Abstract
The ubiquitously expressed nonreceptor tyrosine kinase c-Abl contains three nuclear localization signals, however, it is found in both the nucleus and the cytoplasm of proliferating fibroblasts. A rapid and transient loss of c-Abl from the nucleus is observed upon the initial adhesion of fibroblasts onto a fibronectin matrix, suggesting the possibility of nuclear export [Lewis, J., Baskaran, R., Taagepera, S., Schwartz, M. & Wang, J. (1996) Proc. Natl. Acad. Sci. USA 93, 15174–15179]. Here we show that the C terminus of c-Abl does indeed contain a functional nuclear export signal (NES) with the characteristic leucine-rich motif. The c-Abl NES can functionally complement an NES-defective HIV Rev protein (RevΔ3NI) and can mediate the nuclear export of glutathione-S-transferase. The c-Abl NES function is sensitive to the nuclear export inhibitor leptomycin B. Mutation of a single leucine (L1064A) in the c-Abl NES abrogates export function. The NES-mutated c-Abl, termed c-Abl NES(−), is localized exclusively to the nucleus. Treatment of cells with leptomycin B also leads to the nuclear accumulation of wild-type c-Abl protein. The c-Abl NES(−) is not lost from the nucleus when detached fibroblasts are replated onto fibronectin matrix. Taken together, these results demonstrate that c-Abl shuttles continuously between the nucleus and the cytoplasm and that the rate of nuclear import and export can be modulated by the adherence status of fibroblastic cells.
Keywords: adhesion, integrins, nuclear export signal
Protein tyrosine kinases play important roles in the transduction of extracellular signals. A majority of the known protein tyrosine kinases are localized at or near the plasma membranes, e.g., the receptor-tyrosine kinases or the cytoplasmic tyrosine kinases, Src, JAK, and FAK. The c-Abl tyrosine kinase is unusual in that it is also found in the nucleus (2). The nuclear localization of c-Abl is driven by three nuclear localization signals (NLS) (3). Mutation of all three NLS is required to inhibit the nuclear import of this tyrosine kinase.
Nuclear c-Abl has been implicated in the regulation of gene expression. The c-Abl protein contains three high mobility group-like domains that bind to A+T-rich DNA in a cooperative manner (4). While c-Abl does not select DNA sequences, it can be recruited to specific DNA-binding complexes through protein-protein interactions. For example, c-Abl is shown to interact with the transcription factor RXF1, which binds to the palindromic EP sequence in the hepatitis virus B enhancer (5). Through a direct interaction with the retinoblastoma protein (RB), c-Abl can also be recruited to an E2F-DNA binding complex, both in vitro and in vivo (6). Binding of c-Abl by RB results in the inhibition of c-Abl kinase activity (7). This c-Abl/RB interaction is disrupted at the G1/S transition by the cell cycle-regulated phosphorylation of RB (7), allowing the nuclear c-Abl kinase to become activated as cells commit to S phase (7, 8). In S phase cells, nuclear c-Abl activity is further increased when cells are exposed to DNA damaging agents such as methymelthane sulfonate and ionizing radiation (9). The ionizing radiation-induced activation of c-Abl requires a functional ataxia telangiectasia mutated-kinase, encoded by the gene mutated in the human disease Ataxia telangiectasia (10). The activated nuclear c-Abl tyrosine kinase phosphorylates the C-terminal repeated domain of RNA polymerase II (11–13). Increased tyrosine phosphorylation of RNA polymerase II is observed upon treatment of cells with methymethane sulfonate or ionizing radiation, and this increase in phosphorylation is dependent on c-Abl as well as ataxia telangiectasia mutated (9, 14). Tyrosine phosphorylation of the C-terminal repeated domain can be correlated with increased transcription from several different promoters (13). Taken together, these observations suggest that nuclear c-Abl may participate in the regulation of cell cycle-dependent and DNA damage-induced gene expression.
Although c-Abl contains three NLS, it is not exclusively localized to the nucleus (2, 15). The cytoplasmic pool of c-Abl is not regulated by the cell cycle progression, as RB is nuclear (7, 8). The cytoplasmic c-Abl does associate with F-actin (15, 16). An F-actin binding consensus sequence has been identified at the C terminus of c-Abl, and this sequence has been shown to function in the binding of c-Abl to F-actin (16). The c-Abl protein also contains a G-actin binding site (17). Several cytoplasmic substrates of c-Abl have been identified. These include the SH2/SH3 adaptor protein Crk (18, 19) and the Crk binding protein p130cas (20). Tyrosine phosphorylation of the p130cas protein is dependent on cellular adhesion to the extracellular matrix (ECM) (21). Interestingly, adhesion to the ECM also regulates the c-Abl tyrosine kinase activity (1).
In fibroblasts, detachment from the ECM leads to a loss of c-Abl tyrosine kinase activity, which can be re-activated upon adhesion to fibronectin matrix (1). In addition to the regulation of kinase activity, adhesion to the ECM also affects the subcellular localization of c-Abl. In attached or detached cells, c-Abl is detected in both the cytoplasm and the nucleus, as determined by immunostaining and cell fractionation. When detached cells are replated onto a fibronectin matrix, a transient loss of c-Abl from the nucleus is observed during the first 20 minutes of replating, followed by a rapid reappearance of c-Abl in the nucleus (1). When cells are plated onto poly-l-lysine, which does not stimulate integrin receptors, this effect on the c-Abl localization is not observed. The transient loss of nuclear c-Abl can be explained by two possible mechanisms: either the nuclear c-Abl is rapidly degraded upon reattachment to the ECM, or the nuclear c-Abl is rapidly exported from the nucleus.
The export of macromolecules from the nucleus is an active process. To date, three types of nuclear export signals (NES) in the form of primary amino acid sequences have been identified (22, 23). Among them is the leucine-rich NES first identified in the cellular protein, protein kinase A inhibitor (PKI) (24) and the HIV protein Rev (25). This leucine-rich NES has since been found in other cellular proteins including MEK (26), FxMR1 (27), and zyxin (28). The leucine-rich NES-mediated export is energy dependent and saturable, indicating that specific receptors recognize NES signals and mediate the active export of NES-containing proteins to the cytoplasm. A putative NES-receptor has recently been identified as the CRM1, or exportin-1, protein (29, 30, 40, 41).
In this report, we present evidence that c-Abl does indeed contain a functional NES. We show that c-Abl is exported from the nucleus in response to attachment of cells to the ECM. We also show that c-Abl is continuously shuttling between the nucleus and the cytoplasm. Thus, the subcellular localization of c-Abl is determined by a balance of nuclear import and export, and the dynamic equilibrium between these two processes may determine the biological effects of the c-Abl tyrosine kinase.
MATERIALS AND METHODS
DNA Constructs.
The chloramphenicol acetyltransferase (CAT) reporter plasmid pDM128 has been described (31, 32). The CAT gene is placed within the first intron of the HIV genome next to the RRE sequence. Export of intronic RNA is essential to the expression of CAT activity. The NES-deficient RevΔ3NI construct was described (33). The RevΔ3NI fusions with Abl sequences were generated by PCR-based recombinant DNA methods. The c-Abl NLS(−) was described in (3). The lysine and arginine residues in the three NLS were substituted with glutamine and the c-Abl NLS(−) mutant was shown to be exclusively cytoplasmic (3). The c-Abl NES(−) mutant was constructed by a two-step PCR mutagenesis strategy. The L1064 was changed to alanine through the use of mutagenic primers. Two overlapping PCR fragments spanning the L1064A mutation were combined by a final PCR reaction. The resulting PCR product was fully sequenced and then subcloned into the wild-type (wt) murine c-Abl IV. The c-Abl wt, c-Abl NLS(−), and c-Abl NES(−) were expressed with a retroviral vector pMSCVhyg (34).
Rev Complementation Assay.
293 cells were grown at 37°C in DMEM supplemented with 10% FCS and grown in 10% CO2 293 cells were transfected according to standard protocol for CaPO4-mediated transfection of adherent cells. Assays were done in triplicate in six-well plates. Briefly, a total of 2 μg plasmid DNA in 44 μl of 0.1× TE buffer (1× TE, 10 mM Tris⋅HCl/0.1 mM EDTA) was mixed with 50 μl of 2× HBS buffer (50 mM Hepes, pH 7.05/10 mM KCl/12 mM dextrose/280 mM NaCl/1.5 mM Na2HPO4). CaCl2 (6.8 μl of 2 M) was added dropwise while vortexing the mixture. The mixture was incubated at room temperature for 30 min prior to application to the cells. Approximately 48 hr after transfection the cells were harvested using calcium- and magnesium-free PBS containing 5 mM EDTA. Harvested cells were used for the quantitative β-galactosidase assay and CAT assay. Each transfection typically contained 1 μg of reporter pDM128 DNA, 0.2 μg transactivator DNA, 0.2 μg pCH110 (β-galactosidase internal control) and pUC118 to a total of 2 μg. CAT assays were performed as described (35). All extracts were normalized for β-galactosidase activity.
Microinjection of NIH 3T3 Cells.
One day prior to injection NIH 3T3 cells were plated on 1.0-cm coverslips at 50% confluency. The coverslips were placed in a 3-cm dish containing CO2-deficient medium (GIBCO). LMB-treated cells were cultured with 20 nM LMB for 6 hr before and during injection. Glutathione S-transferase (GST)-Abl NES was made by fusing GST to the c-Abl fragment 1052-EAINKLESNLRELQICPAT-1070. Approximately 50 fl of a mixture of GST-AblNES (≈2 mg/ml) and rhodamine-dextran (Sigma, 1.5 mg/ml) was injected into the nuclei of NIH 3T3 cells using an Eppendorf microinjection system. During injections, cells were incubated at 37°C utilizing a heated stage. After 30 min of incubation at 37°C in an incubator without CO2 the cells were fixed with 4% paraformaldehyde in PBS. GST-AblNES was analyzed by indirect immunofluorescence using a monoclonal anti-GST antibody (Santa Cruz Biotechnology) and confocal microscopy.
Reconstitution of Abl-Deficient 3T3 Cells with c-Abl Localization Mutants.
Bosc23 cells were transfected with the murine stem cell virus (MSCV) c-Abl wt, MSCV c-Abl NLS(−), and MSCV c-Abl NES(−) plasmids using the HBS calcium phosphate method in the presence of 25 μM chloroquine (36). Approximately 48 hr after transfection an infection mixture was made using the retroviral supernatant, fresh DMEM and polybrene (4 μg/ml) and used to infect Abl-deficient 3T3 cells. Stable transfectants were selected for 10 days in media containing 200 μg/ml hygromycin.
Immunofluorescence.
Cells were plated onto coverslips overnight. LMB-treated cells were cultured with 5 nM LMB for 6 hr prior to fixation. Cells were washed 1× PBS and fixed with 3% formaldehyde in Pipes buffer (PB; 100 mM Pipes, pH 6.8/1 mM MgCl2/1 mM EGTA). Cells were permeabilized with 0.5% Nonidet P-40 in PBS and blocked with 10% normal goat serum in PBS. Primary and secondary antibody incubations were for 90 min at room temperature, and antibodies were diluted into the blocking solutions. The antibodies used to visualize c-Abl were the mouse anti-c-Abl, 8E9 (used at 30 μg/ml) and the fluorescein-conjugated sheep anti-mouse secondary antibody (ICN; used at 20 μg/ml). Cells were counterstained with rhodamine-conjugated phalloidin (Molecular Probes) at 0.4 units/ml to label the actin cytoskeleton. For confocal analysis, immunofluorescent samples were scanned with a Bio-Rad MRC 600 laser confocal microscope equipped with a Zeiss 40× objective.
Fibronectin (FN) Attachment Assay.
The FN attachment assays were performed as described (1). Briefly, glass coverslips were coated with FN at 25 μg/ml in PBS for 1 hr at 37°C. Cells were detached with trypsin, washed in defined minimal media (DMEM/Hans F12 medium/0.075% bovine serum albumin/50 μg/liter insulin/50 μg/liter transfernin/0.05 μg/liter sodium selenite/30 μg/ml histidine/0.02μg/mL Mn C12) containing soybean trypsin inhibitor at 250 μg/ml, and plated onto the FN-coated coverslips in defined minimal media containing trypsin inhibitor. At the appropriate timepoints, cells were washed once with PBS and prepared for immunofluorescence as described above.
RESULTS
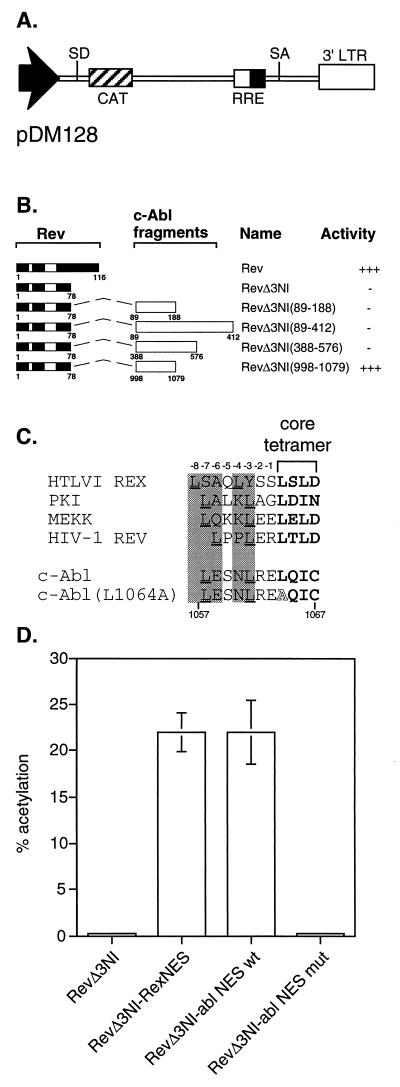
To determine if a functional NES was present in c-Abl, we utilized a complementation assay based on the HIV Rev protein. Rev requires a NES to perform its function of exporting unspliced RNA out of the nucleus. We have previously shown that the function of a NES-mutated Rev can be restored by the fusion of a heterologous NES (32, 37). A related complementation assay has been utilized to identify the NES in the transcription factor TFIIIA (38) and the Fragile-X Mental Retardation Protein (FxMR1) (27). If c-Abl did contain a functional NES, we would be able to demonstrate this by fusing a c-Abl sequence to the NES-mutated Rev (RevΔ3NI). The pDM128 reporter, which contains an intron-disrupted CAT gene (31), was used to assay for Rev function (Fig. 1A). A series of fragments containing c-Abl sequences were fused to the C terminus of RevΔ3NI and tested for their ability to functionally complement the NES-deficiency (Fig. 1B). Among the fusions tested, only the C-terminal region of c-Abl, encompassing amino acids 998-1097 could restore the function of RevΔ3NI, suggesting that an NES may be present at the C terminus of c-Abl.
Figure 1.
Identification of a domain in c-Abl capable of complementing NES-mutated Rev (RevΔ3NI). The Rev activity of exporting unspliced RNA out of the nucleus requires a functional NES. (A) The pDM128 reporter, which contains an intron-disrupted CAT gene, is used to assay for Rev function. (B) A series of fragments of c-Abl were fused to the C terminus of RevΔ3NI and tested for their ability to complement the Rev NES-deficiency. The relative export activity of each derivative is shown. (C) Alignment of selected leucine-rich NES sequences with the c-Abl NES. The core tetramer is shown in boldface type and critical upstream leucines are underlined. Important hydrophobic amino acids, typically leucines, can occupy variable positions upstream of the core tetramer, and these functional positions are indicated with shaded areas. The leucine to alanine substitution at position 1064 is shown in the c-Abl (L1064A) mutant. (D) CAT assay results comparing the function of RevΔ3NI complemented with either the wt c-Abl or the mutated c-Abl (L1064A) NES fragments. Controls include the RevΔ3NI alone or RevΔ3NI complemented with the NES of human T-lymphotropic virus I Rex (amino acids 79–95), representing a positive control. Results shown are CAT activity presented as percent acetylation from a triplicate transfection of 293 cells that was normalized to a cotransfected β-galactosidase internal control. Bars = SEM.
Extensive mutagenesis analysis of the NES of human T-lymphotropic virus I Rex has shown that the Rev-like, or leucine-rich NES, are composed of four hydrophobic amino acids with variable spacing (32, 33). Comparison of the sequence of the complementing region of c-Abl identified a potential leucine-rich sequence at position 1057–1067 of c-Abl (Fig. 1C). To determine the potential role of this putative NES, a mutant was generated where the leucine at position 1064 was mutated to an alanine (L1064A). This type of mutation is known to abrogate the NES function in the Rev complementation assay. The wt and mutant fusion derivatives were then assayed using the pDM128 reporter (Fig. 1D). As a positive control, RevΔ3NI fused to the NES of human T-lymphotropic virus I Rex, which restores the function of RevΔ3NI to Rev NES wt levels, was included in this analysis. The fusion of wt c-Abl 998-1079 efficiently complemented RevΔ3NI to levels seen when RevΔ3NI is fused with the human T-lymphotropic virus I Rex NES (33). The single point mutation (L1064A) completely abrogates the complementation activity of the c-Abl 998-1079 region. This result strongly suggests that the identified leucine-rich sequence is a functional NES.
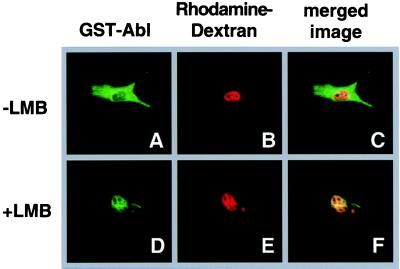
To further demonstrate the function of the c-Abl-NES, c-Abl amino acids 1052–1070 were fused to GST (GST-Abl-1052–1070). The GST-Abl-1052–1070 fusion protein was purified using glutathione affinity chromatography, concentrated and mixed with rhodamine-labeled dextran (rhodamine-dextran), which was used as an injection site marker as it is too large to diffuse through nuclear pores. The GST-Abl-1052–1070/rhodamine-dextran mixture was injected into the nuclei of NIH 3T3 cells (as described in Material and Methods). After a 30-min incubation, the cells were fixed and the GST-Abl-1052–1070 fusion protein was visualized by indirect immunofluorescence using anti-GST antibodies. Thirty minutes after nuclear injection, the GST-Abl-1052–1070 fusion protein was detected in the cytoplasm (Fig. 2A), whereas the rhodamine-dextran microinjection marker remained nuclear (Fig. 2B). Fig. 2C is the merged image of the GST-Abl and rhodamine-dextran, showing little colocalization. This data indicates that the c-Abl NES is capable of exporting a heterologous protein. Pretreatment of cells with LMB, which has been shown to disrupt NES-mediated export by blocking the formation of the NES/CRM1/Ran-GTP complex (29, 39–41), prior to micoinjection inhibited the export of the GST-Abl-1052–1070 protein (Fig. 2 D and F). This result indicates that the c-Abl NES uses the CRM1-mediated nuclear export pathway.
Figure 2.
The C-terminal region of c-Abl can function as an NES when fused to GST. The fusion protein GST-Abl-1052–1070 (labeled GST-Abl) was purified, mixed with the rhodamine-labeled dextran injection marker (rhodamine-dextran), and microinjected into the nuclei of NIH 3T3 cells. After a 30 min incubation at 37°C, cells were fixed and processed for indirect immunofluorescence, and analyzed by confocal microscopy (as described in Materials and Methods). The GST-Abl-1052–1070 fusion protein was visualized using anti-GST antibodies (A and D). (B and E) rhodamine microinjection marker; (C and F) the merged image of GST-Abl and rhodamine-dextran, with yellow indicating colocalization. Cells in D–F were pretreated for 6 hr with 20 nM LMB prior to microinjection.
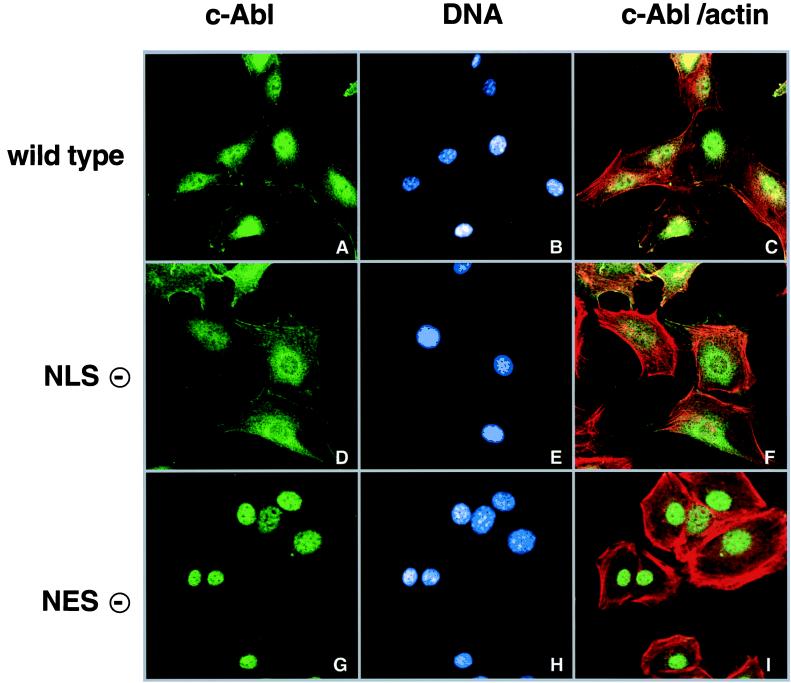
As c-Abl contains three NLS (3), it was unclear why nearly half of the c-Abl is found in the cytoplasm at steady state. To determine if the cytoplasmic localization of c-Abl is the result of nuclear export, we introduced the L1064A mutation into the full length c-Abl protein, with this mutant being designated c-Abl NES(−). The previously described NLS mutant (3) with amino acid substitutions at all three NLS, designated c-Abl NLS(−), was included as a comparison. The wt c-Abl (designated c-Abl wt), c-Abl NLS(−) and c-Abl NES(−) proteins were stably expressed in 3T3 cells derived from an Abl-deficient mouse embryo through retroviral-mediated gene transfer. The subcellular localization of the various c-Abl proteins was determined by indirect immunofluorescence analysis (Fig. 3). Cells were costained with the Hoechst dye to label DNA (shown in blue) and rhodamine-conjugated phalloidin to label F-actin (shown in red). In proliferating 3T3 fibroblasts, c-Abl wt was found in the nucleus and the cytoplasm (Fig. 3A). Colocalization of c-Abl with F-actin was detected at the cell periphery (Fig. 3 A and C). The c-Abl NLS(−) was excluded from the nucleus, with a majority of the c-Abl NLS(−) localizing to the perinuclear region and actin filaments (Fig. 3D). Interestingly, the c-Abl NES(−) mutant was found exclusively in the nucleus, with no detectable association with cytoplasmic F-actin (Fig. 3G). These results indicate the NES is functional in the full-length c-Abl protein, and that this NES is required for the cytoplasmic localization of c-Abl.
Figure 3.
Subcellular localization of c-Abl wt, c-Abl NLS(−) and c-Abl NES(−). The c-Abl proteins were expressed in Abl-deficient 3T3 cells. Cells were plated onto coverslips, fixed and prepared for indirect immunofluorescence. The c-Abl protein was detected using the monoclonal anti-Abl 8E9 antibody (A, D, and G). Cells were counterstained with the Hoechst DNA dye (B, E, and H) and rhodamine-conjugated phalloidin to label actin filaments (C, F, and I). Cells were analyzed by confocal microscopy. C, F and I show a merged image of the c-Abl and actin.
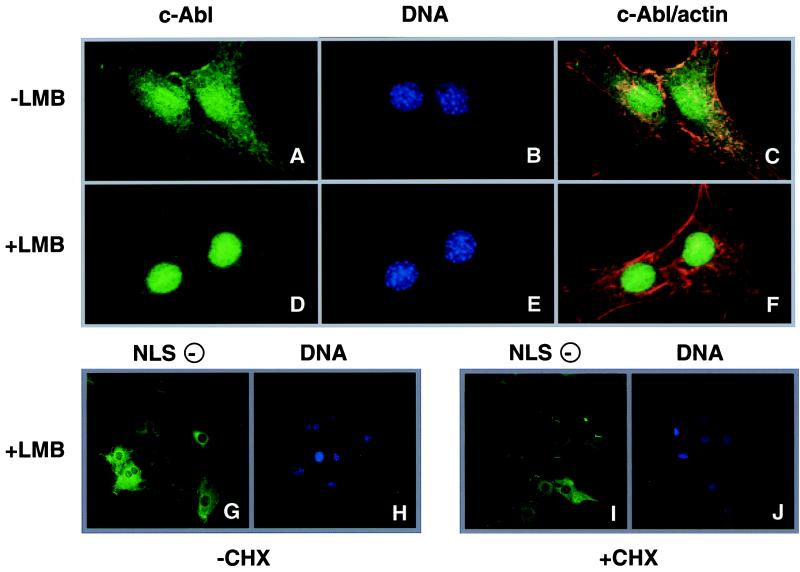
To determine if nuclear export was indeed required for the cytoplasmic localization of c-Abl, cells expressing c-Abl wt were treated with LMB, which blocks CRM1-mediated nuclear export (29, 40, 41, 39), and cycloheximide (CHX), a protein synthesis inhibitor (Fig. 4). CHX is included to eliminate any new c-Abl wt synthesis, allowing the monitoring of the movement of preexisting protein. Without LMB treatment, c-Abl wt is found in the nuclear and cytoplasmic compartments of the cell (Fig. 4A). After a 6-hr incubation with LMB and CHX, c-Abl wt becomes predominantly nuclear (Fig. 4D) and c-Abl wt no longer localizes to the actin filaments (Fig. 4F). As a control, cells expressing c-Abl NLS(−) were similarly treated with LMB in the presence or absence of CHX. LMB treatment did not cause the nuclear accumulation of c-Abl NLS(−) (Fig. 4 G and I), demonstrating that the active nuclear import is required for LMB to alter the subcellular localization of c-Abl. Additionally, LMB treatment, either in the absence (Fig. 4G) or presence (Fig. 4I) of CHX, did not have a significant effect on the overall levels of c-Abl NLS(−) in the cytoplasm. Therefore, the combined treatment with LMB and CHX did not cause a degradation of cytoplasmic c-Abl. The combined results from Figs. 3 and 4 show that the cytoplasmic c-Abl originates from the nucleus as a result of export and that cytoplasmic c-Abl undergoes continuous re-import into the nucleus. Therefore, c-Abl shuttles between the cytoplasm and nucleus in proliferating fibroblasts.
Figure 4.
Treatment of cells with LMB and CHX results in a nuclear accumulation of c-Abl. wt c-Abl expressing cells (panels A–F) were pretreated with CHX for 30 min and then further incubated with (D–F) or without (A–C) 5 nM LMB for 6 hr. Cells were then fixed and stained with the anti-Abl 8E9 antibody (A, D), Hoechst dye (DNA, B and E) and phalloidin (actin). The merged images of the 8E9 and phalloidin staining are shown in C and F. Comparison of D and A or F and C shows that cytoplasmic c-Abl is lost in LMB-treated cells. To test whether CHX+LMB treatment caused a nonspecific loss of cytoplasmic c-Abl, c-Abl NLS(−) expressing cells were similarly treated with 5 nM LMB for 6 hr, either in the presence (G and H) or absence (I and J) of CHX. The anti-Abl stain (G and I) and the Hoechst stain (H and J) were shown. The intensity of Abl stain was not diminished after CHX-LMB treatment, showing that the cytoplasmic pool of Abl is stable under the experimental conditions.
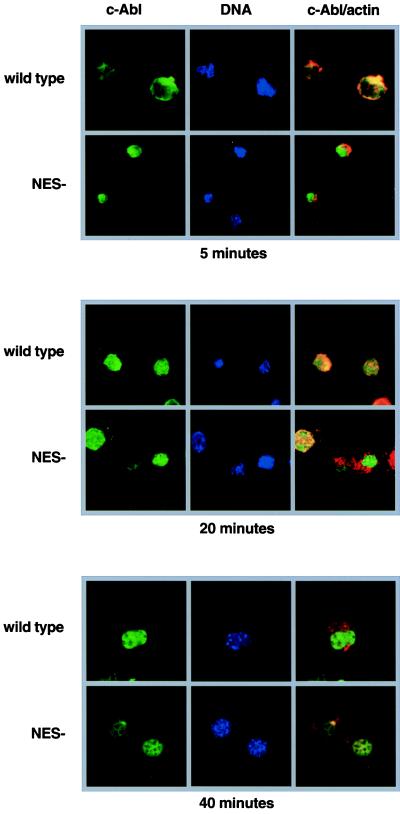
We have previously shown that c-Abl is transiently lost from the nucleus when detached cells were replated onto ECM components such as FN (1). To determine the role of nuclear export in integrin-induced translocation of c-Abl, we performed an attachment assay using cells expressing c-Abl wt or c-Abl NES(−) (Fig. 5). Five minutes after plating onto FN-coated coverslips, the majority of c-Abl wt was found in the cytoplasm. In contrast, the c-Abl NES(−) protein remained nuclear, showing no colocalization with actin. By 20 min, cells that had started spreading showed nuclear c-Abl wt. By 40 min, most cells completed spreading and returned to the adherent fibroblastic morphology. In these cells, the c-Abl wt protein was almost exclusive nuclear. The c-Abl NES(−) mutant remained nuclear throughout this attachment timecourse. Because the c-Abl NES(−) was maintained in the nucleus, adhesion to the ECM is not likely to induce the degradation of nuclear c-Abl protein. In addition, the c-Abl NES must be functional for integrin to induce the translocation of c-Abl. Indeed, pretreatment with LMB also blocked the FN-induced transient translocation of c-Abl wt (data not shown). Thus, a functional export machinery as well as a functional export signal are required for integrin-induced c-Abl movement.
Figure 5.
Mutation of the c-Abl NES prevents the cytoplasmic accumulation of c-Abl induced by cell adhesion. The wt c-Abl (wt) and c-Abl NES(−) proteins were expressed in Abl-deficient 3T3 cells. The cells were trypsinized and plated onto FN-coated coverslips in serum-free medium in the presence of trypsin inhibitor. Cells were fixed at the indicated times and stained with the anti-Abl antibody 8E9 (shown in green), and counterstained with the DNA dye Hoechst (shown in blue) and rhodamine-labeled phalloidin (shown in red). The c-Abl/actin panels show the c-Abl and actin staining patterns merged.
DISCUSSION
c-Abl Contains a Functional NES.
This study has identified a functional NES in the C-terminal region of c-Abl. The NES of c-Abl is capable of restoring nuclear export function to the NES-defective RevΔ3NI protein. A GST-Abl-1052–1070 fusion protein also undergoes active transport out of the nucleus, demonstrating that the c-Abl NES can promote export of heterologous proteins. As shown with the Rex NES (33), mutation of a single hydrophobic amino acid (L1064) in the core tetramer of the c-Abl NES is sufficient to abrogate export activity. We have also found that c-Abl NES function can be blocked by an excess of Rex NES (data not shown), indicating that c-Abl NES likely uses the same export pathway as the Rev-like proteins. Recent work has shown that these Rev-like NES interacts directly with the nuclear export receptor, CRM1 (29, 40, 41). Indeed, LMB, which inhibits the function of CRM1 by blocking formation of the NES/CRM1/Ran-GTP complex, is shown to block the function of the c-Abl-NES. These results suggest that the c-Abl NES is capable of interacting directly with the nuclear export machinery.
Nuclear Export Required for the Cytoplasmic Localization of c-Abl.
The c-Abl NES contributes to the cytoplasmic localization of c-Abl. In proliferating fibroblasts, c-Abl is in both the nuclear and cytoplasmic compartments. A single point mutation within the c-Abl NES results in the exclusive nuclear localization of the c-Abl protein. This result shows that most, if not all, of the c-Abl protein synthesized in the cytoplasm is imported into the nucleus in adherent fibroblasts. Additionally, this result shows that nuclear export is critical for maintaining the cytoplasmic pool of c-Abl. That nuclear export is required for c-Abl to appear in the cytoplasm is also supported by treating cells with the export inhibitor LMB. By treating cells with LMB and the protein synthesis inhibitor CHX, we showed that the cytoplasmic c-Abl can be reimported to the nucleus. Thus, both the import and export processes are required for the observed distribution of c-Abl in the nuclear and cytoplasmic compartments.
While our results indicate that a majority of c-Abl shuttles between the nucleus and cytoplasm, this data does not exclude the possibility that c-Abl localization may also be regulated by retention mechanisms. For example, binding to F-actin may anchor c-Abl and prevent nuclear import. Alternatively, recruitment of c-Abl into specific DNA-binding complexes may prevent nuclear export. Nevertheless, the bulk of c-Abl appears to shuttle between the nuclear and cytoplasmic compartments of proliferating 3T3 fibroblasts.
Biological Implications of the Shuttling of c-Abl.
The c-Abl tyrosine kinase has both nuclear and cytoplasmic functions. As discussed in the Introduction, nuclear c-Abl is likely involved in the regulation of gene expression and the cytoplasmic c-Abl is likely involved in the transduction of adhesion signals. The shuttling of c-Abl between the two compartments may allow the cell to rapidly alter the subcellular locations of c-Abl to emphasize either the nuclear or the cytoplasmic function. For example, the initial adhesion of cells to the extracellular matrix induces the rapid relocalization of nuclear c-Abl into the cytoplasm. Specifically, the translocated c-Abl has been found to localize to early focal adhesions (1), suggesting that c-Abl may be required during the initial formation of focal contacts. The cell may therefore export nuclear c-Abl to increase the cytoplasmic pool size during the early stages of adhesion and cell spreading. Another possible reason for the continuous shuttling of c-Abl may be that c-Abl has the additional function of a carrier that can transport macromolecules between the two subcellular compartments. In this capacity, c-Abl can conceivably carry molecules from the cytoplasm to the nucleus and also in the other direction, from the nucleus to the cytoplasm. The identification of nuclear import as well as export signals within the c-Abl protein suggests that the nuclear-cytoplasmic shuttling of this tyrosine kinase may play a critical role in the regulation of c-Abl biological function.
Acknowledgments
In memory of Laura Whitaker, February 25, 1998. We thank Dr. Fred Gage and Dr. Daniel Peterson for their help with the confocal microscopy, Jayne Stommel for her assistance in the construction of the Rev-Abl fusion expression constructs, and Allison Bocksrucker for her help in preparation of the figures. S.T. was supported by funds from Department of Health and Human Services Fellowship CA69789 and D.M. was supported by funds from the Mack S. Rau Foundation. This work was supported by National Institutes of Health Grant AI35477 to T.J.H. and CA43054 and HL57900 to J.Y.J.W.
ABBREVIATIONS
- FN
fibronectin
- CHX
cycloheximide
- EiCM
extracellular matrix
- CAT
chloramphenicol acetyltransferase
- GST
glutathione S-transferase
- RB
retinoblastoma protein
- LMB
leptomycin B
- NLS
nuclear localization signal
- NES
nuclear export signal
- wt
wild type
- MSCV
murine stem cell virus
References
- 1.Lewis J, Baskaran R, Taagepera S, Schwartz M, Wang J. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Etten R A, Jackson P, Baltimore D. Cell. 1989;58:669–678. doi: 10.1016/0092-8674(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 3.Wen S-T, Jackson P K, Van Etten R A. EMBO J. 1996;15:1583–1595. [PMC free article] [PubMed] [Google Scholar]
- 4.Miao Y, Wang J. J Biol Chem. 1996;271:22823–22830. doi: 10.1074/jbc.271.37.22823. [DOI] [PubMed] [Google Scholar]
- 5.Dikstein R, Agami R, Heffetz D, Shaul Y. Proc Natl Acad Sci USA. 1996;93:2387–2391. doi: 10.1073/pnas.93.6.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch P J, Wang J Y J. Mol Cell Biol. 1995;15:5542–5551. doi: 10.1128/mcb.15.10.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch P J, Wang Y J. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 8.Welch P, Wang J Y J. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Baskaran R, Lea-Chou E, Wood L, Chen Y, Karin M, Wang J. Nature (London) 1996;384:273–276. doi: 10.1038/384273a0. [DOI] [PubMed] [Google Scholar]
- 10.Baskaran R, Wood L, Whitaker L, Canman C, Morgan S, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M, Wang J. Nature (London) 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 11.Baskaran R, Dahmus M, Wang J. Proc Natl Acad Sci USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duyster J, Baskaran R, Wang J Y J. Proc Natl Acad Sci USA. 1995;92:1555–1559. doi: 10.1073/pnas.92.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baskaran R, Chiang G G, Wang J Y J. Mol Cell Biol. 1996;16:3361–3369. doi: 10.1128/mcb.16.7.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskaran R, Chiang G, Mysliwiec T, Krul G, Wang J. J Biol Chem. 1997;272:18905–18909. doi: 10.1074/jbc.272.30.18905. [DOI] [PubMed] [Google Scholar]
- 15.McWhirter J R, Wang J Y J. Mol Cell Biol. 1991;11:1553–1565. doi: 10.1128/mcb.11.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McWhirter J R, Wang J Y J. EMBO J. 1993;12:1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Etten R A, Jackson P K, Baltimore D, Sanders M C, Matsudaira P T, Janmey P A. J Cell Biol. 1994;124:325–340. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren R, Ye Z-S, Baltimore D. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 19.Feller S, Knudsen B, Hanafusa H. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer B J, Hirai H, Sakai R. Curr Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 21.Vuori K, Ruoslahti E. J Biol Chem. 1995;270:22259–22262. doi: 10.1074/jbc.270.38.22259. [DOI] [PubMed] [Google Scholar]
- 22.Ullman K S, Powers M A, Forbes D J. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 23.Nakielny S, Dreyfuss G. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 24.Wen W, Taylor S S, Meinkoth J L. J Biol Chem. 1995;270:2041–2046. doi: 10.1074/jbc.270.5.2041. [DOI] [PubMed] [Google Scholar]
- 25.Fischer K-D, Zmuldzinas A, Gardner S, Barbacid M, Bernstein A, Guidos C. Nature (London) 1995;374:474–477. doi: 10.1038/374474a0. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Gotoh I, Gotoh Y, Nishida E. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 27.Fridell R A, Benson R E, Hua J, Bogerd H P, Cullen B R. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 28.Nix D A, Beckerle M C. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornerod M, Ohno M, Yoshida M, Mattaj I. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 30.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 31.Hope T J, Huang X J, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim F, Beeche A, Hunter J, Chin D, Hope T. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Gene Therapy. 1994;1:136–138. [PubMed] [Google Scholar]
- 35.Donello J E, Beeche A A, Smith G J, Lucero G R, Hope T J. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mancuso V A, Hope T J, Zhu L, Derse D, Phillips T, Parslow T G. J Virol. 1994;68:1998–2001. doi: 10.1128/jvi.68.3.1998-2001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fridell R A, Fischer U, Lührmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff B, Sanglier J J, Wang Y. Chem & Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 40.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 41.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;2788:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]