Abstract
In fission yeast, the rad3 gene product plays a critical role in sensing DNA structure defects and activating damage response pathways. A structural homologue of rad3 in humans (ATR) has been identified based on sequence similarity in the protein kinase domain. General information regarding ATR expression, protein kinase activity, and cellular localization is known, but its function in human cells remains undetermined. In the current study, the ATR protein was examined by gel filtration of protein extracts and was found to exist predominantly as part of a large protein complex. A kinase-inactivated form of the ATR gene was prepared by site-directed mutagenesis and was used in transfection experiments to probe the function of this complex. Introduction of this kinase-dead ATR into a normal fibroblast cell line, an ATM-deficient fibroblast line derived from a patient with ataxia–telangiectasia, or a p53 mutant cell line all resulted in significant losses in cell viability. Clones expressing the kinase-dead ATR displayed increased sensitivity to x-rays and UV and a loss of checkpoint control. We conclude that ATR functions as a critical part of a protein complex that mediates responses to ionizing and UV radiation in human cells. These responses include effects on cell viability and cell cycle checkpoint control.
In eukaryotic cells, unrepaired DNA damage or incomplete replication can trigger checkpoint pathways that result in either a delay in cell cycle progression or programmed cell death (1–3). In the fission yeast, Schizosaccharomyces pombe, the rad3+ gene product plays a critical role in sensing a wide array of DNA structure defects and activating appropriate damage response pathways (4–6). In the budding yeast Saccharomyces cerevisiae, the MEC1 gene product plays a similar role (7–9). The products of the rad3 and MEC1 genes are large, structurally related molecules characterized by the presence of a kinase domain at the carboxyl terminus with significant primary sequence similarity to phosphatidylinositol 3-kinase. This subgroup of protein kinases has been referred to as PIK-related kinases (10).
ATR (ataxia-telangiectasia and rad3+ related) is a PIK kinase identified in humans and mouse by sequence similarity in the protein kinase domain with S. pombe rad3 (6, 11). The ATR gene is expressed ubiquitously but is most highly expressed in testis (12). In meiotic cells, the ATR protein exhibits nuclear localization and is associated with unsynapsed regions of pairing chromosomes (12). Both wild-type ATR immunoprecipitated from mouse testis and a recombinant His-tagged form, affinity-purified after expression in S. cerevisiae, have an associated protein kinase activity (12). Expression of human ATR in yeast can complement the UV sensitivity of esr1–1 (MEC1) mutants in S. cerevisiae but not that of rad3 mutants in S. pombe (6). In human cells, the function of ATR is undetermined, but based on sequence homology and its effects when expressed in S. cerevisiae, it might be expected to overlap with at least some of the functions mediated by Mec1p or Rad3.
S. pombe Rad3 has an associated protein kinase activity. Site-specific mutagenesis of the conserved residue D2249 in the protein kinase domain significantly reduces the kinase activity of Rad3 and results in a phenotype indistinguishable from that of a null mutant with increased radiation sensitivity and hydroxyurea sensitivity. Moderate overexpression of kinase-dead rad3 in wild-type cells leads to a dominant radiation-sensitive phenotype whereas high level expression results in additional phenotypes of slow growth and impaired colony formation (6). The similarity in phenotype between kinase-dead and null mutants of Rad3 suggests that the protein kinase activity is essential for its checkpoint properties and for normal Rad3 function. The additional phenotypes observed upon high level overexpression of kinase-dead Rad3 likely result from disruption of a multisubunit complex that plays additional roles in pathways controlling mitosis. Although direct biochemical demonstration of such a complex has not been reported, two observations suggest its existence. First, Seaton et al. (5) isolated a partial rad3 cDNA by virtue of its ability to complement a rad3 point mutation. This result is consistent with intragenic or α complementation. Second, the existence of a complex is supported by the observation that tagged versions of rad3, when expressed in S. pombe, will homo-multimerize (6).
Several additional mammalian PIK kinases (FRAP/mTOR, DNA-PK, and ATM) have been described in humans. ATM was identified based on its role in the rare, inherited disorder ataxia-telangiectasia (AT) (13). AT is characterized by progressive cerebellar ataxia, immune deficiencies, an increased incidence of cancer, chromosomal instability, and hypersensitivity to ionizing radiation (14). ATM is a nuclear phosphoprotein with protein kinase activity (12). Cells from AT patients, in which the ATM gene is mutated, display reduced survival and an inability to activate cell cycle checkpoints after exposure to ionizing radiation (15).
Despite some overlap in function, it is unlikely that ATM represents a direct human analogue of either rad3+ or MEC1. mec1 and rad3 mutants are sensitive to a wide array of DNA damaging agents including UV, x-rays, alkylating agents, and hydroxyurea whereas AT cells exhibit a specific hypersensitivity to agents, like x-rays, that induce double-strand breaks. Although ATM does exhibit significant primary sequence similarity with MEC1, its sequence is more closely related to another PIK-related kinase gene in S. cerevisiae, TEL1, whose product controls telomere length (16). In S. cerevisiae, mutations in the TEL1 and MEC1 genes act additively such that double mutants are more sensitive to DNA damaging agents than either single mutant (17). These observations raise the possibility that, in humans, the functions executed by Rad3 and/or MEC1p in yeast are partitioned among the products of several genes, including ATM and possibly ATR, and that the products of these genes may cooperate in DNA damage responses.
In the current study, the function of ATR was examined by introducing expression constructs containing a mutated version of the gene, in which the protein kinase domain had been inactivated, into normal human cells, ATM-deficient cells from a patient with AT, and cells expressing only a mutated p53 gene. Expression of these constructs in normal human cells recapitulated the phenotypes induced by introduction of analogous constructs of the rad3 gene into S. pombe, including dominant sensitivity to UV and to ionizing radiation and reduced colony forming ability. Gel filtration analysis of ATR from cell extracts demonstrated that it formed part of a large protein complex in both normal and AT cell lines, consistent with a dominant negative effect of the kinase-dead form. Differential responses observed in normal, AT, and p53 mutant cells upon transfection suggest interaction between several damage response pathways.
MATERIALS AND METHODS
Cell Lines.
The simian virus 40-transformed fibroblast cell lines GM637 (Coriell Institute) and AT3BI (L. Kapp, Stanford Research Institute) and C-33A cervical carcinoma cells (ATCC) were maintained in DMEM supplemented with 15% fetal bovine serum.
Plasmid Constructs.
A full length cDNA clone of the human ATR gene was cloned in the expression plasmid pcDNA3 (Invitrogen). A kinase-dead version of ATR was constructed by site-specific mutagenesis by using the mutagenic oligonucleotide GCGTACATGTAGAaTTCAATTGTCTTTTCA, where the lower case “a” replaced the normally occurring “T” residue, which resulted in the substitution of the critical kinase domain D at position 2494 with E.
Transfections.
Transfections were carried out by using Lipofectin according to instructions provided by the manufacturer (Gibco/BRL). In brief, 2 × 105 cells were plated per 60-mm2 dish and allowed to grow for 24 h. Plasmid DNA (1 to 3 μg) was added to 100 μl of Lipofectin diluted 1:10 in Opti-MEM I (reduced serum) media and incubated for 15 min at room temperature. Cells were washed twice in serum-free DMEM. The Lipofectin/DNA mixture was diluted into 2 ml of Opti-MEM I media, overlaid onto cells, and incubated for 8 h at 37°C in 5% CO2. After incubation, 6 ml of DMEM supplemented with fetal bovine serum was added and plates were incubated for another 48 h. For transient assays of cell viability, cells were harvested at this point, washed, and resuspended in PBS supplemented with 1% fetal bovine serum. Fluorescent cells were counted on a FACSort (Becton Dickinson). For colony-forming assays, cells were harvested 48 h after lipofection, washed, serially diluted, and plated in quadruplicate in the presence of G418 for selection. After 10–14 days, plates were fixed and stained in 50% methanol, 10% glacial acetic acid, and 0.1% Coomassie Blue, and colonies were counted.
Protein Extraction.
Cells were ground in liquid nitrogen with a mortar and pestle, added to 4.4 ml/g of lysis buffer (0.5% Triton X-100/2 mM EDTA/2 mM EGTA/25 mM NaF/0.1 mM sodium vanadate/1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF) 1 μg/ml leupeptin/1 μg/ml pepstatin/2 mM DTT/50 mM sodium phosphate, pH 7.2) and placed in a Dounce homogenizer. The solution was subjected to 24 strokes with the loose fitting pestle and 6 strokes with a tighter fitting pestle. After 30 min on ice, particulates were removed by low speed centrifugation (5 min at 1,000 × g). Aliquots were frozen in liquid nitrogen and stored at −70°C.
Gel Filtration and Western Blotting.
A Superose 6 column (Pharmacia Biotech) was equilibrated in 50 mM Hepes (pH 8.0), 5 mM MgCl2, 0.1 mM sodium vanadate, 25 mM NaF, 2 μg/ml pepstatin A and leupeptin, 5 μg/ml aprotinin, 20 μg/ml calpain I and II, 0.1 mM AEBSF, 5 mM imidazole, and 400 mM NaCl and calibrated with molecular weight standards. Extracts (2.9 mg) were precleared by centrifugation (15 min at 10,000 × g), diluted 1:4 in the same buffer to a volume of 0.7 ml, and loaded on the column, and 0.57-ml fractions were collected. Column fractions (0.085 ml) were separated on an 8% polyacrylamide/SDS gel and transferred to poly(vinylidene difluoride) membrane for 0.5 Amp-hours in 1X Laemli buffer with 20% methanol. Poly(vinylidene difluoride) membranes were blocked in 5% milk, incubated with mAb 224C (at 8 μg/ml in 5% milk), washed extensively with Tris-buffered saline containing 0.5% Tween 20, incubated with goat anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad), washed, and developed by treatment with enhanced chemiluminescence reagents (NEN) followed by autoradiography. mAb 224C was raised to a glutathione S-transferase fusion protein expressing the kinase domain of ATR (AgDH3) (12) as described (18).
Radiation Survival Assays.
Cell survival after irradiation was assayed as described (19). Cells were irradiated at doses of 1, 2, and 3 Gy from a 137Cs source delivering 13.6 Gy/min. After irradiation, cells were plated in quadruplicate at two different cell concentrations, 600 and 1,200 cells/100-mm2 dish, to obtain significant number of colonies at all doses. After 14–21 days, colonies were stained and counted as described for colony-forming assays.
UV Survival Assays.
Cell survival after UV irradiation was assayed essentially as described above for ionizing radiation. Cells were exposed to a 310-nM wavelength UV source for 0, 5, or 10 s. After UV exposure, cells were plated in quadruplicate at two different densities and colonies were stained and counted after 14–21 days.
Cell Cycle Distribution.
Effects of irradiation on cell cycle distribution were assayed as described by Beamish and Lavin (20). After staining with propidium iodide, samples were analyzed on a FACSort. Data were collected with the Cell Quest DNA quantitation program (Becton Dickinson) and analyzed by using by Mac Cycle (Phoenix Flow Systems, San Diego).
RESULTS
ATR Forms Part of a Multi-Protein Complex in Mammalian Cells.
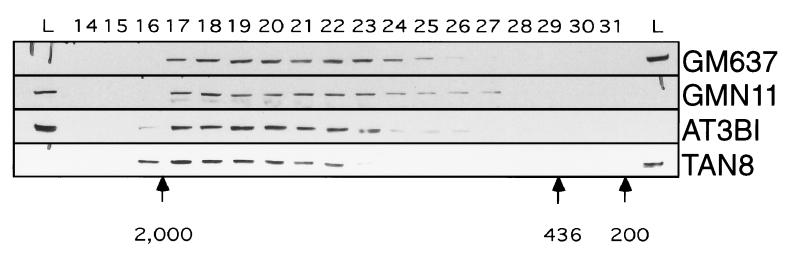
The expression of dominant negative forms of signaling molecules provides an effective means of probing their cellular functions. However, this approach depends on the formation of functionally relevant protein–protein interactions involving the molecule of interest. To validate the dominant negative approach for exploring the function of ATR, we tested for evidence of protein complexes involving native ATR in human cells. Total proteins from a simian virus 40-transformed fibroblast cell line from a normal donor, GM637, and a simian virus 40-transformed cell line from an AT patient, AT3BI, were fractionated by gel chromatography. Proteins from each column fraction were separated by PAGE and immunoblotted with an mAb specific for ATR. As shown in Fig. 1, ATR was found predominantly in complexes of 1,000 kDa average molecular mass in each cell line. The predicted relative molecular mass for ATR is 301.5 kDa, suggesting that ATR was either associated with other proteins or formed a multimeric complex. Comparable results in a normal and AT cell line indicate that ATM, which is not expressed in AT3BI cells, is unlikely to be part of the observed complex. Similar results were obtained when proteins from mouse testis, where ATR is highly expressed, were fractionated, indicating that the observed complexes were not a cell line-specific phenomenon (data not shown).
Figure 1.
Sizing of ATR-containing complexes by gel filtration. Protein extracts prepared as described in Materials and Methods were fractionated by Superose 6 chromatography. Immunoblot analysis was used to determine the elution profile of ATR. Native ATR is ≈300 kDa in size. Elution peaks of molecular size standards are indicated below.
Expression of a Kinase-Inactivated Form of ATR Results in Cell Death.
The detection of protein complexes incorporating ATR suggested that it might be possible to generate a dominant negative mutant of ATR by overexpression of a kinase-inactivated form of the molecule. Accordingly, a full length cDNA of the human ATR gene was cloned in the expression vector pcDNA3. Site-specific modification of the DFG sequence that is highly conserved among the kinase domains of known serine–threonine protein kinases was used to create a mutant form of ATR (ATR.KD) in which the D residue at position 2494 was changed to E. The position and nature of this substitution in ATR.KD was identical to that made in a previous study of S. pombe rad3. In that study, expression of this mutant resulted in significantly reduced in vitro kinase activity in immunoprecipitates and a dominant negative phenotype in rad3+ yeast (6).
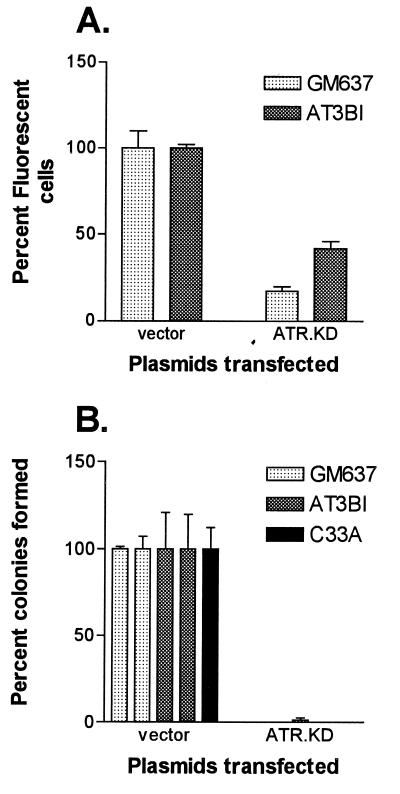
The ATR.KD expression construct was transfected into GM637 and into AT3BI cells. Three separate transient transfections were carried out with each cell line. A limiting amount of an additional plasmid, pTracer, containing the green fluorescent protein (GFP) gene, was added to each transfection as a marker for cells that had taken up exogenous DNA. Differences in the numbers of fluorescent cells were quantitated by FACS to determine the effect of the co-transfected experimental plasmid on cell viability. Transfection of the ATR.KD construct resulted in a 2- to 5-fold reduction in the number of viable fluorescent cells detected in both normal and AT cells (Fig. 2A). This effect on cell viability could not be explained by toxicity or contaminants in the DNA preparation because mock experiments in which all components except lipofectin were added did not result in the same effect.
Figure 2.
Effect of transfection of ATR.KD expression plasmid on cell viability. (A) GM637 and AT3BI cells were transfected transiently in triplicate with a 3:1 molar excess of the indicated constructs relative to a plasmid expressing green fluorescent protein and quantitated by FACS. For each cell line, 10,000 events were collected. Transfection efficiency was 15–25% with vector alone, and results were normalized to these values. (B) Colonies arising from independent transfections with the indicated plasmids were counted after 10–14 days of selection in G418. Average numbers of colonies with vector alone were ≈1,000 for GM637, ≈30 for AT3BI, and ≈65,000 for C-33A. Results were normalized to vector controls.
Additional transfections of GM637 and of AT3BI cells were carried out to obtain stable cloned integrants in which DNA damage responses could be measured. Cells were serially diluted, plated in quadruplicate, and selected in the presence of G418 to provide also a measure of colony-forming ability (Fig. 2B). Introduction of the ATR.KD expression construct into GM637 and AT3BI cells yielded no colonies in an initial transfection. A second transfection resulted in a limited number (<10) of small colonies. No colonies were observed when ATR.KD was transfected into C-33A, a p53 mutant cervical carcinoma cell line (21), despite the fact that these cells had a 50- to 100-fold higher transfection efficiency than either GM637 or AT3BI cells with vector alone.
Upon prolonged G418 selection and propagation, a few of the colonies generated with the ATR.KD expression constructs expanded to a size that would allow cloning. Two clones resulting from transfection of the normal GM637 cell line, GMN11 and GMN15, and one clone from transfection of the AT3BI cell line, TAN8, were isolated. These clones had a distinct slow growth phenotype. To evaluate the effect of expression of the ATR.KD construct on ATR expression and complex formation, protein extracts from GMN11 cells and TAN8 cells were fractionated and immunoblotted for ATR (Fig. 1). Comparable numbers of GM637, GMN11, AT3BI, and TAN8 cells were analyzed. Total ATR levels were reduced slightly relative to parental cell lines in the GMN11 and TAN8 cells, suggesting that high level expression of ATR.KD was not tolerated. As in the parental cell lines GM637 and AT3BI, ATR in GMN11 and TAN8 cells was detected predominantly in higher molecular mass fractions, indicating that the kinase-dead ATR could be incorporated into protein complexes (Fig. 1).
Cloned ATR.KD Transfectants Display Dominant Sensitivity to UV and Ionizing Radiation.
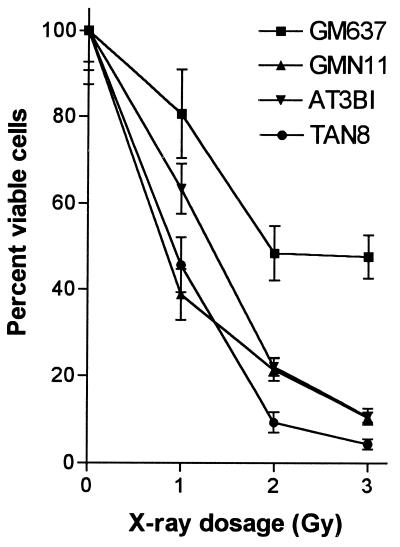
GM637 and AT3BI cells and clones of each of these cell lines transfected with the ATR.KD construct were tested for their sensitivity to ionizing radiation (Fig. 3). As expected for an AT cell line, AT3BI cells were severalfold more sensitive to killing by x-rays than GM637 cells. When irradiated, the GMN11 cell line, expressing the ATR.KD construct in a GM637 background, was as sensitive or more sensitive than AT3BI cells at all doses of x-rays tested. The TAN8 cell line, which expressed ATR.KD on an ATM-deficient background, was as sensitive or more sensitive than GMN11 cells or the parental AT3BI cell line.
Figure 3.
Effect of ionizing radiation on colony-forming ability of ATR.KD-transfected cell lines. Cells were exposed to the indicated doses of x-rays and plated in quadruplicate. Colonies were counted after 10–14 days. Colony counts were normalized to those for unirradiated controls.
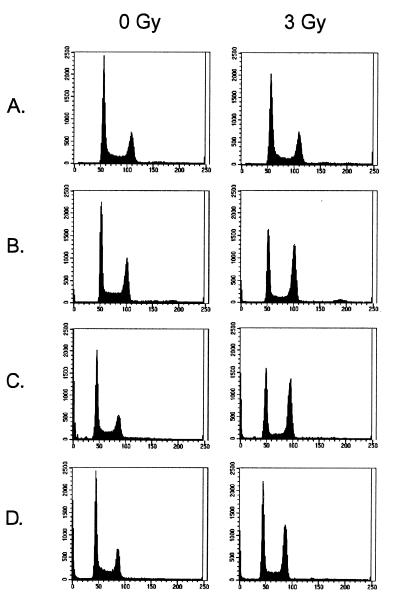
Even in unirradiated cultures, the GMN11 and TAN8 cell lines had plating efficiencies approximately half that of their parental cell lines. Terminal deoxynucleotidyltransferase-mediated UTP end labeling assays on these cell lines failed to reveal significant numbers of apoptotic cells, suggesting that the poor growth of these cultures more likely reflected impaired cell cycle progression rather than activation of programmed cell death. The cell cycle distribution of GMN11 and TAN8 cells and the parental cell lines, unirradiated and 24 h after exposure to 3 Gy of x-rays, was examined by propidium iodide staining for DNA content (Fig. 4). AT cell lines can be discriminated reliably from normal in this assay by a distinctive increase in the number of irradiated cells arrested in G2/M compared with G1, reflecting the inability of AT cells to activate cell cycle checkpoints (20). Even in the unirradiated state, GMN11 cells had a reduced number of cells in G1 compared with the parental GM637 line. There was no significant subgenomic peak that might reflect ongoing programmed cell death in the culture. After irradiation, GMN11 cells displayed a further increase in the frequency of cells in G2/M compared with G1. After irradiation, 42% of GMN11 cells were in G2/M and 31% were in G1 compared with the parental cell line in which 22.7% of cells were in G2 and 46% were in G1. The response of GMN11 cells was comparable to that observed with AT cell lines such as AT3BI. A second cloned ATR.KD transfectant line, GMN15, had a similar response but also displayed a specific accumulation tetraploid cells (8.8%) after irradiation (data not shown). TAN8 cells displayed a typical AT phenotype that could not be discriminated from the parental AT3BI cells.
Figure 4.
Effect of ionizing radiation on cell cycle distribution of ATR.KD-transfected cell lines. Cell lines, unirradiated and irradiated with 3 Gy, were stained with propidium iodide and analyzed for DNA content by FACS. (A) GM637. (B) GMN11. (C) AT3BI. (D) TAN8.
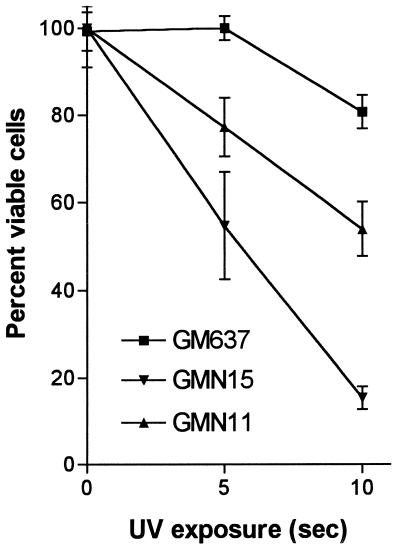
GMN11 and GMN15 cells were tested for sensitivity to UV irradiation, another characteristic phenotype of rad3 mutants in S. pombe. Both cell lines displayed significantly reduced colony-forming ability after exposure to UV compared with the untransfected parental cell line GM637 (Fig. 5).
Figure 5.
Effect of UV radiation on colony-forming ability of ATR.KD-transfected cell lines. Cells were exposed to 310 nM UV for indicated period of time and plated in quadruplicate. Colonies were counted after 10–14 days. Colony counts were normalized to those for unirradiated controls.
DISCUSSION
The primary sequence similarity between the S. pombe rad3 and the human ATR gene suggests the possibility that they share some common functions. However, expression of ATR in S. pombe does not complement a rad3 null mutant, indicating that such overlap, if it exists, is far from complete. Furthermore, some DNA damage response functions mediated by Rad3 in fission yeast are controlled, at least in part, by a second, less-related molecule, ATM, in humans. To clarify the role of ATR in DNA damage responses and its relationship to ATM, we have examined the function of ATR in both normal human cells and cells with defects in DNA damage response pathways, specifically cells lacking functional ATM or p53.
Coexpression of recombinant, affinity-tagged versions of human ATR and S. pombe Rad3 in S. pombe results in their heteromultimerization (6). In the current study, examination of ATR in protein extracts from human fibroblasts fractionated by gel filtration indicated that ATR exists predominantly as part of a higher molecular mass complex. Cloned human ATR expressed and affinity-purified from yeast displays a serine–threonine kinase activity. We have generated a mutated form of ATR in which the kinase domain was inactivated by site-specific mutagenesis at the conserved residue D2494. Expression of this mutant form of ATR in all cell lines examined resulted in dramatic losses in viability. Surviving cells had a dominant phenotype of radiation and UV sensitivity, slow growth, and impaired cell cycle checkpoint control in response to radiation, a phenotype comparable to that observed when the analogous mutant form of Rad3 is expressed in wild-type S. pombe. Recently, Cliby et al. (22) have reported similar results by using a doxycycline-inducible kinase-dead ATR construct. The combination of biochemical evidence from chromatography of ATR-containing complexes and genetic evidence from the expression of a kinase-dead form of ATR strongly suggests that (i) some of the functions mediated by ATR require that it have an intrinsic protein kinase activity, (ii) the active form of ATR exists as part of a higher molecular mass complex, (iii) this complex plays a role in mediating responses to cellular damage induced by either UV or ionizing radiation, and (iv) maintenance of this ATR-containing complex may be essential for cell viability. It remains to be determined whether the active form of the ATR-containing complex is a homomultimer or, more likely, a complex including additional, unrelated subunits such as human isoforms of S. pombe Rad1+, Rad17+, Rad9+, and Hus1+ (3).
A comparison of the effects of ATR.KD expression in normal cells and cells with defects in damage response pathways provided additional insights into the role of ATR in human cells. Expression of the kinase-dead form of ATR in normal and ATM-deficient cells resulted in significant cell death in transient assays that was even more pronounced after drug selection. The drastic reduction in colony-forming ability resulting from ATR.KD expression was not dependent on the presence of functional p53 because no viable colonies were obtained when ATR.KD was transfected into p53 mutant C-33A cells. Indeed, because the absolute numbers of colonies obtained in transfections with vector alone were more than 50-fold higher in C-33A cells than in GM637 cells, the cell-killing effect of ATR.KD actually may be enhanced in the absence of functional p53.
The relationship of ATR and ATM in human cells is of particular interest given the reported synergistic effects of mec1/tel1 double mutants on radiosensitivity in S. cerevisiae (17). In the current study, normal fibroblasts expressing ATR.KD (GMN11) had both reduced survival and a loss of cell cycle checkpoint control after irradiation, resulting in a phenotype quite similar to that of AT cells. However, expression of ATR.KD in AT cells did not significantly enhance either their radiosensitivity or radiation-induced loss of cell cycle checkpoint control. The similarity of the cellular phenotype in AT cells, ATR.KD-expressing normal cells, and ATR.KD-expressing AT cells, including poor growth kinetics, radiation sensitivity, and loss of G1 checkpoint control in response to ionizing radiation, suggests a possible linear pathway relationship with ATR located downstream of ATM. Consistent with this hypothesis, we have observed that overexpression of a normal ATR gene by transfection results in a 2- to 3-fold increase in the normally poor colony-forming ability of AT cells (J.W., unpublished work). Furthermore, the degree of phosphorylation and the protein kinase activity of atr is reduced signficantly in testes from atm−/− mice (K.S.K. and M.F.H., unpublished work). However, expression of a kinase-dead form of ATR also results in increased UV sensitivity, a phenotype not observed in AT cells. This increased sensitivity to UV may be mediated, in part, through other proteins that interact with ATR as part of a multiprotein complex, consistent with our detection of ATR in higher molecular mass complexes in human cells. ATM is unlikely to be part of such a complex because similar results were obtained in gel filtration experiments in normal and AT cells. Thus, ATR and ATM null cells with common genetic backgrounds will be needed to clarify further their, likely complex, functional relationship.
Acknowledgments
We thank Dr. Kevin Shaw for technical assistance with the chromatography experiments. This work was supported, in part, by a grant from the National Institutes of Health (CA57569) to P.C. and by ICOS Corporation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: AT, ataxia-telangiectasia.
References
- 1.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Carr A M, Hoekstra M F. Trends Cell Biol. 1995;5:32–40. doi: 10.1016/s0962-8924(00)88934-5. [DOI] [PubMed] [Google Scholar]
- 3.Carr A M. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- 4.Jiminez G, Yucel J, Rowley R, Subramani S. Proc Natl Acad Sci USA. 1992;87:4952–4956. doi: 10.1073/pnas.89.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seaton B L, Yucel J, Sunnerhagen P, Subramani S. Gene. 1992;119:83–89. doi: 10.1016/0378-1119(92)90069-2. [DOI] [PubMed] [Google Scholar]
- 6.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 7.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 8.Kato R, Ogawa H. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulovich A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 10.Keith C T, Schreiber S L. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 11.Cimprich K A, Shin T B, Keith C T, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keegan K S, Holtzman D A, Plug A W, Christenson E R, Brainerd E E, Flaggs G, Bentley N J, Taylor E M, Meyn M S, Moss S B, et al. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 13.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 14.Gatti R A, Boder E, Vinters H V, Sparkes R S, Norman A, Lange K. Medicine. 1991;70:99–117. [PubMed] [Google Scholar]
- 15.Meyn M S. Cancer Res. 1996;55:5991–6001. [PubMed] [Google Scholar]
- 16.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 17.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 18.Tjoelker L W, Seyfried C E, Eddy R L, Jr, Byers M G, Shows T B, Calderon J, Schreiber R B, Gray P W. Biochemistry. 1994;33:3229–3236. doi: 10.1021/bi00177a013. [DOI] [PubMed] [Google Scholar]
- 19.Murnane J, Fuller L F, Painter R B. Exp Cell Res. 1985;158:119–126. doi: 10.1016/0014-4827(85)90437-9. [DOI] [PubMed] [Google Scholar]
- 20.Beamish H, Lavin M F. Int J Radiat Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- 21.Crook T, Wrede D, Vousden K H. Oncogene. 1991;6:873–875. [PubMed] [Google Scholar]
- 22.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]