fiore P P, Kraus M H. Mol Cell Biol. 1994;14:492–500. doi: 10.1128/mcb.14.1.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marte B, Graus-Porta D, Jeschke M, Fabbro D, Hynes N E, Taverna D. Oncogene. 1995;10:167–175. [PubMed] [Google Scholar]
- 48.Elenius K, Paul S, Allison G, Sun J, Klagsbrun M. EMBO J. 1997;16:1268–1278. doi: 10.1093/emboj/16.6.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen B, Green J M, Foy L, Fell H P. J Biol Chem. 1996;271:4813–4818. doi: 10.1074/jbc.271.9.4813. [DOI] [PubMed] [Google Scholar]
- 50.Lemmon M A, Bu Z, Ladbury J E, Zhou M, Pinchasi D, Lax I, Engelman D, Schlessinger J. EMBO J. 1997;16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tazhar E, Pinkas-Kramarski R, Moyer J D, Klapper L N, Alroy I, Levkowitz G, Shelly M, Henis S, Eisenstein M, Ratzkin B J, Sela M, Andrews G C, Yarden Y. EMBO J. 1997;16:4938–4950. doi: 10.1093/emboj/16.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bacus S S, Stancovske I, Huberman E, Chin D, Hurwitz., Mills G B, Ullrich A, Sela M, Yarden Y. Cancer Res. 1992;52:2580–2589. [PubMed] [Google Scholar]
- 53.Bacus S S, Gudkov A V, Zelnick C R, Chin D, Stern R, Stancovski I, Peles E, Ben-Baruch N, Farbstein H, Lupu R, Wen D, Sela M, Yarden Y. Cancer Res. 1993;53:5251–5261. [PubMed] [Google Scholar]
- 54.Guy P, Platke J V, Cantley L, Cerione R A, Carraway K I., III Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinkas-Kramarski R, Eilam R, Spiegler O, Lavi S, Liu N, Chang D, Wen D, Schwartz M, Yarden Y. Proc Natl Acad Sci USA. 1994;91:9387–9391. doi: 10.1073/pnas.91.20.9387. [DOI] [PMC free article] [PubMed] [Google Scholar]