Abstract
Many bacterial pathogens of plants and animals use a type III secretion system to deliver diverse virulence-associated ‘effector’ proteins into the host cell1. The mechanisms by which these effectors act are mostly unknown; however, they often promote disease by suppressing host immunity2. One type III effector, AvrPtoB, expressed by the plant pathogen Pseudomonas syringae pv. tomato, has a carboxy-terminal domain that is an E3 ubiquitin ligase3. Deletion of this domain allows an amino-terminal region of AvrPtoB (AvrPtoB1–387) to be detected by certain tomato varieties leading to immunity-associated programmed cell death4. Here we show that a host kinase, Fen, physically interacts with AvrPtoB1–387 and is responsible for activating the plant immune response. The AvrPtoB E3 ligase specifically ubiquitinates Fen and promotes its degradation in a proteasome-dependent manner. This degradation leads to disease susceptibility in Fen-expressing tomato lines. Various wild species of tomato were found to exhibit immunity in response to AvrPtoB1–387 and nbot to full-length AvrPtoB. Thus, by acquiring an E3 ligase domain, AvrPtoB has thwarted a highly conserved host resistance mechanism.
Pseudomonas syringae pv. tomato (Pst) causes bacterial speck disease of tomato (Solanum lycopersicum) by using its type III secretion system to deliver about 30 effectors into the plant cell5. Tomato varieties that are immune to speck disease express the Pto kinase that detects either of two Pst effector proteins, AvrPto or AvrPtoB. This detection involves the physical interaction of Pto with AvrPto or AvrPtoB and results in the rapid activation of an array of host defence responses including localized programmed cell death (PCD)6. The Pto gene was isolated from the wild tomato species S. pimpinellifolium and is a member of a small clustered gene family6,7 (Fig. 1a). Embedded within this gene cluster is the Prf gene that encodes a leucine-rich repeat-containing protein that physically interacts with Pto and is required for Pto-mediated immunity6,8. Another member of the Pto gene family encodes the Fen kinase, which shares 80% amino acid identity with Pto but does not recognize AvrPto or AvrPtoB9,10.
Figure 1. The Fen kinase is responsible for the Rsb phenotype.
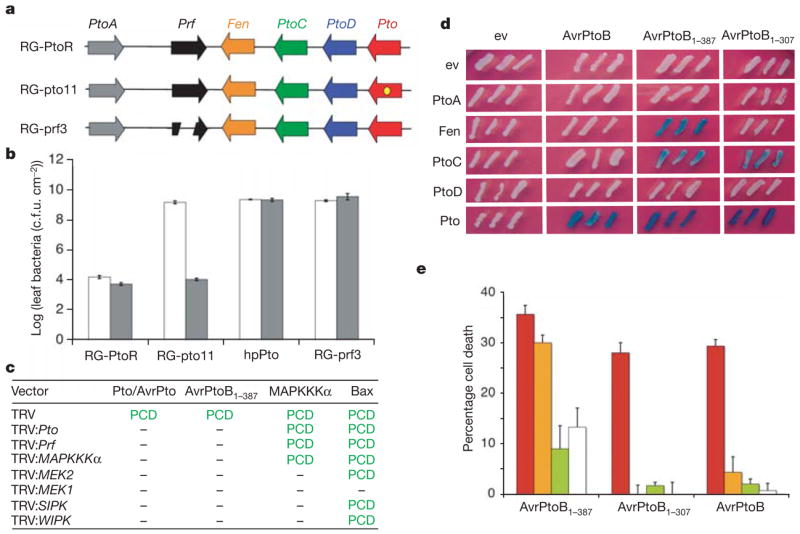
a, Genome organization of the Pto family and Prf from Solanum pimpinellifolium. Rio Grande-PtoR (RG-PtoR) plants are wild type6, RG-pto11 plants have a deleterious point mutation (yellow dot) in Pto, and RG-prf3 plants have a deletion in Prf6. b, Growth of Pst strains delivering AvrPtoB (white) or AvrPtoB1–509 (grey) in RG-PtoR, RG-pto11, RG-prf3 and RG-PtoR(hpPto) leaves. Error bars represent s.e.m. (n = 6). c.f.u., colony-forming units. c, Response after transient expression of Pto with AvrPto, AvrPtoB1–387, mitogen-activated protein kinase kinase kinase (MAPKKK)α or Bax in N. benthamiana leaves silenced for genes in the Pto pathway14 (listed at the left) (see Supplementary Fig. 2). MEK, MAP kinase/ERK kinase; SIPK, salicylic-acid-induced protein kinase; WIPK, wound-induced protein kinase; dashes indicate no PCD. d, Pto family members or empty vector (ev) tested for interaction with forms of AvrPtoB in the yeast two-hybrid system. Blue patches show positive interactions. e, Response of RG-PtoS protoplasts after coexpression of AvrPtoB1–387 with Pto (red columns), Fen (orange columns), PtoC (green columns) or empty vector (white columns). Data are presented as percentage cell death in experimental samples after subtraction of cell death percentages occurring with empty vector. Error bars represent s.e.m. (n = 3).
AvrPtoB is a modular protein4. An N-terminal region (AvrPtoB1–307) is sufficient to elicit Pto/Prf-mediated PCD, whereas a C-terminal region (AvrPtoB388–553) contains a domain that is an E3 ubiquitin ligase3,11. Two truncations of AvrPtoB (AvrPtoB1–509 and AvrPtoB1–387) lacking the E3 ligase domain, or AvrPtoB point mutants compromised in E3 ligase activity, are detected by tomato varieties lacking the Pto kinase and by a tobacco species, Nicotiana benthamiana3,4. This Pto-independent detection leads to PCD, is dependent on Prf and is referred to as Rsb (for ‘resistance suppressed by AvrPtoB C terminus’)4. AvrPtoB1–307 does not elicit Rsb immunity, indicating that a domain between amino acid residues 308 and 387 is required for Rsb-associated recognition (Supplementary Fig. 1). Here we identify the host protein responsible for Rsb and explain the mechanism by which AvrPtoB E3 ligase activity suppresses this recognition.
Dependence on Prf indicated that Rsb might involve a member of the Pto family. Four members of the Pto gene family in S. pimpinellifolium are transcribed in leaves (Fen, PtoC, PtoD and Pto), of which only two, Pto and Fen, encode active kinases12. The kinase activity of Pto is necessary for immunity, leading us to propose that the protein responsible for Rsb would also be an active kinase6. To determine whether a Pto family member is involved in Rsb immunity, we examined a tomato line, RG-PtoR(hpPto), that is a stable transformant knocked down for expression of the Pto gene family by RNA-mediated interference13. RG-PtoR(hpPto) plants were inoculated with Pst DC3000 strains delivering either AvrPtoB or AvrPtoB1–509 (Fig. 1b). In leaves of RG-PtoR(hpPto) plants, both of these strains reached populations similar to that of the control susceptible line, RG-prf3, indicating a complete loss of Pto-mediated immunity and the Rsb phenotype. As expected, RG-PtoR plants were resistant to both Pst strains, whereas RG-pto11, which lacks Pto, showed it had the Rsb phenotype by being resistant to only the strain delivering AvrPtoB1–509. By using virus-induced gene silencing in N. benthamiana, we found that knocked-down expression of the Pto gene family, Prf, or previously described components of the Pto-mediated PCD-associated signalling pathway compromised the Rsb phenotype14 (Fig. 1c, and Supplementary Fig. 2). These results implicated one or more members of the Pto family in Rsb immunity.
There is a strict correlation between immunity conferred by Pto and the ability of this kinase to interact with AvrPtoB in the yeast two-hybrid system10. We proposed that the host protein responsible for Rsb would interact with AvrPtoB1–387 and not with the non-Rsb-eliciting fragment, AvrPtoB1–307. Using the yeast two-hybrid system we tested the Pto family members for interaction with AvrPtoB, AvrPtoB1–387 or AvrPtoB1–307 (Fig. 1d). Of these, only Fen interacted exclusively with AvrPtoB1–387. PtoC interacted with AvrPtoB1–387 but also with AvrPtoB1–307. Pto, as expected, interacted with AvrPtoB and both AvrPtoB truncations, whereas PtoA and PtoD showed no interactions. Western blots confirmed the expression of each of the Pto family proteins and AvrPtoB proteins (Fig. 1d, and Supplementary Fig. 3).
To determine whether Fen or PtoC activates immunity, we expressed each of these proteins with AvrPtoB, AvrPtoB1–387 or AvrPtoB1–307 in protoplasts of tomato RG-PtoS or N. benthamiana (Fig. 1e, Supplementary Fig. 4A). Fen activated PCD when expressed with AvrPtoB1–387 but not with AvrPtoB or AvrPtoB1–307, whereas PtoC was unable to initiate PCD when expressed with any of these AvrPtoB proteins. Western blots confirmed expression of the Pto family proteins (Fig. 1e, and Supplementary Fig. 4B). As expected, Pto activated PCD in tomato when expressed with AvrPtoB, AvrPtoB1–387 or AvrPtoB1–307. The inability of PtoC to activate effector-elicited PCD, its interaction with the non-Rsb eliciting fragment AvrPtoB1–307 and its lack of kinase activity12 excluded a function for this protein in Rsb. Taken together, these data indicate that Fen is responsible for Rsb immunity.
It was possible that the lack of interaction between Fen and AvrPtoB in yeast (Fig. 1d) involved the E3 ligase activity of AvrPtoB. We therefore tested whether Fen interacted with E3 ligase-deficient AvrPtoB mutants. The AvrPtoB-Quad protein (Quad) contains four mutations in critical lysine residues11, and a second mutant, E2BS, has three point mutations at predicted E2-binding sites3. Both AvrPtoB mutants interacted with Fen and PtoC, indicating that the E3 ligase activity interferes with certain protein interactions (Fig. 2a). As expected, the altered proteins interacted with Pto and were unable to interact with PtoA or PtoD.
Figure 2. Fen is ubiquitinated in the presence of AvrPtoB.
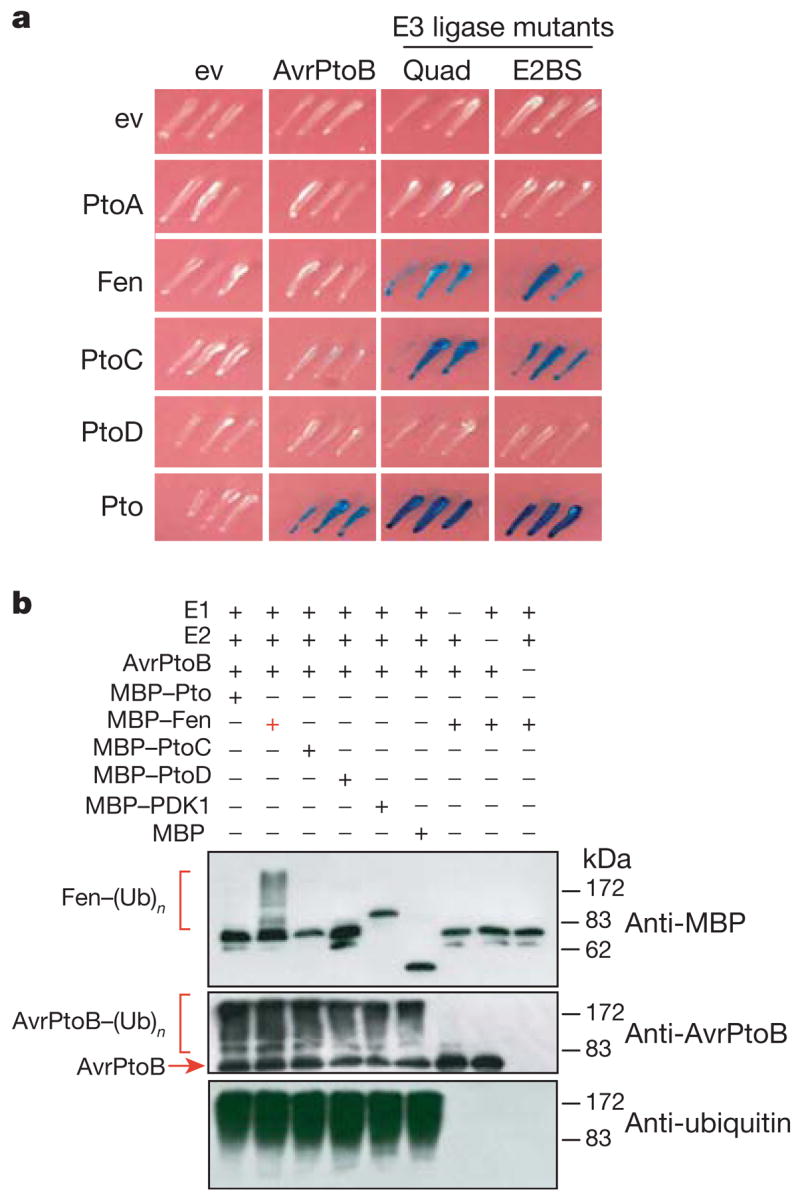
a, Pto family members or empty vector (ev) tested for interaction with AvrPtoB, or AvrPtoB proteins with compromised E3 ligase activity (Quad and E2BS) in the yeast two-hybrid system. Blue patches show positive interactions. b, Immunoblots of an in vitro ubiquitination assay with recombinant E1, E2, AvrPtoB, ubiquitin (Ub) and MBP fusions of the kinases shown. Polyubiquitination of Fen is indicated by (Fen–(Ub)n). Polyubiquitinated forms of AvrPtoB (AvrPtoB–(Ub)n) confirmed E3 ligase activity. An anti-Ub antibody detected the incorporation of ubiquitin in each assay.
We proposed that the AvrPtoB E3 ligase might ubiquitinate Fen to disrupt recognition of the AvrPtoB N-terminal region. To test this possibility, ubiquitination assays were performed in vitro with Fen and AvrPtoB and a series of controls (Fig. 2b). Among five kinases tested, only Fen was ubiquitinated in the presence of AvrPtoB, as indicated by the appearance of high-molecular-mass Fen proteins (Fig. 2b). Absence of E1, E2 or AvrPtoB proteins in the assay abolished the high-molecular-mass forms of Fen (Fig. 2b). In particular, Pto was not ubiquitinated by AvrPtoB in this assay. Fen ubiquitination might be due to unique ubiquitination sites (namely lysine residues) in this kinase. There are only five lysine residues that are present in Fen but absent from Pto and PtoC (Lys 70, Lys 72, Lys 155, Lys 253 and Lys 290; Supplementary Fig. 5A). Of these, Lys 70 and Lys 72 are close to the ATP-binding site (Lys 69) of Fen, raising the possibility that this region might be targeted for ubiquitination. However, arginine substitutions at any one of the five lysine residues had no effect on Fen ubiquitination (Supplementary Fig. 5B), suggesting that either multiple lysine residues are ubiquitinated or other structural differences between Pto and Fen account for the differential ubiquitination.
Ubiquitination of Fen by AvrPtoB raised the possibility that, in the plant cell, AvrPtoB E3 ligase activity might target Fen for degradation. To test this, we expressed Fen, PtoC or Pto with AvrPtoB or Quad proteins in RG-prf3 tomato protoplasts and assessed protein abundance. Fen accumulated poorly in the presence of AvrPtoB, reaching only about 35% of the abundance with the Quad protein (Fig. 3a). In contrast, Pto and PtoC accumulated in the presence of AvrPtoB to levels comparable to their abundance with the Quad protein (Fig. 3a).
Figure 3. In the presence of AvrPtoB, Fen is degraded in a proteasome-dependent manner.

a, Immunoblot to detect accumulation of Fen, Pto or PtoC in RG-prf3 protoplasts in the presence of AvrPtoB or Quad. Visualization of ribulose 1,5-bisphosphate carboxylase–oxygenase (Rubisco) subunits by Coomassie blue confirmed equal loading. The Fen panel shows a roughly 20-s exposure, in contrast with about 10 s for the others; however, the images are from the same experiment. HA, haemagglutinin. b, As in a after treatment with MG132 (minus, dimethylsulphoxide (DMSO) only; plus, with 50 μM MG132). c, As in a with the addition of general plant protease inhibitor (PPI) cocktail (minus, DMSO only; plus, 0.2% PPI).
Ubiquitination often marks a protein for degradation by means of the 26S proteasome. If AvrPtoB targets Fen for degradation, then inhibition of the proteasome should allow Fen to accumulate in the presence of AvrPtoB. In RG-prf3 protoplasts, we expressed Fen or Pto with AvrPtoB or Quad in the presence of MG132, a proteasome inhibitor. Treatment with MG132 resulted in a roughly 80% increase in Fen accumulation when expressed with AvrPtoB (Fig. 3b). MG132 had no effect on Fen coexpressed with Quad or with Pto coexpressed with AvrPtoB or Quad. Similar results were seen with a second proteasome inhibitor, MG115 (data not shown). We tested the effect of a general plant protease inhibitor cocktail on Fen abundance and found no effect on Fen accumulation (Fig. 3c). These results support a function for the AvrPtoB E3 ligase in proteasome-dependent degradation of the Fen kinase.
The specific targeting of Fen by AvrPtoB to suppress Rsb immunity suggested a selective advantage for the N-terminal region of AvrPtoB to acquire and maintain the E3 ligase domain. We therefore examined whether Rsb immunity is conserved among cultivated and wild species of tomato. Cultivars Moneymaker (MM), VFNT Cherry (VFNT-C), and Ailsa Craig (AC) were tested for the presence of Rsb by infiltrating their leaves with two Rsb-eliciting Pst strains (DC3000ΔavrPtoB plus AvrPtoB1–387, or DC3000ΔavrPtoB plus Quad) or two non-Rsb-eliciting strains (DC3000ΔavrPtoB plus AvrPtoB, or DC3000ΔavrPtoB plus AvrPtoB1–307). RG-PtoR, RG-pto11 and RG-prf3 lines were included as controls. Rsb immunity was activated in MM, VFNT-C and AC and, as expected, in RG-pto11 (Fig. 4a).
Figure 4. Rsb is present in many cultivated and wild species of tomato.
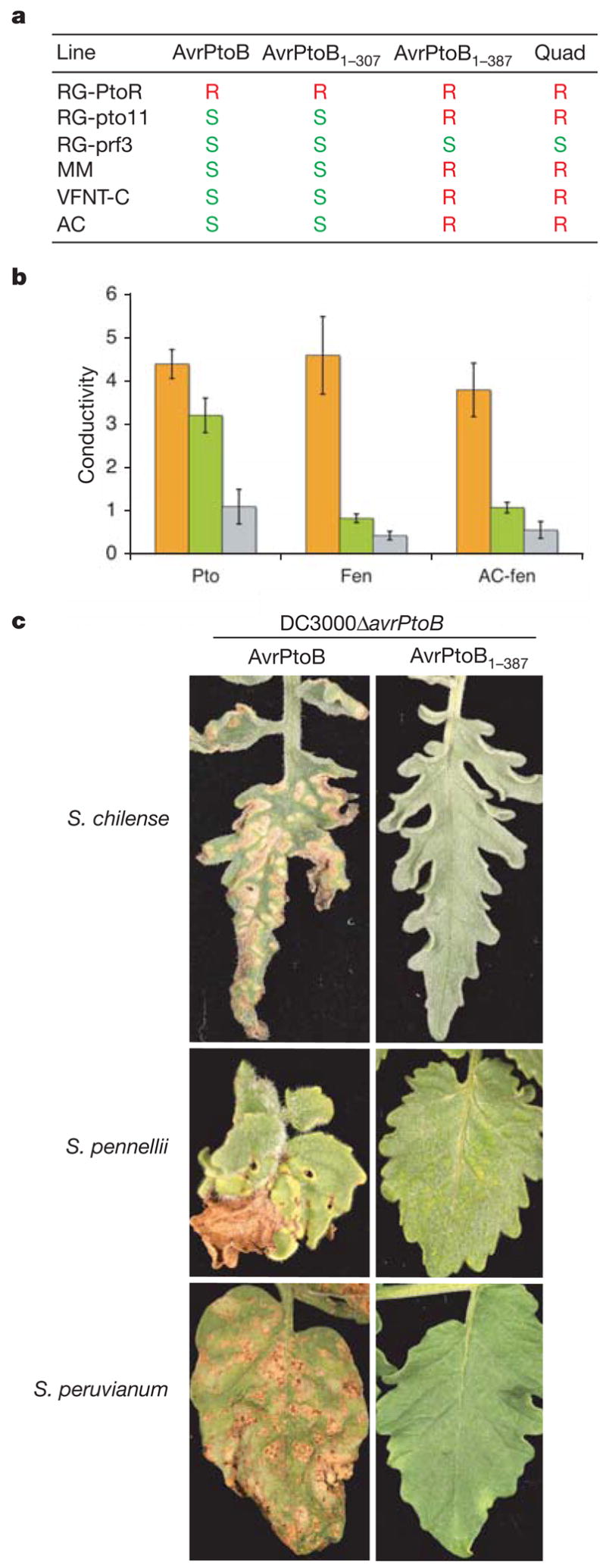
a, Inoculation of cultivated tomato varieties MM, VFNT-C or AC with Pst strains. R and S indicate resistant or susceptible plants, respectively. b, Electrolyte leakage after coexpression of AC-fen, Fen or Pto with AvrPtoB1–387 (orange), AvrPtoB1–307 (green) or AvrPtoB (grey). The ratio of conductivity of samples coexpressing AvrPtoB1–307, AvrPtoB1–387 or AvrPtoB versus empty vector control is shown. Error bars represent s.e.m. (n = 3). c, Leaves from 3 of 21 wild tomato species shown to exhibit Rsb immunity in response to inoculation with Pst delivering AvrPtoB1–387 (see Supplementary Table 1): left, disease; right, Rsb immunity.
To test whether Rsb immunity was activated by a S. lycopersicum orthologue of Fen, we examined a previously described Fen allele from AC (AC-fen)15. As assessed by ion leakage, expression in N. benthamiana leaves of AC-fen with AvrPtoB1–387, but not with AvrPtoB1–307, caused PCD comparable to the expression of Pto or Fen with AvrPtoB1–387, (Fig. 4b). As expected, only Pto initiated PCD when expressed with AvrPtoB1–307. Orthologues of Fen can therefore confer Rsb immunity. A screen of 37 accessions of wild species of tomato revealed that 21 express Rsb immunity but only 5 seem to have Pto-mediated immunity (Supplementary Table 1; Fig. 4c shows three examples). The wide occurrence of Rsb is consistent with phylogenetic analyses indicating that the Fen gene arose before the Pto gene in the Solanum species16 and suggests that the E3 ligase activity of AvrPtoB evolved to circumvent a broadly conserved host resistance mechanism.
The interaction of AvrPtoB1–387 with Fen kinases might indicate that the effector evolved to interfere with a possible function of these kinases in basal defence. In fact, E3 ligase-deficient AvrPtoB proteins are able to suppress certain basal defences17,18. We therefore compared the growth of a type III secretion-defective Pst strain (DC3000ΔhrcQ-U (ref. 19)) in wild-type tomato plants and in a line overexpressing Fen (Supplementary Fig. 6). Similar overexpression experiments have demonstrated a function for the Arabidopsis protein RIN4 as a negative regulator of basal defence20. However, we found no effect of Fen overexpression on growth of the DC3000ΔhrcQ-U strain (Supplementary Fig. 6).
Molecular mimicry of host proteins by bacterial pathogens is common; however, so far only a handful of bacterial proteins are known to manipulate the host ubiquitination system3,21–23. Of these, AvrPtoB, Salmonella SopA, the Shigella IpaH family and Salmonella SspH1 have intrinsic E3 ubiquitin ligase activity, potentially releasing them from dependence on host proteins to secure substrate specificity22,23. Interestingly, IpaH9.8 and SspH1 also ubiquitinate protein kinases, although the relevance of this activity to the host–pathogen interaction has not been established24. Overexpression of the AvrPtoB E3 ligase in leaves suppresses PCD induced by the pro-apoptotic protein Bax and by many, but not all, plant resistance proteins4,17,18,25. AvrPtoB also suppresses PCD in yeast4. Our present data do not explain these general PCD suppression activities; they may be due to a second, highly conserved target of AvrPtoB or to overexpression of the effector. However, our data fully account for the immunity suppression observed with AvrPtoB-expressing Pseudomonas strains on infection of tomato leaves lacking Pto3,4,11. Furthermore, our results suggest that Pto evolved not only to recognize AvrPto and AvrPtoB but also to be invulnerable to AvrPtoB-mediated ubiquitination and subsequent degradation.
Our results can be viewed in the context of the evolutionary processes that may have shaped this pathogen–host interaction (Supplementary Fig. 7). It is known that the N-terminal region of AvrPtoB is able to suppress host basal defences17,18. This region promotes pathogen virulence and therefore an N-terminal-only form of AvrPtoB might have existed independently of the C-terminal E3 ligase domain. We now know that Fen binds the AvrPtoB1–387 region to negate basal defence suppression, activate defence signalling and confer immunity on the host. Immunity mediated by Fen and by Pto requires Prf, suggesting that Prf evolved to function with a progenitor of the Pto family. It is possible this progenitor, unlike Fen, had a function in basal defence. An alternative is that members of the Pto family evolved together with Prf solely for effector-triggered immunity. To counter recognition by Fen, disrupt immunity-associated PCD and restore basal defence suppression activity, the N-terminal region of AvrPtoB may have acquired an E3 ligase domain to mediate the degradation of Fen and Fen orthologues. A similar ability to disrupt immunity has been reported for AvrRpt2, which disrupts signalling by the resistance protein RPM1 in Arabidopsis by cleaving an RPM1-interacting protein, RIN4 (ref. 26). It is possible that some AvrPtoB truncated proteins evolved to evade Fen recognition. For example, P. syringae pv. maculicola expresses an AvrPtoB homologue (HopPmaL27) that lacks both the E3 ligase domain and part of the Fen recognition determinant (residues 307–387). HopPmaL elicits immunity in response to Pto but not Fen28. Pto seems to have evolved to counter both of these strategies. It remains to be determined how Pto evades AvrPtoB-mediated ubiquitination.
METHODS SUMMARY
Plant inoculations
Tomato plants were vacuum infiltrated with Pst (about 5.5 × 104 colony-forming units per ml) suspended in 10 mM MgCl2 and 0.00002% Silwet. Bacterial leaf populations were measured from three plants per treatment, three days after infiltration.
Virus-induced gene silencing (VIGS) and Agrobacterium-mediated transient expression
VIGS was induced by using the tobacco rattle virus vector delivered by Agrobacterium tumefaciens14. For transient gene expression, A. tumefaciens was used to deliver a 35S cauliflower mosaic virus expression cassette (pTEX)4. Ion leakage was measured two days after infiltration with A. tumefaciens.
Yeast two-hybrid assay
A LexA-based two-hybrid system was used to test for interactions between Pto family members in the bait vector and AvrPtoB or AvrPtoB mutants or truncations in the prey vector.
In vitro ubiquitination assay
The in vitro ubiquitination reactions were performed with recombinant maltose-binding protein (MPB)-tagged Pto family proteins, His6-tagged E1, His6-tagged E2, ubiquitin and glutathione S-transferase (GST)-tagged AvrPtoB.
Protoplast bioassays
Protoplasts were isolated from seedling leaves and transformed with pTEX by using polyethylene glycol. Protoplast viability was determined by staining with Evans Blue.
Immunoblotting quantification
After detection of immunolabelled proteins by chemiluminescence, signal intensities were quantified by the Storm blot imaging system and quantified using ImageQuant TL software.
Plant material
Accessions of wild species of tomato were obtained from the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu/).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank K. Munkvold and X. Tang for critical review of the manuscript; B. Randall for assistance with plant inoculations; J. Cohn and P. Pascuzzi for generation and characterization of the RG-PtoR(hpPto) line; J. Li and X. Tang for unpublished data; S. Collier for technical assistance; and our greenhouse staff for plant care. This work was supported, in part, by the NIH, the NSF and the Triad Foundation (G.B.M.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 2.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nature Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 4.Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 2003;22:60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 7.Martin GB, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 8.Mucyn TS, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin GB, et al. A member of the Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994;6:1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 11.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci USA. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JH, et al. Functional analysis of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol Plant Microbe Interact. 2002;15:281–291. doi: 10.1094/MPMI.2002.15.3.281. [DOI] [PubMed] [Google Scholar]
- 13.Pascuzzi PE. PhD thesis. Cornell Univ; 2006. Structure-based functional analyses of Pseudomonas type III effector protein AvrPto and evaluation of putative virulence targets in tomato. [Google Scholar]
- 14.del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004;23:3072–3082. doi: 10.1038/sj.emboj.7600283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Y, Loh YT, Zhou J, Martin GB. Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato and encode active protein kinases. Plant Cell. 1997;9:61–73. doi: 10.1105/tpc.9.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riely BK, Martin GB. Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc Natl Acad Sci USA. 2001;98:2059–2064. doi: 10.1073/pnas.98.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He P, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 18.de Torres M, et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 19.Badel JL, Shimizu R, Oh HS, Collmer A. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact. 2006;19:99–111. doi: 10.1094/MPMI-19-0099. [DOI] [PubMed] [Google Scholar]
- 20.Kim MG, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 22.Angot A, Vergunst A, Genin S, Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathogens. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rytkonen A, Holden DW. Bacterial interference of urbiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Jamir Y, et al. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 2004;37:554–565. doi: 10.1046/j.1365-313x.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman DS, et al. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 28.Lin NC, Abramovitch RB, Kim YJ, Martin GB. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl Environ Microbiol. 2006;72:702–712. doi: 10.1128/AEM.72.1.702-712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.


