Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor through which organochlorine contaminants including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), and some polycyclic aromatic hydrocarbons induce toxicity and altered gene expression. Atlantic salmon has multiple AHR genes, of which two belong to the AHR1 clade and four belong to the AHR2 clade. The four AHR2 forms (α, β, γ, δ) are more highly expressed than the AHR1 (α, β,) forms and all six AHRs are highly similar in pairs, likely originating from a whole-genome duplication in the salmonid ancestor. It has been speculated that having multiple AHRs contributes to the very high sensitivity of salmonid species to TCDD and related chemicals. To test the hypothesis that all four salmon AHR2 proteins are expressed and functional, we measured mRNA transcription for each AHR2 inside several tissues, cloned the cDNAs and evaluated the functional properties of the expressed proteins. Analysis by real-time PCR revealed that the receptors showed differences in transcript levels among salmon tissues and that in general AHR2α was transcribed at higher levels than the other three AHR2s. Velocity sedimentation analysis showed that all four in vitro-expressed AHR2 proteins exhibit specific, high-affinity binding of [3H]TCDD. When expressed in COS-7 cells, all four AHR2 proteins were able to drive the expression of a reporter gene under control of murine CYP1A1 enhancer elements. From EC50 values determined in TCDD concentration-response experiments, all four salmon AHR2s show similar sensitivity to TCDD. In summary, all four Atlantic salmon AHR2 appear to function in AHR-mediated signaling, suggesting that all four proteins are involved in TCDD-mediated toxicity.
1. Introduction
Salmonid fishes are among the most sensitive vertebrates to the effects of halogenated aromatic hydrocarbons (HAHs) such as polychlorinated dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs) and biphenyls (PCBs) (Elonen et al. 1998). Many of these chemicals occur at elevated levels in farmed and wild salmon (Isosaari et al. 2006;Jacobs et al. 2002;Shaw et al. 2006). Studies on various salmonid species have identified several tissues as primary targets for TCDD-induced pathologic lesions (e.g. lymphomyeloid and epithelial tissue) with exposure resulting in severe cardiovascular impairments and lesions in brain, retina and liver (Spitsbergen et al. 1988;Spitsbergen et al. 1991;Walker et al. 1991;Walker et al. 1996;Walker and Peterson 1991). Rainbow trout (Oncorhynchus mykiss) sac fry exposed to TCDD suffer from yolk sac edema, hemorrhage, craniofacial malformation, and growth retardation, ultimately resulting in mortality (Hornung et al. 1999). Salmonid early life stages are especially sensitive to exposure, and LC50 values suggest that salmonids could be up to 50 times more sensitive to the highly toxic 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) than other fishes and the most sensitive mammals (Hahn 2001).
Induced toxicity from planar HAHs is mediated by the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor that resides in the cytoplasm of vertebrate cells. After exposure, the AHR binds to the HAH and initiates a signaling pathway that includes interactions with the aryl hydrocarbon receptor nuclear translocator (ARNT) and translocation to the cell nucleus, where it activates the transcription of responsive genes including those encoding biotransformation enzymes, such as cytochrome P450s (CYPs) (Nebert and Dalton 2006).
The vertebrate AHR genes are divided into two distinct evolutionary lineages, AHR1 and AHR2 (Hahn et al. 1997). Mammals have a single AHR gene which belongs to the AHR1 lineage. Fishes have, in general, more AHR genes than other vertebrates because they have retained AHR2 genes (which were lost in the lineage leading to mammals) and because of a fish-specific whole-genome duplication event in their early evolutionary past (Hahn et al. 2006). The ancestor to the modern salmonid fish species later underwent an additional genome duplication 25–100 million years ago further duplicating their genes (Allendorf and Thorgaard 1984). This evolutionary scenario gained further support by the discovery of a high number of AHR genes in the Atlantic salmon (Salmo salar) genome, all highly similar in pairs. This species has at least six AHR genes, of which two are AHR1s (α, β) and four are AHR2s (α, β, γ, δ) (Hansson et al. 2003; Hansson et al. 2004). These six AHRs include three paralogous gene pairs (AHR1α and β; AHR2α and β; AHR2γ and δ) that are similar within their coding regions (94%, 84% and 92% amino acid identity within each set of the paralogs). The Atlantic salmon AHR2α and AHR2β genes are considered to be the orthologs of the two rainbow trout AHR2 genes previously characterized for ligand-binding and transactivation (Abnet et al. 1999b;Abnet et al. 1999a;Pollenz et al. 2002). Why as many as six AHR genes have been retained in Atlantic salmon is not fully understood. However, previous analyses of the salmon AHRs suggest that the two AHR1 genes are expressed at much lower levels than the four AHR2s (Hansson et al. 2004). This is consistent with other studies indicating that AHR2 is the predominant form in fish (Abnet et al. 1999a;Karchner et al. 1999;Roy and Wirgin 1997;Tanguay et al. 1999). Also, in zebrafish (Danio rerio) the AHR1 forms are either inactive (AHR1a; (Andreasen et al. 2002a)) or have reduced TCDD sensitivity as compared with AHR2 (AHR1b (Karchner et al. 2005)). Furthermore, in zebrafish the single AHR2 has been shown to mediate CYP1A induction and developmental toxicity (Prasch et al. 2003). We thus focused on the four Atlantic salmon AHR2s for our analyses in this paper.
Several questions about the role of salmonid AHRs in HAH-induced toxicity remain to be addressed. It is currently unknown if all four Atlantic salmon AHR2 genes encode functional proteins that are capable of binding TCDD with high affinity and mediating the biochemical and toxic responses observed after exposure to HAHs, and whether one or more of these receptors contributes differentially to the effects of HAHs. The very high sensitivity to HAHs seen specifically in salmonids may be related to the fact that they have more AHR forms than many other fish species. In order to investigate this relationship further and to get a deeper understanding of these duplicated genes’ functions and their role in the activation of biochemical and toxic responses to HAH exposure, we have cloned all four AHR2 cDNAs from Atlantic salmon, expressed the encoded proteins in vitro, and analyzed their abilities to bind TCDD and activate transcription.
2. Materials & Methods
2.1. Chemicals
The radiolabeled TCDD ([3H]TCDD, 35 Ci/mmol, >99% radiochemical purity) was obtained from Chemsyn Science Laboratories (Lenexa, KS), [35S]methionine was purchased from Amersham Biosciences (Piscataway, NJ), and TCDD was obtained from Ultra Scientific (Hope, RI). All other chemicals were from Sigma (St. Louis, MO).
2.2. Tissues and RNA isolation
Tissues were sampled from Atlantic salmon captured in the Baltic Sea, as described earlier (Hansson et al. 2006). Total RNA was isolated using RNA STAT-60 (Tel-Test B, Friendswood, TX) or TRIzol® Reagent (Gibco BRL/Life Technologies, Gaithersburg, MD) according to the manufacturer’s instructions. Poly(A)+RNA was purified using oligo(dT) spin columns (5 Prime -> 3 Prime, Boulder, CO). All RNA samples were stored at −80°C. Purity was measured in a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE) and was re-confirmed immediately prior to use for RT–PCR. For isolation of RNA from COS-7 cells transfected with either AHR2α or AHR2γ construct, medium was removed and total RNA was extracted with RNA STAT-60 (Tel-Test, Inc. Friendswood, TX) according to the manufacturer’s instructions. Three wells of cells transfected with the same construct were pooled and subjected to extraction. Isolated RNA was then used as template for reverse-transcription to cDNA, which subsequently was used in real-time PCR analyses.
2.3. Reverse-transcription
cDNA template for amplification of full-length AHR fragments was synthesized from two salmon individuals, using 1 µg of poly(A)+ RNA extracted from brain, heart and liver. The samples were reverse transcribed using the Omniscript cDNA Synthesis Kit from Qiagen (Valencia, CA). For the analyses of tissue-specific transcription we used 2 µg of poly(A)+ RNA extracted from blood, brain, heart, liver, muscle, spleen, and ovary/testis from four individual salmon of which two were ripe females and two were males. For these analyses, cDNA was synthesized with SuperScript™ II Reverse Transcriptase (Invitrogen, Carlsbad, CA). For measurements of AHR transcription in COS-7 cells transfected with salmon AHR2 expression constructs, we used ~2 µg of total RNA. These samples were reverse transcribed using the Qiagen Omniscript cDNA Synthesis Kit.
2.4. Oligonucleotide primers
Primers were synthesized by Invitrogen (Carlsbad, CA) or Sigma-Genosys (The Woodlands, TX) and were used as described. Primer sequences are listed in Table 1.
TABLE 1.
Primer sequences used in the PCR reactions and accession numbers of the genes
| GENE | SEQUENCE | ACCESSION NR. |
|---|---|---|
| Full-length PCRs: | ||
| AHR2α | 5′- CCCTTGTTAATTTGAGGACTATCTCGGAGG-3′; | AY219864 |
| 5′- CCATCATTTTTACACCGTAACATGTGACTC-3′; | ||
| AHR2β | 5′- TTTGAACATTTTATAGCCGATTTACAGCAGA-3′; | AY219865 |
| 5′- CCATTTTTACCTCGTAAGGCAAAGTGACG -3′; | ||
| AHR2γ | 5′- CAATAAGAGGGACACGTATAGGATAGTCT-3′; | AY052499 |
| 5′- GTATCACAACAAATTCCATTCAGCCGTGGA-3′; | ||
| AHR2δ | 5′- GAATAAGAGGGAAACGTATTGGATAGTAT-3′; | AF495590 |
| 5′- GTATCTTAACAAATTCCATTCAGCCGCAG-3′; | ||
| Real-time PCR: | ||
| AHR2α | 5′- TACCTGGCTGGACGCAACA-3′; | AY219864 |
| 5′- AGCCCCCCTTGTCAATGGA-3′; | ||
| AHR2β | 5′- GCTGCCTCCTCGACAACTCTTC-3′- | AY219865 |
| 5′- GCTGTCCAGAACCCGTCCATT-3′; | ||
| AHR2γ | 5′- AAGCAGGACCAGGCCATCT-3′; | AY052499 |
| 5′- AACGTGCCTAGTGATTCCTGG | ||
| AHR2δ | 5′- CCCCTCTCTGGACATAGCTGAT-3′; | AF495590 |
| 5′- AATGTGCCTGGTGATTCCTGG-3′; | ||
| b-actin | 5′- CGCCGCCTCCTCTTCCTC-3′; | AF012125 |
| 5′- CGGTATGGAGTCTTGCGGTATC-3′- | ||
| rtARNTb | 5′- TCAGACATGGTTCCCACCTG-3′; | U73841 |
| 5′- CCACAAACAGGAAGCCATCA-3′; | ||
2.5. Real-time PCR
Primers for real-time PCR were designed manually to span intron/exon junctions (known from comparisons to mammalian AHR sequences and salmon AHR characterizations by M. Hansson) and to ensure high specificity for each AHR2 gene (Table 1).
Analyses of transcription levels in salmon tissues were performed in an MX300P thermocycler (Stratagene, La Jolla, CA) using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA). PCR amplifications were carried out in 25 µl reaction volumes that contained 2 µl of diluted (1:3) cDNA, 50 ng of each primer, 0.25 µl ROX reference dye, 12.5 µl supermix and H2O. Standard curves were prepared for each of the AHR2 genes as well as the Atlantic salmon β-actin gene in six 10-fold dilution series from cloned fragments. The PCR conditions were 50 °C for 2 min, 95 °C for 2 min, followed by 95 °C for 15 s/64 °C for 30 s (40 cycles) for all genes. At the end of each PCR run, the PCR products were subjected to melt curve analysis to ensure that only a single product was amplified. We used three technical replicates of each RNA sample. AHR2 transcription data were quantified based on threshold cycle values in comparison to the standard curve and were averaged and normalized to values of β-actin.
Measurements of AHR2α and AHR2γ transcript levels in transfected COS-7 cells were performed in an iQ5 thermocycler (Bio-Rad Laboratories, Hercules, CA) with the iQ SYBR Green Supermix (Bio-Rad). Transcription of the rainbow trout ARNTb gene served as the internal control. Six tenfold dilutions of cloned salmon AHR2α and AHR2γ fragments as well as the rainbow trout ARNTb were used as standards. Standards were included in each run and equal volumes of standard and cDNA template (1 µl) were used in the PCRs. PCR amplifications were in 25 µl reaction volumes containing 1 µl cDNA from COS-7 cells, 50 ng each primer, 12 µl supermix and H2O. The PCR conditions were 95 °C for 3 min, followed by 95°C for 10 s/61°C for 30 s (40 cycles) ending with a melt curve analysis for all samples. We used three technical replicates of each sample. AHR2 transcription data were quantified based on threshold cycle values and were averaged and normalized to values of the rainbow trout ARNTb.
2.6. Cloning and DNA sequencing
All PCR products were cloned into the TOPO vector (Invitrogen). PCR products were sequenced by the University of Maine DNA Sequencing Facility (Orono, ME), or at the Bay Paul Center Sequencing Facility (Marine Biological Laboratory, Woods Hole, MA).
2.7. Expression constructs
Full-length Atlantic salmon AHR2α, β, γ, and δ cDNAs were amplified using primers listed in Table 1, cloned into the TOPO TA vector, and sequenced. The AHR2 α and β cDNAs were excised from the TOPO vector with NotI/SpeI and inserted into the same cut sites in the pcDNA3.1 vector. The AHR2 γ and δ cDNAs were excised from the TOPO vector with BamHI/NotI and inserted into a pcDNA3.1 vector that had been cut with the same enzymes.
Expression vectors for rainbow trout AHR2 α and β in pBK-CMV were gifts from Dr. R. Peterson (University of Wisconsin, Madison, WI, USA). The rainbow trout AHR2 α and β cDNAs were subcloned into the pcDNA 3.1 expression vector using EcoRI/NotI. The plasmid pGudLuc 6.1 containing the firefly luciferase reporter under the control of an MMTV promoter regulated by four AHREs from the murine CYP1A1 promoter (Long et al. 1998), was a gift from Dr. M. Denison (University of California, Davis, CA, USA). The rainbow trout ARNTb was a gift from Dr. Pollenz (University of South Florida, FL, USA).
2.8. In vitro expression of protein, ligand-binding assay and velocity sedimentation
The TnT-Quick Coupled Reticulocyte Lysate System (Promega, Madison, WI) was used to synthesize 35S-methionine-labeled or -unlabeled proteins from 1 µg salmon-AHR2 expression construct according to the manufacturer's instructions. Five µl of the [35S]methionine-labeled TnT reactions were subjected to SDS-polyacrylamide gel electrophoresis followed by fluorography to visualize labeled proteins. The [35S]-labeled salmon AHR2 proteins were quantified by scintillation counting of excised gel fragments.
TCDD-ligand binding for all four AHR2 proteins was measured using unlabeled proteins by velocity sedimentation on sucrose gradients in a vertical tube rotor according to the method described by Karchner et al. (1999). For each protein, a single TnT reaction (50 µl) was diluted 1:1 with MEEDMG buffer, incubated overnight at 4°C with [3H]TCDD (8 nM), and fractionated on 10–30% sucrose gradients. The binding of [3H]TCDD to unprogrammed lysate (UPL) was also assessed, as a measure of nonspecific binding. Radioligand concentrations were verified by sampling each tube for total counts. [14C]Ovalbumin (3.6 S) and [14C]catalase (11.3 S) were used as sedimentation markers.
2.9. Transfection and luciferase reporter assay
To evaluate the ability of Atlantic salmon AHRs to activate transcription, we performed transient transfection experiments using the mammalian COS-7 cell line (derived from monkey kidney). COS-7 cells were purchased from the American Type Culture Collection (Manassas, VA). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) with 10% fetal calf serum (Sigma) at 37°C under 5% CO2. Cells were plated at 3 × 104 cells per well in 48-well plates. Transfections with salmon AHR2 constructs were carried out 24 h later, in triplicate. We diluted the AHR constructs as well as the Lipofectamine 2000 reagent (Invitrogen) in serum-free DMEM prior to transfections. For each well, a total of ~300 ng of DNA was mixed with 1 µl of Lipofectamine 2000 and added to cells in DMEM with serum. Five h after transfections, cells were dosed with TCDD (10 nM except for concentration-response experiments where TCDD concentration varied between 0 and 10 nM) or DMSO vehicle (0.5% final concentration). Renilla luciferase (pRL-TK; Promega) was used as the transfection control. Transfected DNA amounts were 5 ng to 30 ng of each Atlantic salmon AHR2 construct, 5 ng of rainbow trout AHR2, 25 ng of pSPORTMAHR (mouse AHR, a gift of Dr. C. Bradfield, Univ. of Wisconsin), 50 ng of rainbow trout pcDNA-ARNTb, 20 ng of pGudLuc 6.1, and 3 ng of pRL-TK. DNA content was kept constant (300 ng) by the addition of empty pcDNA vector. We lysed the transfected cells 18 h after the TCDD or DMSO treatment and measured luciferase activities using the Dual Luciferase Assay (Promega) in a TD 20/20 Luminometer (Turner Designs, Sunnyvale, CA). The final transactivation values were expressed as a ratio of the firefly luciferase units to the Renilla luciferase units.
For measurements of AHR2 mRNA levels in transfected COS-7 cells, we transfected cells (grown under the same circumstances as described above) with 50 ng of rtARNTb and either 5 ng or 30 ng of the AHR2α or γ constructs approximately 18 hours before RNA isolations took place. The cells were dosed with either 10 nM TCDD or DMSO approximately 5 hours after transfection. Cells were then left for 13 hours in the incubator, pooled and subjected to RNA-isolation (see separate section).
EC50 values for induction of luciferase were determined by nonlinear regression analysis using Prism version 3 software (GraphPad, San Diego, CA). The data for expression of each AHR2 gene were transformed to fractional response by subtracting the lowest level of expression from the expression at each concentration of TCDD and then dividing each value by the maximal expression for that gene; resulting in fractional response values ranging from 0 (no induction) to 1 (maximal induction). Transformed data from all four AHR2 dose-response experiments were fitted to the equation Y =Bottom+(Top-Bottom)/{1+ 10^((log EC50 − X)×HillSlope)), where X is the logarithm of TCDD concentration and Y is the normalized luciferase activity, and the slope was allowed to vary. The EC50 values are presented as mean values ± standard deviation (SD).
3. Results
3.1. AHR2 tissue-specific expression
We used real-time RT-PCR to measure mRNA transcription of all four salmon AHR2 forms in salmon brain, heart, liver, muscle, spleen, blood and ovary or testis (Fig. 1). The highest levels were seen for AHR2α followed by AHR2γ>AHR2β>AHR2δ. AHR2α was generally transcribed at high levels for all tissues (except blood) but was highest in liver, muscle and testis. Transcription levels of AHR2β were highest in the heart. Levels of AHR2γ were highest in heart and muscle. AHR2δ levels were generally very low compared to the other three AHR2s; levels in heart and muscle were higher than in the other tissues.
Figure 1.
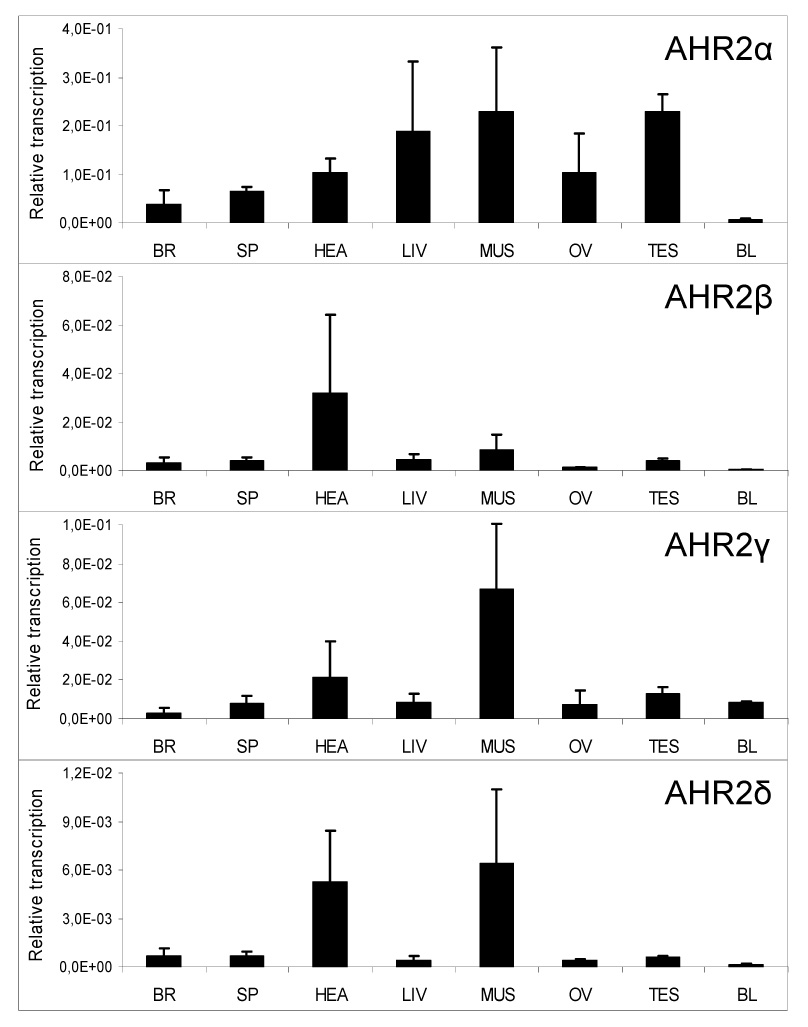
A comparison of expression of multiple forms of AHR2 among different tissues of Atlantic salmon that were collected from the Baltic Sea. Real-time RT-PCR using gene-specific primers was used to analyze levels of AHR2α, β, γ, and δ mRNA in tissues of four Atlantic salmon (two females and two males). Tissues analyzed were brain (BR), spleen (SP), heart (HEA), liver (LIV), muscle (MUS), ovary (OV), testis (TES) and blood (BL). AHR2 levels are relative values, normalized to levels of β-actin, and are expressed as mean ± SD. Note differences in relative transcript levels (y-axis) among the genes.
3.2. cDNA cloning and sequencing
To perform functional characterization of Atlantic salmon AHRs, it was necessary first to generate full-length expression constructs for each salmon AHR2 isoform. This was accomplished by amplifying the full-length cDNAs using the cDNA sequences determined previously (Hansson et al. 2003;Hansson et al. 2004) and the primers listed in Table 1. The full-length AHR2α, AHR2β, AHR2γ and AHR2δ cDNAs were amplified from the brain, liver, heart, and brain, respectively. The inserted fragments in the TOPO clones were all sequenced, confirming that the fragments did not contain any changes in the protein coding region caused by errors during the PCR amplifications.
3.3. In vitro synthesis of AHR2 proteins and specific binding of [3H]TCDD
We synthesized the four Atlantic salmon AHR2 proteins by in vitro transcription and translation, labeling them with [35S]methionine to confirm protein expression and to demonstrate that proteins of the predicted size were synthesized from all four constructs (Fig. 2). These analyses revealed a weaker expression of the AHR2α and β proteins compared to γ and δ. Prolonging exposure of the film up to 12 hours increased the intensity of all four bands (data not shown) but the AHR2 γ and δ bands were consistently stronger than the AHR2α and β bands.
Figure 2.
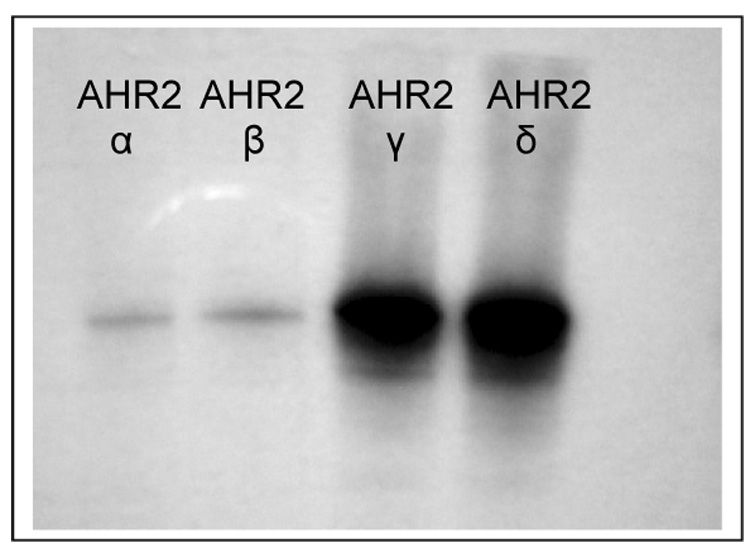
In vitro transcription/translation (TnT) of Atlantic salmon AHR2α, AHR2β, AHR2γ and AHR2δ. Constructs (1 µg) were expressed by TnT T7 to synthesize [35S]methionine-labeled proteins. Reactions were performed together using the same batch of reticulocyte lysate and same concentration of [35S]methionine. The TnT reactions were analyzed by gel electrophoresis, followed by fluorography for 2 hours.
Scintillation counting of excised gel fragments confirmed the lower expression from the AHR2α and β constructs. Specifically, analysis of five µl of the [35S]methionine-labeled TnT reactions for each expression construct yielded (mean±SD from three separate experiments) 39.7 (±5.2) fmol AHR2α, 34.5 (±7.2) fmol AHR2β, 416 (±25.2) fmol AHR2γ and 320 (±39.2) fmol AHR2δ.
We synthesized unlabeled salmon AHR2 proteins by in vitro transcription and translation to evaluate their [3H]TCDD-binding abilities by velocity sedimentation analysis (Fig. 3). This method has been used previously to examine the binding of [3H]TCDD to rainbow trout AHR2s as well as various mammalian and piscine AHRs (Abnet et al. 1999a;Karchner et al. 1999;Karchner et al. 2002). We observed specific binding of [3H]TCDD (8 nM) to all four Atlantic salmon AHR2s, with peaks between fractions 11 to 20. No specific binding was observed with lysate programmed with an empty expression vector. The amount of specific binding obtained with the salmon AHR2s was 10 fmol/reaction (AHR2α), 12.4 fmol/reaction (AHR2β), 37 fmol/reaction (AHR2γ) and 36 fmol/reaction (AHR2δ). Thus, the amount of specific [3H]TCDD binding was lower for AHR2α and β than for AHR2γ and δ; this difference is consistent with, but of a lower magnitude than, the differential protein expression as determined from [35S]methionine-labeled proteins‥
Figure 3.
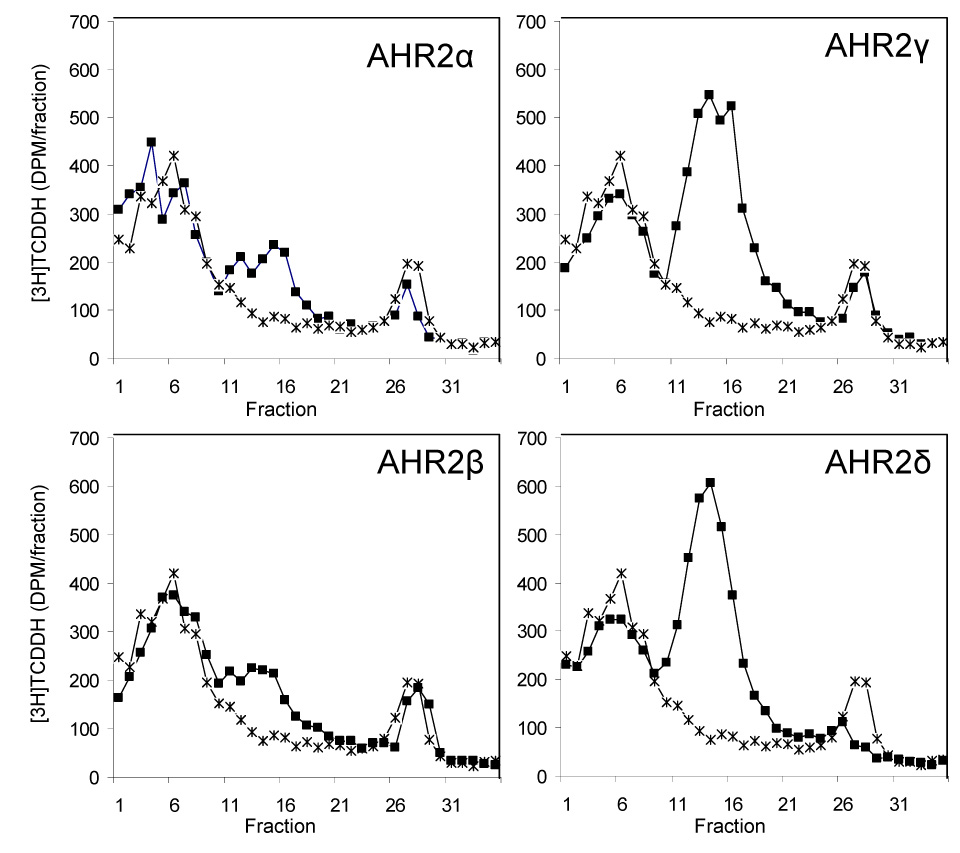
Velocity sedimentation analysis of ligand binding ([3H]TCDD) by the four Atlantic salmon AHR2s: AHR2α, AHR2β, AHR2γ, and AHR2δ. For each protein, a diluted TnT reaction was incubated with [3H]TCDD (8 nM) (filled squares), and fractionated on 10–30% sucrose gradients. The binding of [3H]TCDD to unprogrammed lysate (UPL) was also assessed, as a measure of nonspecific binding (stars). The sedimentation markers [14C]ovalbumin (3.6 S) and [14C]catalase (11.3 S) eluted at fractions 3–4 and 15–16, respectively. Specific binding is the difference between total binding (radioligand binding to expressed protein) and nonspecific binding (radioligand binding to UPL). The amount of specific binding obtained was 10 fmol/reaction (α), 12.4 fmol/reaction (β), 37 fmol/reaction (γ) and 36 fmol/reaction (δ). It should be noted that all the reactions were done using the same amount of plasmid (1 ug) and that under those conditions the salmon AHR2 plasmids express different amounts of protein (as shown in Fig. 2).
3.4. AHR2 transcriptional activity in COS-7 cells
We assessed the ability of the four AHR2s to activate transcription of a reporter gene under control of AHR response elements after transient transfection into cultured cells. We used COS-7 cells because they express very low levels of endogenous AHR and have been widely used to characterize AHR proteins from a variety of species (Abnet et al. 1999b;Karchner et al. 2002;Karchner et al. 2005). The salmon AHR2s were transiently expressed in COS-7 cells along with the rainbow trout ARNT2b, and a luciferase reporter gene under the control of AHREs.
In DMSO-treated cells, transfection of AHR2 expression construct resulted in ~10-fold increases of the basal level of luciferase expression for the AHR2γ and δ constructs and to a lesser degree (~3-fold increase) the AHR2 α and β constructs (Fig. 4), as seen with many other AHRs (Jensen and Hahn 2001; Karchner et al. 2002). Luciferase expression was induced in all cells treated with TCDD (10 nM) as compared with control cells treated with DMSO (Fig. 4). Exposure of the cells to TCDD caused 3- up to 5-fold increases in luciferase expression in AHR2-transfected cells.
Figure 4.
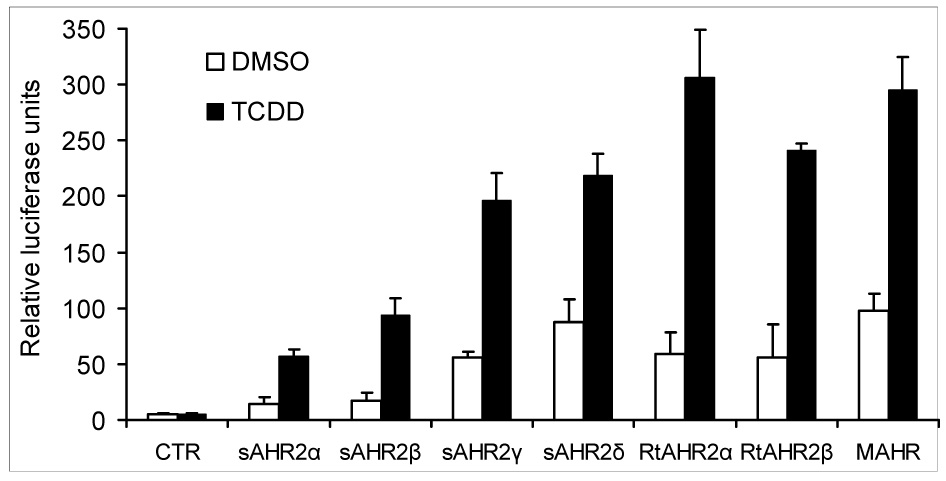
TCDD-induced reporter gene activation in COS-7 cells transfected with rainbow trout ARNTb together with Atlantic salmon AHR2α, AHR2β, AHR2γ, AHR2δ, rainbow trout AHR2α or AHR2β or mouse AHR (MAHR) construct and propagated for 6 hours. The amount of expression construct used was 30 ng for the Atlantic salmon AHR2α and AHR2β, 5 ng for the Atlantic salmon AHR2γ, AHR2δ, rainbow trout AHR2α or AHR2β and 25 ng for the mouse AHR. Cells were then treated with 10 nM TCDD or 0.5% DMSO for 18 hours and evaluated for luciferase activity. Relative luciferase units were calculated by normalizing firefly luciferase activity to the transfection control Renilla luciferase. Each data point represents the mean of triplicate wells and error bars represent S.D. Results shown are representative of three independent experiments.
The basal and TCDD-induced luciferase activities were much greater in cells transfected with AHR2γ or AHR2δ than in cells transfected with the same amount of DNA encoding AHR2α or AHR2β. To determine if this difference was the result of differential expression of AHR2 mRNAs in the transfected cells, we performed real-time RT-PCR to measure transcripts of AHR2α (representing AHR2α and AHR2β) and AHR2γ (representing AHR2γ and AHR2δ) 13 hours after transfection. COS-7 cells transfected with the AHR2γ expression vector expressed much higher levels of AHR2 transcript than cells transfected with equivalent amounts of AHR2α (Fig.5). Consistent with previous results obtained by in vitro transcription and translation, the AHR2α expression level was more than 10 times lower than that of AHR2γ. In contrast, the expression of co-transfected rtARNTb was similar in all wells (data not shown), and thus ARNTb served as an appropriate control for these analyses.
Figure 5.
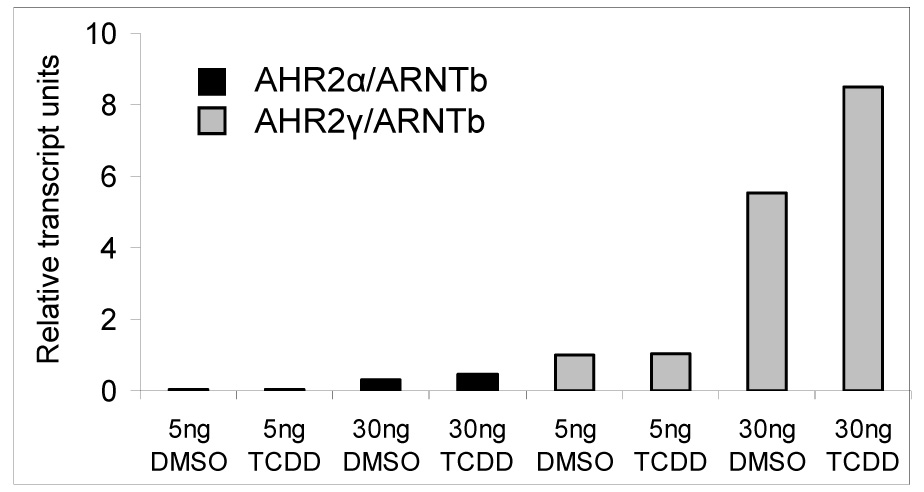
Measurement of AHR2 transcripts in COS-7 cells transfected with either Atlantic salmon AHR2α or AHR2γ together with the rainbow trout ARNTb (rtARNTb) in the presence of absence of TCDD. Levels of AHR2 transcripts were measured using real-time RT-PCR and normalized to levels of rtARNTb transcripts.
3.5. Concentration-response experiments
To determine the sensitivity of each salmon AHR2 in relation to each other, we compared the transactivation abilities of the four AHR2s in COS-7 cells exposed to increasing concentrations of TCDD (0.01–10 nM; Figure 6). Because of the low in vitro expression of AHR2α/AHR2β compared to AHR2γ/AHR2δ, we used greater amounts of AHR2α and AHR2β constructs as compared to AHR2γ and AHR2δ constructs for these analyses (see Figure 6).
Figure 6.
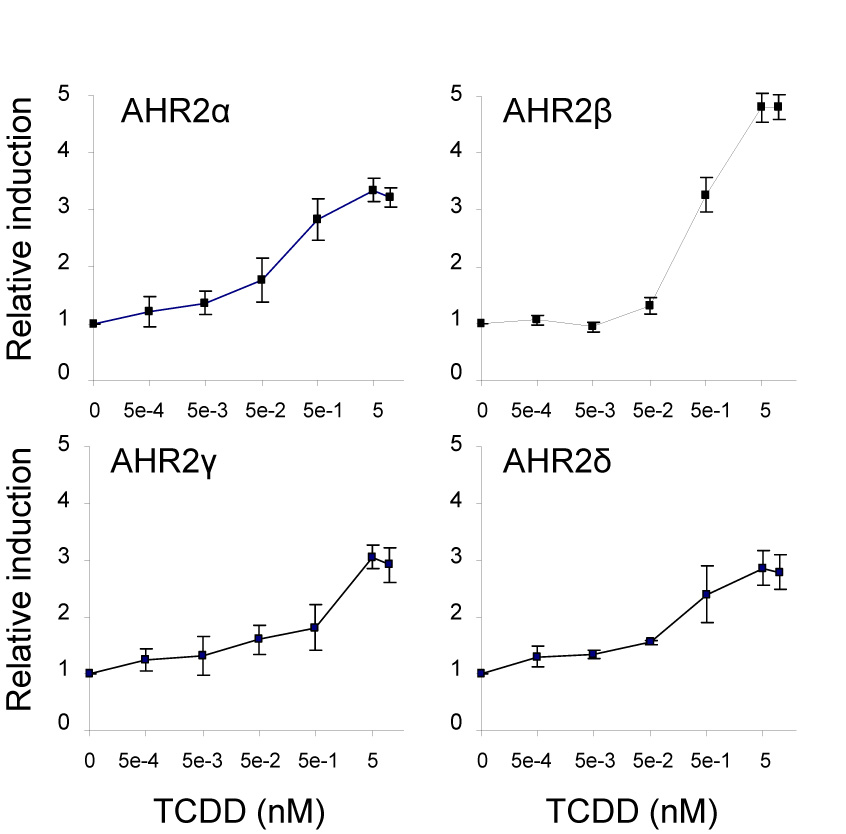
Concentration-response curves for TCDD-induced reporter gene activation in COS-7 cells transfected with each of the Atlantic salmon AHR2 constructs: α, β (50 ng each), γ or δ (10 ng each) and cotransfected with pGudLuc6.1 (20 ng), pRL-TK (3 ng) and rainbow trout ARNTb (50 ng) expression construct. The cells were exposed to increasing concentrations of TCDD (0–10 nM). Relative luciferase units were determined by normalizing the firefly luciferase activity to the transfection control Renilla luciferase activity. For each AHR2, the relative fold induction values were then calculated by dividing the luciferase ratio for each TCDD-treated well by the mean ratio for DMSO-treated wells. All data are represented as the means ± standard deviation (SD) for three wells. The mean DMSO and TCDD (10 nM) luciferase ratios for each AHR2 were as follows: 16,3±5.4 and 51.6±6.1 (AHR2α); 20.8±6.5 and 99.9±14.1 (AHR2β); 55.1±2.1 and 161±9.9 (AHR2γ); 66.7±21.9 and 186±30.9 (AHR2δ). The mean fold-induction for DMSO and TCDD (10 nM) were 3.2 (α); 4.8 (β), 2.9 (γ) and 2.8 (δ), respectively. EC50 values were obtained by nonlinear regression. The calculated EC50 values for each AHR2 were: 0.10 nM±0.035 (AHR2α), 0.40 nM±0.01 (AHR2β), 0.52 nM±0.43 (AHR2γ) and 0.24 nM±0.035 (AHR2δ).
All of the AHR2 forms exhibited TCDD-inducible transactivation of luciferase expression. AHR2β appeared to be the most responsive of the four proteins to activation by TCDD with a maximum 5.5-fold induction compared to 3.7-fold induction for AHR2α, 3.4-fold for AHR2γ and 3.1-fold induction for AHR2δ.
For calculations of EC50 values we made the assumption that induction of luciferase reached a plateau by 10 nM TCDD; in support of this, luciferase activities did not differ between the 5 and 10 nM concentrations (Fig. 6). We then transformed the data into fractional responses (Evans et al. 2005;Poland and Glover 1975) and determined EC50 values by nonlinear regression. To calculate fractional responses for each AHR2, the lowest level of expression (usually DMSO control) was subtracted from the expression at each concentration of TCDD and the resulting values were divided by the maximal luciferase expression for that AHR2; thus, fractional response values ranged from 0 (no induction) to 1 (maximal induction). The EC50 values for the salmon AHR2 proteins were (mean±SD from four separate experiments, with range of values in parentheses): AHR2α, 0.10 nM±0.035 (0.062–0.13 nM); AHR2 β, 0.40 nM±0.01 (0.39–0.41 nM); AHR2γ, 0.52 nM±0.43 (0.10–0.96 nM); and AHR2δ, 0.24 nM±0.035 (0.21–0.28 nM).
4. Discussion
To identify the mechanistic basis of the hypersensitivity of salmonids to planar HAHs,, it is essential to determine which of the six AHRs identified previously (Hansson et al. 2003;Hansson et al. 2004) mediate the typical TCDD-responses in salmon. As an initial step in this assessment, we evaluated the functionality of the four Atlantic salmon AHR2 isoforms. Here, we present results from several functional analyses showing that all four Atlantic salmon AHR2s are transcriptionally active and exhibit high-affinity binding to TCDD. The analyses clearly demonstrate that this tetraploid species expresses all four of its AHR2s. Real-time RT-PCR measurements suggest that these four AHR2s are, however, differentially expressed in tissues of adult salmon (Fig.1). In most tissues, AHR2α mRNA is transcribed at higher levels than any of the other three AHR2s, with transcript levels for the AHR2δ, in contrast, being very low. This suggests that the receptors have distinct physiological functions and that AHR2α plays a more active role in mediating effects of PHAHs than the other three receptors. The EC50 values for induction of reporter gene expression also suggest that AHR2α is the most sensitive of the four AHR2s, and that AHR2β is the least sensitive. The observed difference in concentration response relationships between these two Atlantic salmon AHR2s is consistent with analyses of the two rainbow trout AHR2 orthologs, which previously have been demonstrated to differ in responsiveness to TCDD (Abnet et al. 1999a;Andreasen et al. 2002c;Pollenz et al. 2002). EC50 values for the rtAHR2s are ~0.1 nM (α) and ~1 nM (β), comparable to the observed values for the Atlantic salmon AHR2α (~0.1 nM) and AHR2β (~0.4 nM). Rainbow trout orthologs of the Atlantic salmon AHR2γ and δ have not been characterized but evidence suggests that there is at least one more AHR2 gene in rainbow trout (Hansson et al. 2003). The EC50 values for the four salmon AHR2s are also comparable to that determined earlier for the zebrafish AHR2 (0.7 nM) analyzed under similar conditions (Karchner et al. 2005).
The Atlantic salmon AHR2γ and δ are transcribed at relatively low levels in salmon and transcribed primarily in heart and muscle. Heart was also identified as the primary site of AHR2β expression. Several studies have identified the heart as a principal target organ for TCDD toxicity in fishes, resulting in altered gene expression and cardiovascular impairments (Carney et al. 2006;Handley-Goldstone et al. 2005;Hornung et al. 1999;Spitsbergen et al. 1991). The expression of all four salmon AHR2 forms in heart could indicate that this organ is at higher risk of toxicity after TCDD exposure. Because fish embryos appear to be even more sensitive than adults to TCDD-induced cardiovascular toxicity, it will be important to measure the expression of these AHR2 forms during development in salmon. Why salmon muscle tissue contains comparatively high levels of the AHR2s is unknown. In other fish species, AHR2 expression is inducible by TCDD (Andreasen et al. 2002b;Tanguay et al. 1999). The AHR2 levels were measured in tissues taken from Baltic Sea salmon, where individuals are exposed to very high levels of PCBs and other organochlorine pollutants (Hansson et al. 2006). These contaminants are stored inside the fatty muscle tissues but whether this contributes to the observed relatively high expression of the AHR2 genes in muscle of these fish is not known. Future analyses using salmon from pristine environments will help answer how AHR expression is regulated in individuals from non-exposed salmon populations.
All four salmon AHR2 proteins exhibit specific, high-affinity binding of TCDD as assessed by velocity sedimentation analysis (Fig. 3). This is consistent with other analyzed fish AHR2 proteins and allows us to suggest that all the salmon AHR2s are capable of mediating the biochemical and toxic effects of TCDD. However, to confirm these results we investigated whether each AHR2 protein could drive the expression of a reporter gene under control of AHRE sequences from the murine CYP1A1 promotor. These experiments (Fig.4) clearly demonstrated that all four salmon AHR2 proteins can act together with the rainbow trout ARNTb to stimulate specific, TCDD- dependent interactions with the mammalian AHRE, resulting in transcriptional activation of the reporter gene.
One of the interesting results was that the four AHR cDNAs show differences in expression in vitro and in cultured cells. This was revealed initially in in vitro transcription and translation reactions (Fig.2). The AHR2γ and δ constructs consistently were expressed at levels ~10-fold higher than AHR2α and β. The underlying reason for this difference is unknown but could involve differential transcription, translation, or protein stability, related to sequence differences in untranslated regions, at the translational start site, or in the amino acid sequences of the encoded proteins. The expression constructs included different amounts of 5′-untranslated region: ~100 bp in the AHR2α expression construct but only ~40 bp for AHR2β, γ and δ. The untranslated regions are very similar between the AHR2α and AHR2β as well as between the AHR2γ and AHR2δ genes, but there is very little sequence similarity between the two pairs of paralogous genes. The translational start sites for the salmon AHR2s all deviate from the typical Kozak consensus ACCATGG-sequence (α: AACATGT; β: ACCATGT; γ and δ: ATTATGT).
In the transient transfection assays, the AHR2γ and δ constructs consistently supported higher levels of luciferase transcription (basal and TCDD-induced) than equivalent amounts of AHR2α and β expression constructs. Currently available antibodies against fish AHRs (Merson et al. 2006) do not recognize any of the salmon AHR2s (Hansson MC, unpublished results) so it was not possible to measure the AHR2 protein content in transfected COS-7 cells. However, real-time RT-PCR measurements of AHR2 transcripts in cells transfected with either 5 ng or 30ng AHR2α or 5ng or 30 ng AHR2γ demonstrated that the differences in luciferase transcription corresponded to differences in the expression of the transfected AHR2 constructs. Levels of AHR2γ transcripts were >10 times higher than those of AHR2α transcripts at both 5 ng and 30 ng of transfected expression construct. This suggests that more AHR2 protein is being synthesized in the cells transfected with AHR2γ and AHR2δ than in cells transfected with AHR2α and AHR2β, but that the differences occur at the level of transcription or mRNA stability rather than at the level of translation or protein stability. The exact sequence differences responsible for the differential transcription remain to be determined.
Previous transfection experiments testing the rainbow trout AHR2α and AHR2β have utilized COS-7 cells or the E36 hamster cell line. It has been argued that E36 cells are more suitable for testing the properties of fish proteins because they are grown at the lower temperature of 30°C instead of the 37°C required for the COS-7 (Pollenz et al. 2002). However, our salmon AHR2 proteins were synthesized at 30°C in our in vitro transcription and translation reactions, with expression patterns similar to what was observed in the COS-7 cells (approximately 10-fold expression difference). Thus, we conclude that the differences in expression that we observed are most likely temperature-independent. In the future, we hope to test the Atlantic salmon AHRs with a salmon ARNT(s), which so far has not been characterized.
The evolutionary history of the salmon AHR2s explains the presence of multiple genes in this species but not why four separate AHR2 proteins appear to have very similar functional qualities. In salmonids, the observed rate of gene silencing after the whole-genome duplication event occurred 25–100 million years ago has been slower than predicted by theoretical models, and this has been suggested to be due to retained tetrasomic segregation (Ferguson and Allendorf 1991). It is likely that the four AHR2 genes have been retained due to their acquisition of highly specialized and adaptive functions and that the AHR2 proteins have distinct physiological roles (Lynch and Conery 2000). The AHR2 isoform- and tissue-specific differences in gene expression support this scenario. The AHRs’ functions are also likely to be affected by their partners in toxicity mediation and regulation, especially ARNT but also the aryl hydrocarbon receptor repressor (AHRR) genes, which have been demonstrated to effectively repress activation of AHR target genes in fish (Evans et al. 2005;Karchner et al. 2002;Roy et al. 2006). How many ARNT and AHRR genes exist in Atlantic salmon is currently unknown. More information about these AHR-associated genes and proteins will in the future add to our understanding about how toxicity from planar HAHs is mediated and regulated in salmon.
In summary, we found that the four Atlantic salmon AHR2 are transcribed differently among tissues with levels of AHR2α being the highest. All four AHR2 genes and their proteins are functional in AHR-mediated signaling in studying TCDD-exposed cells. Thus, the high sensitivity to TCDD seen in salmonids may be, at least in part, a consequence of having multiple functional AHR genes.
Acknowledgements
We thank Diana Franks for assistance in the lab and especially with the velocity-sedimentation analyses and Dr. Sibel Karchner for help with the COS-7 experiments. We also thank Drs. R. Pollenz, R. Peterson, M. Denison, and C. Bradfield for expression constructs, and three anonymous reviewers for helpful comments on the manuscript. This project was, in part, funded by grants to Dr. M. Hansson from The Swedish Research Council (Vetenskapsrådet) and The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas), and by grants R01ES006272 and P42 ES007381 to M. Hahn from the U.S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Abnet CC, Tanguay RL, Hahn ME, Heideman W, Peterson RE. Two forms of aryl hydrocarbon receptor type 2 in rainbow trout (Oncorhynchus mykiss). Evidence for differential expression and enhancer specificity. J.Biol. Chem. 1999a;274:15159–15166. doi: 10.1074/jbc.274.21.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abnet CC, Tanguay RL, Heideman W, Peterson RE. Transactivation activity of human, zebrafish, and rainbow trout aryl hydrocarbon receptors expressed in COS-7 cells: greater insight into species differences in toxic potency of polychlorinated dibenzo-p-dioxin, dibenzofuran, and biphenyl congeners. Toxicol.Appl.Pharmacol. 1999b;159:41–51. doi: 10.1006/taap.1999.8719. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Thorgaard GH. Tetraploidy and the evolution of salmonid fishes. In: Turner BJ, editor. Evolutionary genetics of fishes. New York: Plenum Press; 1984. pp. 1–53. [Google Scholar]
- Andreasen EA, Hahn ME, Heideman W, Peterson RE, Tanguay RL. The Zebrafish (Danio rerio) Aryl Hydrocarbon Receptor Type 1 Is a Novel Vertebrate Receptor. Mol.Pharmacol. 2002a;62:234–249. doi: 10.1124/mol.62.2.234. [DOI] [PubMed] [Google Scholar]
- Andreasen EA, Spitsbergen JM, Tanguay RL, Stegeman JJ, Heideman W, Peterson RE. Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol.Sci. 2002b;68:403–419. doi: 10.1093/toxsci/68.2.403. [DOI] [PubMed] [Google Scholar]
- Andreasen EA, Tanguay RL, Peterson RE, Heideman W. Identification of a critical amino acid in the aryl hydrocarbon receptor. J.Biol.Chem. 2002c;277:13210–13218. doi: 10.1074/jbc.M200073200. [DOI] [PubMed] [Google Scholar]
- Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol.Pharmacol. 2006;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- Elonen GE, Spehar RL, Holcombe GW, Johnson RD, Fernandez JD, Erickson RJ, Tietge JE, Cook PM. Comparative toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin to seven freshwater fish species during early life-stage development. Environ.Toxicol.Chem. 1998;17:472–483. [Google Scholar]
- Evans BR, Karchner SI, Franks DG, Hahn ME. Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: structure, function, evolution, and AHR-dependent regulation in vivo. Arch.Biochem.Biophys. 2005;441:151–167. doi: 10.1016/j.abb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Ferguson MM, Allendorf FW. Evolution of the fish genome. In: Hochachka PW, Mommsen TP, editors. Phylogenetic and biochemical perspectives. Amsterdam: Elsevier Science publishers B.V; 1991. pp. 25–42. [Google Scholar]
- Hahn ME. Dioxin toxicology and the aryl hydrocarbon receptor: insights from fish and other non-traditional models. Mar.Biotechnol.(NY) 2001;3:S224–S238. doi: 10.1007/s10126-001-0045-y. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Evans BR, Franks DG, Merson RR, Lapseritis JM. Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: insights from comparative genomics. J.Exp.Zoolog.A Comp Exp.Biol. 2006;305:693–706. doi: 10.1002/jez.a.323. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. PNAS. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley-Goldstone HM, Grow MW, Stegeman JJ. Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol.Sci. 2005;85:683–693. doi: 10.1093/toxsci/kfi116. [DOI] [PubMed] [Google Scholar]
- Hansson MC, Persson ME, Larsson P, Kjellman C, von Schantz T. Polychlorinated biphenyl load, aryl hydrocarbon receptor, and cytochrome P4501A1 induction in a wild population of Atlantic salmon (Salmo salar) from the Baltic Sea. Environ.Toxicol.Chem. 2006;25:2197–2207. doi: 10.1897/05-416r.1. [DOI] [PubMed] [Google Scholar]
- Hansson MC, Wittzell H, Persson K, von Schantz T. Characterization of two distinct aryl hydrocarbon receptor (AhR2) genes in Atlantic salmon (Salmo salar) and evidence for multiple AhR2 gene lineages in salmonid fish. Gene. 2003;303:197–206. doi: 10.1016/s0378-1119(02)01178-2. [DOI] [PubMed] [Google Scholar]
- Hansson MC, Wittzell H, Persson K, von Schantz T. Unprecedented genomic diversity of AhR1 and AhR2 genes in Atlantic salmon (Salmo salar L.) Aquat.Toxicol. 2004;68:219–232. doi: 10.1016/j.aquatox.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Hornung MW, Spitsbergen JM, Peterson RE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters cardiovascular and craniofacial development and function in sac fry of rainbow trout (Onchorhynchus mykiss) Toxicol.Sci. 1999;47:40–51. doi: 10.1093/toxsci/47.1.40. [DOI] [PubMed] [Google Scholar]
- Isosaari P, Hallikainen A, Kiviranta H, Vuorinen PJ, Parmanne R, Koistinen J, Vartiainen T. Polychlorinated dibenzo-p-dioxins, dibenzofurans, biphenyls, naphthalenes and polybrominated diphenyl ethers in the edible fish caught from the Baltic Sea and lakes in Finland. Environ.Pollut. 2006;141:213–225. doi: 10.1016/j.envpol.2005.08.055. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Ferrario J, Byrne C. Investigation of polychlorinated dibenzo-p-dioxins, dibenzo-p-furans and selected coplanar biphenyls in Scottish farmed Atlantic salmon (Salmo salar) Chemosphere. 2002;47:183–191. doi: 10.1016/s0045-6535(01)00201-6. [DOI] [PubMed] [Google Scholar]
- Jensen BA, Hahn ME. cDNA Cloning and characterization of a high affinity aryl hydrocarbon receptor in a Cetacean, the Beluga, Delphinapterus leucas. Toxicol.Sci. 2001;64:41–56. doi: 10.1093/toxsci/64.1.41. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Hahn ME. AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: tandem arrangement of ahr1b and ahr2 genes. Biochem.J. 2005;392:153–161. doi: 10.1042/BJ20050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Powell WH, Hahn ME. Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AHR Repressor, AHR1, and AHR2. J Biol.Chem. 2002;277:6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Powell WH, Hahn ME. Identification and functional characterization of two highly divergent aryl hydrocarbon receptors (AHR1 and AHR2) in the teleost Fundulus heteroclitus. J Biol.Chem. 1999;274:33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- Long WP, Pray-Grant M, Tsai JC, Perdew GH. Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol.Pharmacol. 1998;53:691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Merson RR, Franks DG, Karchner SI, Hahn ME. Development and characterization of polyclonal antibodies against the aryl hydrocarbon receptor protein family (AHR1, AHR2, and AHR repressor) of Atlantic killifish Fundulus heteroclitus. Comp Biochem.Physiol C.Toxicol.Pharmacol. 2006;142:85–94. doi: 10.1016/j.cbpc.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat.Rev.Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- Poland A, Glover E. Genetic expression of aryl hydrocarbon hydroxylase by 2,3,7,8-tetrachlorodibenzo-p-dioxin: Evidence for a receptor mutation in genetically non-responsive mice. Mol.Pharmacol. 1975;11:389–398. [Google Scholar]
- Pollenz RS, Necela B, Marks-Sojka K. Analysis of rainbow trout Ah receptor protein isoforms in cell culture reveals conservation of function in Ah receptor-mediated signal transduction. Biochem Pharmacol. 2002;64:49–60. doi: 10.1016/s0006-2952(02)01061-4. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol.Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- Roy NK, Courtenay SC, Chambers RC, Wirgin II. Characterization of the aryl hydrocarbon receptor repressor and a comparison of its expression in Atlantic tomcod from resistant and sensitive populations. Environ.Toxicol.Chem. 2006;25:560–571. doi: 10.1897/05-347r.1. [DOI] [PubMed] [Google Scholar]
- Roy NK, Wirgin I. Characterization of the aromatic hydrocarbon receptor gene and its expression in Atlantic tomcod. Arch.Biochem.Biophys. 1997;344:373–386. doi: 10.1006/abbi.1997.0238. [DOI] [PubMed] [Google Scholar]
- Shaw SD, Brenner D, Berger ML, Carpenter DO, Hong CS, Kannan K. PCBs, PCDD/Fs, and organochlorine pesticides in farmed Atlantic salmon from Maine, eastern Canada, and Norway, and wild salmon from Alaska. Environ.Sci.Technol. 2006;40:5347–5354. doi: 10.1021/es061006c. [DOI] [PubMed] [Google Scholar]
- Spitsbergen JM, Kleeman JM, Peterson RE. Morphologic lesions and acute toxicty in rainbow trout (Salmo gairneri) treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. J.Toicol.Environ.Health. 1988;23:333–358. doi: 10.1080/15287398809531119. [DOI] [PubMed] [Google Scholar]
- Spitsbergen JM, Walker MK, Olson JR, Peterson RE. Pathologic alterations in early life stages of lake trout, Salvelinus namaycush, exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin as fertilized eggs. Aquat.Toxicol. 1991;19:41–72. [Google Scholar]
- Tanguay RL, Abnett CC, Heideman W, Peterson RE. Cloning and characterization of the zebrafish (Danio rerio) aryl hydrocarbon receptor. Biochim.Biophys.Acta. 1999;1444:35–48. doi: 10.1016/s0167-4781(98)00252-8. [DOI] [PubMed] [Google Scholar]
- Walker MK, Cook PM, Butterworth BC, Zabel EW, Peterson RE. Potency of a complex mixture of polychloronated dibenzo-p-dioxin, dibenzofuran, and biphenyl congeners compared to 2,3,7,8-tetrachlorodibenzo-p-dioxin in causing fish early life stage mortality. Fund.Appl.Toxicol. 1996;30:178–186. doi: 10.1006/faat.1996.0054. [DOI] [PubMed] [Google Scholar]
- Walker MK, Peterson RE. Potencies of polychlorinated dibenzo-p-dioxin, dibenzofuran, and biphenyl congeners relative to 2,3,7,8-tetrachlodibenzo-p-dioxin, for producing early life stage mortality in rainbow trout (Oncorhynchus mykiss) Aquat.Toxicol. 1991;21:219–238. [Google Scholar]
- Walker MK, Spitsbergen JM, Olson JR, Peterson RE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) toxicity during early life stage development of lake trout (Salvelinus namaycush) Can.J.Fish.Aquat.Sci. 1991;48:875–883. [Google Scholar]


