Abstract
The establishment of dorsal–ventral polarity in the oocyte involves two sets of genes. One set belongs to the gurken-torpedo signaling pathway and affects the development of the egg chorion as well as the polarity of the embryo. The second set of genes affects only the dorsal–ventral polarity of the embryo but not the eggshell. gastrulation defective is one of the earliest acting of this second set of maternally required genes. We have cloned and characterized the gastrulation defective gene and determined that it encodes a protein structurally related to the serine protease superfamily, which also includes the Snake, Easter, and Nudel proteins. These data provide additional support for the involvement of a protease cascade in generating an asymmetric signal (i.e., asymmetric Spätzle activity) during establishment of dorsal–ventral polarity in the Drosophila embryo.
Keywords: dorsal–ventral patterning, protease cascade
Dorsal–ventral polarity in the Drosophila embryo is specified by localized activation of a ligand, Spätzle (SPZ), that ultimately results in a ventral to dorsal gradient of dorsal (DL) protein in the blastoderm nuclei (1, 2). This gradient leads to differential expression of zygotic genes that specify different cell fates along the dorsal–ventral axis of the embryo. The DL nuclear gradient is established by the localized activation of the Toll (TL) signaling pathway (3, 4). Although the TL receptor is distributed uniformly in the plasma membrane, genes upstream of TL act to asymmetrically activate the ligand SPZ (5, 6). At least seven genes are required for correct activation of TL on the ventral side of the egg (7, 8). Loss of function mutations in these genes lead to embryos that completely lack ventral structures and are dorsalized. Among these seven, nudel (ndl), pipe (pip), and windbeutel (wind) are required in the somatic follicle cells that surround the oocyte (7, 9, 10) whereas the four remaining genes, gastrulation defective (gd), snake (snk), easter (ea), and spz, are required in the germ line (7, 11, 12). EA and SNK both share structural homology with extracellular, trypsin-like, serine proteases (8, 13–16). Epistasis studies suggest a pathway in which ndl and gd act immediately upstream of snk, ea, spz, and tl (15, 16). The NDL protein encodes an extracellular matrix-like molecule that may be anchored in the vitelline membrane (10). The wind gene encodes a protein with motifs similar to a vertebrate endoplasmic reticulum protein of unknown function (17). The pip gene has not been characterized molecularly.
GD is critical for generating the asymmetric presentation of SPZ. Injection of perivitelline (PV) fluid from embryos lacking both maternal and zygotic DL protein can rescue snk and ea embryos but cannot rescue gd embryos, indicating that GD activity is not present in the fluid whereas SNK and EA activities are (8). Ventral cuticle elements can be rescued in gd mutants as well as in snk and ea embryos by injection of fluid containing SPZ. However, rescue of gd embryos is unique in that the polarity of the rescued embryos depends on the site of injection. This pliability of gd embryo polarity, the insolubility of the GD protein, and the temperature-sensitive period of gd function (12) suggest that gd may be the first of the maternally required genes, acting in concert with the somatically expressed genes (ndl, pip, wind) to cause the localized activation of SPZ (8, 10, 18, 19).
Here, we report the cloning and characterization of the gd gene. We show that GD displays significant structural similarity to trypsin-like serine proteases although there are some important differences. We propose that GD acts early to establish a localized complex involving other putative proteases EA, SNK, and NDL that leads to localized activation of SPZ.
MATERIALS AND METHODS
Drosophila Stocks and Mapping.
Mutations used are described in refs. 12, 20–22. Map positions were assigned based on the following: 97 of 129 recombinants between miniature (m) and furrowed (fw) occurred distal to l(1)13 (cacophony) placing cac at 36.66; 5 of 8 recombinants between l(1)13 and fw were distal to gd4, placing gd at 36.78; 2 of 10 gd/fw recombinants were distal to twisted gastrulation (tsg), placing tsg at 36.79. Thus, gd is located proximal to cac and distal but very close to tsg.
Molecular Procedures.
Genomic DNA libraries (23, 24) were probed with EcoRI fragments isolated from a microdissection library (prepared by A.P.M., unpublished work) in the European Molecular Biology Laboratory 4 phage vector. Phage were purified and fragments subcloned for further walking by using standard methods (25). A plasmid cDNA library from 0- to 4-h-old embryos (26) was probed with a 1.3-kb HindIII genomic subclone that hybridized to the only RNA in the region that was expressed exclusively in females (≈23.9–25.2; Fig. 1), and the longest was sequenced.
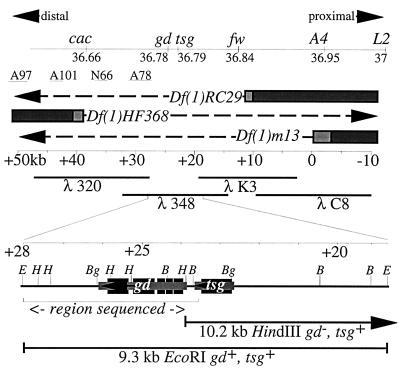
Figure 1.
Correlation of the genetic and molecular maps and the transcripts of gd and tsg. The top line displays the recombination distances in centimorgans between the loci in this region (cf. Materials and Methods). They are depicted according to their orientation along the X chromosome with proximal to the right. The approximate locations of breakpoints for several inversions were determined by Southern blotting as follows: In A97 (51.9/49.3kb), In A101 (44.5/42.5), In N66 (38.1/36.1) and In A78 (32/27.6). The bars represent DNA that is present in the deficiency chromosomes Df(1)HF368, Df(1)m13, and Df(1)RC29. The lightly shaded areas represent the uncertainty of the breakpoints, and the dashes indicate the DNA which is missing. The portion of the chromosomal walk used to identify the gd locus is shown immediately below the breakpoints with the extent of overlapping λ phage clones 320, 348, K3, and C8 indicated in kilobases. The positions of the 9.3-kb EcoRI and 10.2-kb HindIII transformation subclones are shown expanded below the chromosome walk as is the region that was sequenced. The sequenced region abuts other sequenced regions from the tsg gene and beyond. A partial restriction map of the 9.3-kb EcoRI subclone is displayed with E = EcoRI, H = HindIII, Bg = BglII, and B = BamHI. The extent and orientation of the 2.1-kb gd and 1.0-kb tsg transcripts are shown. The smaller lightly shaded blocks indicate the primary transcripts, and the protein coding regions are shown as larger darker blocks.
RNA was prepared from collections of developmental stages observed to be at least 95% free of older stages and extracted as described (27). Poly(A)-containing RNA was prepared by the batch isolation method (25).
Restriction endonuclease digests were performed in TA buffer (28). DNA sequencing was performed as described in the Sequenase kit of United States Biochemical. The entire gd cDNA and 9.4-kb genomic subclone were sequenced on both strands by primer walking. No differences were observed between the genomic and cDNA sequences other than introns. Other routine DNA manipulations were performed according to standard methods (25, 29).
In situ hybridizations were performed by using digoxygenin RNA probes prepared as described by the manufacturer (Boehringer Mannheim). Ovaries were prepared as described by Tautz and Pfeifle (30) by using a final probe concentration of ≈10 ng/μl in a 50-μl volume at 55°C.
RESULTS
Chromosomal Location of gd.
The gd locus fails to complement Df(1)KA10, placing it in the 11A region (12) on the X chromosome. Three other deficiencies Df(1)HF368 (11A2–3;11B9), Df(1)m13 (10C-D;11A3–5), and Df(1)RC29 (11A1–4; 11A4–5) also fail to complement cac, gd, tsg, and fw, placing all four loci between polytene bands 11A2 and 11A5. The relative order of these genes was determined by recombination mapping (cf. Materials and Methods; Fig. 1).
Molecular cloning of gd and transcript identification.
EcoRl inserts from a microdissection library were used to select genomic clones from a Canton S genomic library (23). The 11A breakpoint of In(1)N66 (7A7–8;11A3–5) is sufficiently close to gd to affect its expression (12); therefore, a chromosomal walk (31) was initiated from a genomic clone (λC8) that hybridized in situ both proximal to the In(1)N66 11A breakpoint and distal to the constriction in 11A7,8 (21). A total of 100 kb of overlapping genomic DNA fragments was isolated (21) (Fig. 1). The breakpoints of Df(1)HF368, Df(1)m13, and Df(1)RC29, as well as the location of inversion breakpoints that affected gd expression, were identified by Southern blotting (Fig. 1). This focused attention on a 33-kb region defined by the distal breakpoint of Df(1)HF368 and the proximal breakpoint of Df(1)RC29 (21) (Fig. 1), which corresponds to the cytogenetic location of gd determined above.
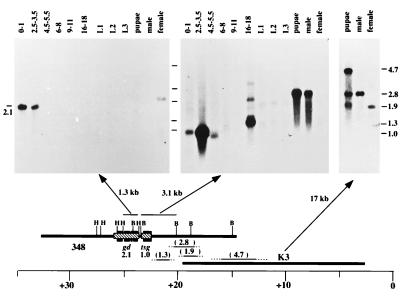
Phage clones spanning this 33-kb region were used to probe developmental Northern blots containing RNA from different stages of the Drosophila life cycle. We identified six transcripts of different sizes (1, 1.3, 1.9, 2.1, 2.8, and 4.7 kb; Fig. 2). Only the 2.1-kb transcript was found exclusively in ovaries and early embryos and thus fit the developmental profile expected for gd (12). To further localize these transcripts, similar RNA blots were probed with subclones across the region of highest transcript density (21). This analysis determined the following transcript order (2.1, 1, 1.3, 2.8, 1.9, and 4.7 kb) and localized the 2.1-kb gd candidate mRNA to a region spanning the 1.3-kb HindIII fragment (24–25.3) (Fig. 2). To determine whether any mutations in the gd locus might lead to restriction fragment length polymorphisms, Southern blots containing HindIII digested DNA from homozygous gd mutant females were probed with the DNA from the λ348 clone. Two gd alleles lacked a 0.6-kb HindIII fragment, which is one of the two restriction fragments that hybridized to the 2.1-kb transcript. In the gd12 mutation, this fragment is absent, whereas in gd6, both the 0.6- and the 1.3-kb fragments are missing and a new fragment appears at ≈1.9 kb in length (not shown). Immediately proximal to the 2.1-kb transcript is a 1.0-kb transcript expressed only in early embryos (Fig. 2). This transcript corresponds to the tsg locus (32).
Figure 2.
Transcript map of the gd region. mRNA from stages throughout the entire life cycle were probed. The results obtained with three probes are shown here (i.e., a 1.3-kb HindIII and 3.1-kb BamHI fragments and the entire K3 phage DNA). The 2.1-kb transcript seen in early embryos (0–3.5 h) and faintly in females is the only transcript in the region with a developmental pattern consistent with the genetics of gd. It is the only transcript detected by the 1.3-kb HindIII fragment. The band in females and embryos is of similar size although this result is distorted here by a curve in the migration front. The 3.1-kb BamHI probe hybridizes to three bands whereas the entire K3 phage hybridizes to the 2.8-, the 1.9-, and a 4.7-kb transcript. Additional probing with other subclones permitted ordering the transcripts as shown (21). The tsg transcript has been identified by transformation (32). The 4.7-kb transcript likely corresponds to the furrowed gene (Corces, V.G., Johns Hopkins Univ., personal communication). The 2.8-kb transcript appears specific to males, and the 1.9-kb transcript is specific to females. From the hybridization pattern, we infer that these are likely alternative splicing variants of a single gene. The 1.3-kb transcript may have a large splicing variant as well. We have not yet identified any genetic lesions that may correspond to these transcription units.
Germ-Line Transformation.
A 9.3-kb EcoRI fragment spanning the putative gd transcript as well as tsg was subcloned into the Casper transformation vector (33) and was used to transform a w1118 fly stock (34, 35). w+ transformant lines were tested for their ability to complement gd and tsg mutations. Normally, gd8 homozygous females produce eggs that do not hatch into larvae, but each independent transformant line caused homozygous gd8 females to produce viable embryos. The percentage of eggs that hatched ranged from 67 to 90%. In addition, this transformation construct also rescued tsg mutations. Thus, all of the essential information for both gd and tsg expression resides in the 9.3-kb EcoRI fragment. A second transformation vector containing a 10.5-kb HindIII fragment that partially overlaps the 9.3-kb EcoRl fragment rescued tsg but not gd (36), thus limiting the extent of the gd locus to ≈4.5-kb (Fig. 1). The only transcript encoded by this region is the 2.1-kb transcript, which was designated as the gd transcript based on the correlation of expression period with genetic analysis, its location within two overlapping transformation constructs, its position just distal to the tsg locus, and two restriction fragment length polymorphisms associated with gd mutants.
Structure of the gd Gene.
Genomic clones spanning the gd region were used to probe a cDNA library (26). The longest cDNA clone isolated is the same length as the mRNA detected on Northern blots (21) and is likely to be nearly full length. Both the cDNA clone and the 9.3-kb EcoRI genomic subclone from which it is derived were sequenced. The cDNA sequence revealed a single long ORF beginning with the first ATG in the sequence. The cDNA contained 30 bp of 5′ untranslated sequence that is located 241 bp downstream from the poly(A) site of the tsg gene. The ORF is followed by 247 bp of 3′ untranslated sequence containing multiple stop codons in all frames and terminates in a poly(A) tail. The only consensus poly(A) addition signal (AATAAA) in the region is located 37 bp before the end of the gd cDNA. Two AACAAA motifs are located further downstream at bp 2626 and 3218, but we have seen no evidence on RNA blots that either of these is used as a polyadenylation site. Comparison of the cDNA and the genomic sequence reveals the presence of four introns. We have found no evidence for alternative splicing.
The gd cDNA sequence predicts a 59-kDa polypeptide. This was confirmed by in vitro translation of transcripts from the cDNA clone, which yielded a protein of ≈60 kDa with a pI of 6.94 as determined by two dimensional PAGE (not shown). The amino-terminal residues are hydrophobic (37), suggesting a secretory leader sequence that would be cleaved after amino acid 23 (38). Because GD activity is not freely diffusible in the PV fluid, the GD protein sequence was examined for potential anchoring sites. Two hydrophobic domains are notable, one near the middle of the protein that could represent a membrane spanning region and the second, a hydrophobic tail, that could act as a membrane anchor. However, each of these domains contains charged amino acid residues. The protein contains three putative N-linked glycosylation sites (NXS/T). There are no RGD amino acid sequences that might interact with extracellular components nor any WIID or LDL repeats, as seen in the NDL protein (10).
gd Encodes a Protein with Similarity to Serine Proteases.
A blast search suggested that GD is related to the family of serine proteases (39). The most closely related proteins are factor IX of the mammalian clotting cascade, two crab coagulation factors, and a urokinase-type plasminogen activator. Sequence alignment of GD with these proteins and chymotrypsin shows that GD shares the 3 amino acids of the catalytic triad (H, D, S) and all but one of the cysteine bridges (Fig. 3). In addition, several other structural features that identify serine proteases (40) are present. However, GD also has some features that are atypical of the basic serine protease family but that are seen in other related proteins. For example, it contains a putative activating cleavage site that is not typical, and the conserved D adjacent to the active site S is replaced by I and a short insert of amino acids immediately adjacent. There is a small acidic region N-terminal to the catalytic site, which is a feature shared by SNK but not EA.
Figure 3.
Alignment of GD with related proteins. (A) Diagram of the gene structure. Region shown in the alignment below is the Ser protease domain with diagonal lines. A hydrophobic secretion signal appears at the N terminus and a hydrophobic tail at the C terminus. Putative N-linked glycosylation sites (NXT/S) are noted by inverted Ys. (B) Each protein is shown beginning with the activating cleavage site (|) and showing the catalytic portion of the protein. Amino acid numbers of the Drosophila protein are shown above the alignment. The putative cleavage site listed for GD is atypical. The H D S residues of the catalytic triad (marked with ∗) are conserved in GD as are most of the Cys bridges with the exception of the penultimate C indicated by (?). A number of other residues are conserved as well. Residues shown in bold correspond to some of the key residues that define distinct structural domains of serine proteases (40). Molecular modeling suggests that the sequence differences seen here could be accommodated into a folded protein that preserved the catalytic site and the substrate groove. The sequences used in this alignment are chicken urokinase P15120, human factor IX clotting enzyme P00740, horseshoe crab proclotting enzyme precursor P21902, crab coagulation factor B A48050, and Bos taurus chymotrypsin National Center for Biotechnology Information sequence ID: 442949.
gd Is Expressed in Both Follicle Cells and the Germ Line.
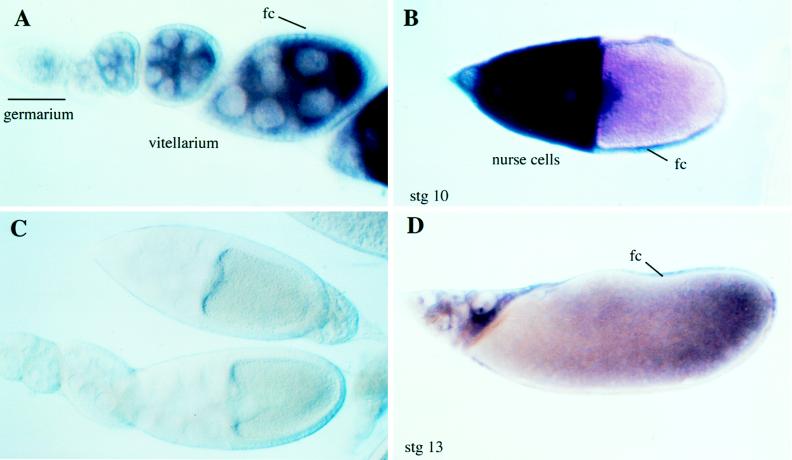
In situ hybridization using antisense riboprobes from the gd locus indicates that expression of gd begins in previtellogenic stages. gd mRNA is seen in the germ line-derived nurse cells of the germarium (Fig. 4A). Expression continues throughout oogenesis with transcripts from the nurse cells accumulating in the oocytes of the vitellarium (Fig. 4A). Of interest, at about stage 10, gd expression can be detected in the surrounding follicle cells, although the level of signal is lower than the intense signal seen in the nurse cells at that stage (Fig. 4B). In some stage 10 oocytes, accumulation of mRNA appears to be somewhat graded along the dorsal–ventral axis with marginally higher levels of mRNA in the ventral follicle cells (Fig. 4B). By stage 13, residual mRNA remains in the shrinking nurse cells and in the follicle cells surrounding the oocyte (Fig. 4D).
Figure 4.
In situ hybridization to gd mRNA in ovaries. (A) gd mRNA first appears in the germarium. Levels continue to build in the nurse cells and oocytes of the vitellarium. Expression also begins to appear in the follicle cells (fc) as the oocytes move into stage 10. (B) Extensive transcription in both nurse cells and in follicle cells is seen in stage 10 oocytes. In oocytes where the D/V axis can be inferred from the position of the oocyte nucleus, a rough ventral to dorsal gradient of transcript often can be detected. (D) By stage 13, residual gd transcripts remain in the nurse cells as well as the surrounding follicle cells. (C) Sense strand control probe shows no detectable background staining.
GD Protein Is Processed.
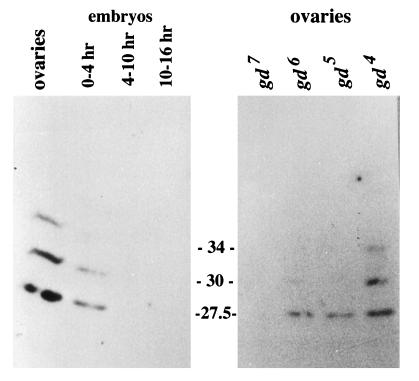
An EcoRV restriction fragment from the gd cDNA (amino acids 347–548) was cloned in-frame into pBS SK and used to express protein. Bacterially derived protein was harvested from inclusion bodies and used as an immunogen in rabbits. Western blots of extracts from ovaries and early embryos revealed three cross reacting bands (34, 30, and 27.5 kDa) that were smaller than the 60-kDa full length protein observed in in vitro translation experiments (Fig. 5). These bands were not detected by the preimmune serum. Furthermore, all three peptides were completely absent in extracts from gd7 females whereas extracts of gd5 and gd6 lacked the two larger bands and gd4 possessed all three. Extracts of carefully staged embryos and ovaries showed that the crossreacting polypeptides are most abundant in the ovaries, that the level of protein decreases from the moment of egg laying, and that it is essentially gone by 4 h (Fig. 5).
Figure 5.
Physical characterization of the GD protein. (A) Western blot of protein extracts from OreR females and staged embryos probed with antisera raised against the GD peptide. The peptide produced by in vitro translation of synthetic gd mRNA is 60 kDa (not shown), and the peptides detected in vivo are smaller. (B) Western blot of protein extracts from ovaries of various gd mutant females suggests that some gd alleles affect the protein detected by this antiserum.
DISCUSSION
There are two sets of genes involved in establishing the dorsal–ventral polarity of the mature oocyte. The first set (reviewed in refs. 41 and 42) includes genes in the gurken-torpedo signaling pathway, and mutations in these genes lead to defective development of the chorion and in most instances to loss of ventrally derived pattern elements. The second set of genes (called the “dorsal group genes”) functions slightly later in development, and mutations in these genes affect only the dorsal–ventral polarity of the embryo but not the eggshell. GD is the earliest acting germ line-dependent member of the second group.
GD as a Secreted Serine Protease.
Three of the dorsal group genes, SNK, EA, and NDL, are related to serine proteases. The work reported here identifies a fourth serine protease-related dorsal group protein, GD. This observation raises the possibility of a signal amplification cascade of sequential protease activation similar to that occurring in blood clotting (reviewed in ref. 43).
The gd protein possesses the catalytic triad (H, D, S) characteristic of serine proteases. Although the homology to serine proteases suggests a proteolytic role, several significant differences between the sequence of GD and other members of the chymotrypsin family of serine proteases suggest that GD may behave atypically. Most eukaryotic serine proteases undergo an activation process after zymogen cleavage in which the α amino group of the catalytic domain forms a salt bridge to an aspartate residue (D) located adjacent to the active site serine (44). In GD, this aspartic acid residue is replaced by isoleucine, making it unlikely that the protease could be activated via the classical mechanism. However, several bacterial members of the α-lytic endopeptidase family (e.g., subtilisin) contain residues other than D (e.g., T) adjacent to the active serine (45), and complement component C2 contains E instead of D (46). Typically, activation involves cleavage after a positively charged amino acid (R or K) followed by two branch chain amino acids and a highly conserved G. The N-terminal residue is most commonly I but may be L, V, or M (45, 47). One putative cleavage site for GD based on position relative to conserved motifs (e.g., G-WPW and CGGTSLV) is S ITRG. Although this putative site has S instead of R or K and has G in the 4th position rather than the 3rd, cleavage at this site would produce a peptide of 30.5 kDa that could possibly correspond to the 30-kDa species observed on the Western blots. The extensive heteroallelic complementation exhibited by gd is consistent with the possibility that the GD protein may function as a multimer (12) perhaps during an activation process.
There is precedence for unusual activating cleavage sites in other proteases. The plasminogen activator of the vampire bat is activated by cleavage between a histidine followed by serine (i.e., H∧STGG) (48). The crab coagulation factor is cleaved at the sequence I∧IAGG (Fig. 3). Complement factors C2 and B are cleaved by a novel mechanism between R and K (49). GD has a sequence motif beginning at amino acid 136 (i.e., R∧KLSFIPDKKSSLLLD… ) that resembles the complement B cleavage region (underlined residues). However, cleavage at this R K site would produce a 43-kDa protein, which we do not observe on Western blots. Additional processing events that may further reduce the size make the potential use of this site difficult to analyze. An alternative is that GD is not activated by cleavage. The rhesus monkey apolipoprotein is inactive as a protease, lacking the active site S. Human apolipoprotein contains the complete H-D-S triad, but activation apparently would require cleavage of an S-I bond. Thus far, no proteolytic activity associated with human apolipoprotein has been reported. Other examples of inactive serine proteases containing a complete triad include mouse α-NGF (50) and bovine procarboxypeptidase A subunit in which the lack of proteolytic activity has been attributed to the absence of two N-terminal hydrophobic residues (45, 47). Thus, one possibility is that GD cannot undergo an activating cleavage and consequently does not show proteolytic activity, perhaps like α-NGF or procarboxypeptidase. An alternative possibility is that GD is indeed proteolytically active but undergoes an atypical activation event and apparently other processing events, yielding three different polypeptides. In either case, it appears unlikely that GD is activated by a standard mechanism.
GD Is Central to Establishing Asymmetric Signaling of Toll.
The dorsal–ventral asymmetry of the embryo is established by a signal that is transmitted through the uniformly distributed TL receptor, resulting in a graded relocation of DL into the nuclei of the blastoderm embryo (3, 4). Injection experiments involving the use of dominant active EA (15) and SNK (16), as well as injection of PV from dl embryos into gd mutant embryos, lead to production of ventral elements at the site of injection rather than in the normal ventral region (8). These data suggest that D/V polarity is established by asymmetric presentation of the TL ligand to the oocyte. PV fluid from dl embryos (thought to be depleted of SPZ ligand because of the presence of the TL receptor) can rescue D/V polarity in snk and ea embryos. This same PV fluid cannot restore normal ventral structures to either gd or ndl embryos. In contrast, injection of PV fluid from Tl mutant embryos (thought to contain active SPZ ligand) into gd embryos produces ventral structures at the site of injection (8). The same fluid injected into snk or ea embryos produces embryos with normal polarity independent of the site of injection (8). This result strongly suggests that GD is a key component required for establishing the localized activation of SPZ and thus the asymmetric activation of TL. Because the temperature-sensitive period for both NDL and GD action includes a period before fertilization when the initial D/V asymmetry is known to be established (12), it is possible that these two gene products cooperate to form a localized anchor for a SPZ activating complex.
EA and SNK both share significant structural homology with extracellular trypsin-like serine proteases (13, 14). Experiments using dominant active forms of SNK and EA show that SNK activates EA, which in turn activates the SPZ ligand. In combination with the somatically expressed genes, wind, pip, and ndl, GD activates SNK in a location-dependent manner that marks the future ventral cells (15, 51). The recent cloning of ndl indicates that NDL is a large (≈350 kDa) extracellular glyco-protein with motifs suggesting that it might bind to extracellular matrix as well as to other proteins. NDL also contains a serine protease catalytic domain (10). The occurrence of several protease-like proteins both upstream (NDL) and downstream (SNK; EA) of GD may be responsible for the multiply processed GD peptides observed on Western blots. The physical location of the NDL protein (including whether it is incorporated as a component of the vitelline membrane) is not yet known, although the fragility of the ndl embryos suggests that NDL may be required for stability of the vitelline membrane (10). gd mRNA is expressed in follicle cells beginning at about stage 10 and may be graded in a ventral to dorsal manner, although it is expressed uniformly in the nurse cell/oocyte complex. The location of GD remains to be determined.
Four of the five currently identified genes needed for asymmetric activation of the SPZ ligand encode secreted members of the serine protease superfamily. We propose that GD functions as part of an anchored complex that triggers a proteolytic activation hierarchy involving NDL, SNK, and EA, resulting in localized activation of SPZ ligand and asymmetry along the D/V axis.
Acknowledgments
We fondly acknowledge the work and assistance of the late George Lefevre whose extensive genetic characterization of this region provided the foundation for this study. We are indebted to R. DeLotto for sharing his unpublished results and for fruitful discussions, to R. Bradshaw for informative discussions about proteases, and to H. Theisen and K. Arora for comments on the manuscript. We gratefully acknowledge the support of National Science Foundation Grants DC8615701 to K.D.K. and J.L.M. and DCB8904047 to K.D.K. and National Institutes of Health Grants GM28972 and PO1 HD27173 to J.L.M. and HD17607 to A.P.M. A.P.M. acknowledges the assistance of Dr. Jan-Erik Edström and the European Molecular Biology Organization course at the European Molecular Biology Laboratory where the microdissection of region 11A was accomplished. We gratefully acknowledge the computational support of GenBank, the University of California Irvine office of academic computing, and the resources of the National Drosophila Stock Center in Bloomington, IN.
ABBREVIATIONS
- SPZ
Spätzle
- DL
dorsal
- TL
Toll
- NDL
nudel
- gd
gastrulation defective
- snk
snake
- ea
easter
- tsg
twisted gastrulation
- PV
perivitelline
Footnotes
References
- 1.Morisato D, Anderson K V. Annu Rev Genet. 1995;29:371–399. doi: 10.1146/annurev.ge.29.120195.002103. [DOI] [PubMed] [Google Scholar]
- 2.Ray R P, Schupbach T. Genes Dev. 1996;10:1711–1723. doi: 10.1101/gad.10.14.1711. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto C, Hudson K L, Anderson K V. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto C, Gerttula S, Anderson K V. Development (Cambridge, UK) 1991;111:1021–1028. doi: 10.1242/dev.111.4.1021. [DOI] [PubMed] [Google Scholar]
- 5.Morisato D, Anderson K V. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-x. [DOI] [PubMed] [Google Scholar]
- 6.Schneider D S, Jin Y, Morisato D, Anderson K V. Development (Cambridge, UK) 1994;120:1243–1250. doi: 10.1242/dev.120.5.1243. [DOI] [PubMed] [Google Scholar]
- 7.Stein D, Roth S, Vogelsang E, Nüsslein-Volhard C. Cell. 1991;65:725–735. doi: 10.1016/0092-8674(91)90381-8. [DOI] [PubMed] [Google Scholar]
- 8.Stein D, Nüsslein-Volhard C. Cell. 1992;68:429–440. doi: 10.1016/0092-8674(92)90181-b. [DOI] [PubMed] [Google Scholar]
- 9.Manseau L J, Schupbach T. Genes Dev. 1989;3:1437–1452. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- 10.Hong C C, Hashimoto C. Cell. 1995;82:785–794. doi: 10.1016/0092-8674(95)90475-1. [DOI] [PubMed] [Google Scholar]
- 11.Seifert E, Muller-Holtkamp F, Marcey D, Jäckle H. Roux’s Arch Dev Biol. 1987;196:78–82. doi: 10.1007/BF00402028. [DOI] [PubMed] [Google Scholar]
- 12.Konrad K D, Goralski T J, Mahowald A P. Dev Biology. 1988;127:133–142. doi: 10.1016/0012-1606(88)90195-9. [DOI] [PubMed] [Google Scholar]
- 13.DeLotto R, Spierer P. Nature (London) 1986;323:688–692. doi: 10.1038/323688a0. [DOI] [PubMed] [Google Scholar]
- 14.Chasan R, Anderson K V. Cell. 1989;56:391–400. doi: 10.1016/0092-8674(89)90242-0. [DOI] [PubMed] [Google Scholar]
- 15.Chasan R, Jin Y, Anderson K V. Development (Cambridge, UK) 1992;115:607–616. doi: 10.1242/dev.115.2.607. [DOI] [PubMed] [Google Scholar]
- 16.Smith C L, DeLotto R. Nature (London) 1994;368:548–551. doi: 10.1038/368548a0. [DOI] [PubMed] [Google Scholar]
- 17.Konsolaki M, Schupbach T. Genes Dev. 1998;12:120–131. doi: 10.1101/gad.12.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson K V, Nüsslein-Volhard C. Gametogenesis and the Early Embryo. New York: Liss; 1986. pp. 177–194. [Google Scholar]
- 19.Schüpbach T, Wieschaus E. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefevre G., Jr Genetics. 1971;67:497–513. doi: 10.1093/genetics/67.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goralski T J. Ph.D. Thesis. Bloomington, IN: Indiana University; 1985. [Google Scholar]
- 22.Lindsley D L, Zimm G G. The Genome of Drosphila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 23.Maniatis T, Hardison R C, Lacy E, Lauer J, O’Connell C, Quon D, Sim D K, Efstratiadis A. Cell. 1978;15:687–699. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 24.Scott M P, Weiner A J, Hazelrigg T I, Polisky B A, Pirrotta V, Scalenghe F, Kaufman T C. Cell. 1983;35:763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 26.Brown N H, Kafatos F C. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 27.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 28.O’Farrell P H, Kutter E, Nakanishi M. Mol Gen Genet. 1980;179:421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 30.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 31.Bender W, Spierer P, Hogness D S. J Mol Biol. 1983;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- 32.Mason E D, Konrad K D, Webb C D, Marsh J L. Genes Dev. 1994;8:1489–1501. doi: 10.1101/gad.8.13.1489. [DOI] [PubMed] [Google Scholar]
- 33.Pirrotta V. In: Vectors: A Survey of Molecular Cloning Vectors and Their Uses. Rodriguez R L, Denhart D T, editors. London: Butterworths; 1988. pp. 437–466. [Google Scholar]
- 34.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 35.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni S J, Hall J C. Genetics. 1987;115:461–475. doi: 10.1093/genetics/115.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.von Heinje G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Greer J. Proteins Struct Funct Genet. 1990;77:317–334. [Google Scholar]
- 41.Lehmann R. Cell. 1995;83:353–356. doi: 10.1016/0092-8674(95)90111-6. [DOI] [PubMed] [Google Scholar]
- 42.Morgan M M, Mahowald A P. Arch Insect Biochem Physiol. 1996;33:211–230. doi: 10.1002/(SICI)1520-6327(1996)33:3/4<211::AID-ARCH4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 43.Hecht P M, Anderson K V. Trends Cell Biol. 1992;2:197–202. doi: 10.1016/0962-8924(92)90246-j. [DOI] [PubMed] [Google Scholar]
- 44.Bode W, Huber R. FEBS Lett. 1978;90:265–269. doi: 10.1016/0014-5793(78)80382-2. [DOI] [PubMed] [Google Scholar]
- 45.Rawlings N D, Barrett A J. Methods Enzymol. 1994;244:19–61. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mole J E, Anderson J K, Davison E A, Woods D E. J Biol Chem. 1984;259:3407–3412. [PubMed] [Google Scholar]
- 47.Rawlings N D, Barrett A J. Methods Enzymol. 1994;244:205–218. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kratzschmar J, Haendler B, Langer G, Boidol W, Bringmann P, Alagon A, Donner P, Schleuning W D. Gene. 1991;105:229–237. doi: 10.1016/0378-1119(91)90155-5. [DOI] [PubMed] [Google Scholar]
- 49.Kjalke M, Welinder K G, Koch C. J Immunol. 1993;151:4147–4152. [PubMed] [Google Scholar]
- 50.Isackson P J, Ullrich A, Bradshaw R A. Biochemistry. 1984;23:5997–6002. doi: 10.1021/bi00320a015. [DOI] [PubMed] [Google Scholar]
- 51.Smith C, Giordano H, DeLotto R. Genetics. 1994;136:1355–1365. doi: 10.1093/genetics/136.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]