Abstract
The Ets family consists of a large number of evolutionarily conserved transcription factors, many of which have been implicated in tumor progression. Extensive studies on this family of proteins have focused so far mainly on the biochemical properties and cellular functions of individual factors. Since most of the Ets factors can bind to the core consensus DNA sequence GGAA/T in vitro, it has been a challenge to differentiate redundant from specific functions of various Ets proteins in vivo. Recent findings, however, suggest that such apparent redundancy may in fact be a central component of a network of differentially regulated specific Ets factors, resulting in distinct biological and pathological consequences. The programmed “Ets conversion” appears to play a critical role during tumor progression, especially in control of cellular changes during epithelial–mesenchymal transition and metastasis. Coordination of multiple Ets gene functions also mediates interactions between tumor and stromal cells. As such, these new insights may provide a novel view of the Ets gene family as well as a focal point for studying the complex biological control involved in tumor progression.
Keywords: Ets, transcriptional regulation, ECM, cancer, invasion, metastasis, EMT, epithelium, stroma
The Ets family of proteins consists of a large number of evolutionarily conserved transcription factors. There are 25 human and 26 murine Ets family members. Ets factors control specific genes that perform critical roles in diverse processes, including cell proliferation, apoptosis, differentiation, lymphoid cell development, angiogenesis, and invasiveness [Sementchenko and Watson, 2000]. To date, studies in the field have largely focused on individual Ets factors and have indeed yielded valuable, albeit fragmented, information. Recent findings, on the other hand, have suggested that several Ets factors may participate in a coordinated program that modulates cell migration and invasiveness, thus affecting tumor progression toward metastasis. In this review, we will examine the recent progress in Ets biology and provide a forum for discussion on a systemic view of Ets functions. Detailed descriptions of Ets proteins and their functions have been provided in several recent reviews [Watson and Seth, 2000; Watson et al., 2001; Dittmer, 2003; Oikawa and Yamada, 2003].
Cancer results from a multi-step series of genetic changes that lead to essential alterations in cell physiology such as loss of growth controls and normal apoptotic response as well as sustained angiogenesis, invasion, and metastasis [Hanahan and Weinberg, 2000]. While it is known that most human tumors are derived from epithelial cells that have undergone multiple genetic alterations [Hanahan and Weinberg, 2000], it is also becoming clear that the alterations in the tumor micro-environment are necessary for tumor progression [Bissell et al., 2002]. For example, stromal cell functions are required for survival and proliferation of epithelial cells. In addition, specific interactions between tumor epithelial cells and endothelial cells are also required for neo-vascularization of the tumor. Accumulated data have shown that Ets factors control pathways not only in epithelial cells, but also within the stromal compartment and the precise regulation of Ets functions in epithelial and stromal cells affects their interaction, both in normal development and in cancer.
The Ets Gene Family
Identification of the v-ets oncogene of the avian leukemia virus, E26, in 1983, led to the discovery of a large family of conserved genes, isolated from phylogenetically divergent species from Drosophila and C. elegans to human [Watson and Seth, 2000; Watson et al., 2001; Oikawa and Yamada, 2003]. Ets proteins are transcription factors that share a conserved winged helix-turn-helix DNA binding domain (ETS domain), recognizing unique DNA sequences containing GGAA/T (Ets binding sites, EBS). Several Ets proteins also contain the Pointed (PNT) domain, important for protein–protein interaction. Ets factors act as positive or negative regulators of the expression of genes that are involved in various biological processes, including control of cellular proliferation, differentiation, hematopoiesis, apoptosis, tissue remodeling, angiogenesis, and transformation [Sementchenko and Watson, 2000; Watson and Seth, 2000; Watson et al., 2001; Dittmer, 2003; Oikawa and Yamada, 2003]. Our recent literature survey allowed identification of over 200 Ets target genes [Sementchenko and Watson, 2000], and the number of genes regulated via EBS is constantly increasing. This number of Ets target genes is between those previously estimated for p53 (200–300 target genes) and for the hormone receptor family (50–100 genes).
Modulation of Ets Function
Protein–protein interaction
Combinatorial interactions of Ets factors with other proteins, including other sequence specific transcription factors, result in context-dependent transcriptional regulation and define target gene specificity (reviewed in [Li et al., 2000a]). Depending on the precise sequence context, binding of an Ets protein near other transcription factors results in higher affinity interaction and synergistic repression or activation of specific target genes. A more recent proteomic approach has identified several Ets-associated proteins (EAPs) that modulate Ets activities through different mechanisms such as blocking DNA binding, subnuclear sequestering, and inhibiting synergistic interaction with co-factors [Li et al., 2000b; Pei et al., 2003].
Regulation by signaling and post-translational modification
Many Ets family transcription factors are end effector molecules of multiple signal transduction pathways. It has been well established that their function can be controlled by phosphorylation-mediated effects on DNA binding, protein–protein interaction, transcriptional activation, and subcellular localization (reviewed in [Yordy and Muise-Helmericks, 2000]). The best characterized Ets modulators are the mitogen-activated protein kinases (MAPK): Erk, JNK, and p38. Erks are activated in response to mitogenic signals, while JNKs and p38/SAPKs (stress-activated protein kinases) respond to stress signals. Specific Ets factors are substrates for some or all of these signal transduction pathways. MAP kinase phosphorylation of ELK1 in its carboxyl-terminal transactivation domain leads to enhanced DNA binding and transcriptional activity. ER81 and ERM are targets of the Ras/Raf/Mek/Erk signaling cascade, whereas SpiB is phosphorylated both by Erks and JNK. Phosphorylation of a MAPK site adjacent to the PNT domain has been shown to positively regulate transcriptional activities of Ets1 and Ets2. Phosphorylation can also affect the sub-cellular localization of Ets proteins. For example, the subcellular localization and function of ERF is controlled by Ras/MAP kinase-mediated signal transduction pathway. Upon mitogenic stimulation ERF is immediately phosphorylated and exported to the cytoplasm. Upon growth factor deprivation ERF is rapidly dephosphorylated and transported back into the nucleus. Another well-studied example of phosphorylation-mediated nuclear localization is the antagonist pair of Drosophila Pointed (Pnt) and Yan proteins. Upon receptor tyrosine kinase (RTK) activation and MAPK phosphorylation, the repressor Yan is exported from the nucleus and degraded. Concurrently, Pnt (the Ets1 homolog) is activated by the same kinase cascade.
Until recently, Ets factors have been considered mainly effectors of the RTK signaling. However, since it has become clear that integrin signaling is also a major contributor to intracellular activity of Erks [Liotta and Kohn, 2001; Comoglio et al., 2003], either through focal adhesion kinase or as co-receptors of various RTKs, the possibility of Ets activities regulated by cell adhesion deserves more attention. Indeed, it has been shown that Erk nuclear translocation and phosphorylation of Elk-1 can be induced by integrin-mediated adhesion [Aplin et al., 2001].
In addition to MAPK-mediated phosphorylation, another well-characterized post-translational modification is mediated by Ca2+/calmodulin-dependent protein kinases (CaMKs), in response to calcium flux. In particular, CaMKII targets six serine residues adjacent to the DNA binding domain (ETS domain) of Ets1. Phosphorylation of these serines is correlated with reduced DNA binding activity due at least in part to stabilization of an autoinhibitory structure.
More recently, it was demonstrated that ER81 was acetylated and activated by p300 and P/CAF in response to HER2/Neu signaling [Goel and Janknecht, 2003]. We also found that Ets1 was modified by acetylation, in response to TGFβ signaling [Czuwara-Ladykowska et al., 2002]. Although a general consensus, if any, of the consequences of acetylation on Ets stability, interaction with other proteins, subcellular localization, DNA-binding affinity, and target gene selectivity remains to be determined, these critical findings broaden the scope of Ets functions in terms of their roles in responding to a wider spectrum of micro-environmental cues. We have also noted that post-translational modification of Ets factors is cell-type specific (M. Trojanowska and D. Watson, unpublished results) and this observation has important implications for the studies of Ets factor functions in epithelial versus stromal compartments.
Ets Target Genes
The highly conserved DNA binding domain of the Ets factors, which is reflected by their universal ability to bind the core DNA target GGAA/T in vitro, makes identifying true target genes of individual Ets factors both critical and challenging. The presence of closely related, but functionally divergent Ets factors such as Ets1, Ets2, and Fli1 suggests that individual Ets members may have evolved unique roles, manifested through the control of specific target genes. Subtle differences in target sites, availability of tissue-specific co-factors, as well as their own tissue distribution and differential response to external signals may contribute to their distinct functions. As such, several key areas are critical for understanding what defines an Ets target gene: first, the functional importance of the EBS must be demonstrated by mutagenesis. Second, the specific Ets factor or factors responsible for transcriptional control of specific target genes need to be identified. One important experimental approach for identifying Ets targets is the creation of knockout mice lacking the function of a single or multiple family members. Analysis of these mice will allow for identification of genes whose expression or repression is dependent upon an Ets family member. To date, 13 of the 26 murine Ets genes (PU.1, SpiB, Ets1, Ets2, Tel, Fli1, PEA3, MEF, ER81, ERM, ESE1, ESE3, and SpiC) have been targeted by homologous recombination. These studies demonstrate important roles for Ets genes in embryogenesis and hematopoiesis [Bartel et al., 2000; Oikawa and Yamada, 2003]. Furthermore, specific in vivo targets for Ets genes have been identified based on the knockout mice: for example, c-mpl [Kawada et al., 2001] and Tie2 [Hart et al., 2000] have reduced expression in knockout Fli1 mice. Third, it is becoming increasingly evident that cellular context defines the direction and magnitude of response to Ets factors. Indeed, recent efforts have lead to discovery of tissue-specific co-factors that modulate transcriptional regulation by Ets factors [Li et al., 2000a,b; Oikawa and Yamada, 2003; Pei et al., 2003].
These considerations, when coupled with improved target-identification methodologies such as chromatin-immunoprecipitation (ChIP) [Wells and Farnham, 2002] accompanied by microarray analysis should greatly improve the confidence in assigning physiologically relevant target genes to specific Ets factors.
Using these improved methodologies, several new insights of Ets functions have emerged. Increased expression of repressor Ets factor METS during late macrophage differentiation, accompanied by reduced expression of activator Ets2, can lead to downregulation of cell cycle control genes through the same EBS [Klappacher et al., 2002]. This study demonstrates that the “redundant” DNA binding capacity of different Ets factors is in fact key to a cell differentiation program. We suspect that such “Ets conversion” programs may represent a new paradigm for physiological and pathological transitions (Fig. 1). Additionally, although it was originally observed that specific Ets factors function either as positive or negative regulators of transcription, it is now quite evident that the same Ets factor may function in either fashion, reflecting promoter and cell context specificity. For example, the putative tumor suppressor Ets factor PDEF is a positive regulator of Maspin and a negative regulator of uPA [Feldman et al., 2003]. Fli1 can function as an activator of TN-C, while acting as a repressor of collagen I (Fig. 1). Furthermore, different Ets factors may have reciprocal functions: Ets1 and Fli1 differentially regulate the collagen α2(I) promoter [Czuwara-Ladykowska et al., 2001; Kubo et al., 2003]. Thus, the precise balance or “regulated imbalance” between cancer/metastasis-promoting and-inhibiting Ets factor activities, which differentially regulate specific target genes, contributes to tissue homeostasis or tumor progression, respectively. Figure 1 provides some specific examples that illustrate the Ets conversion program in promoting epithelial–mesenchymal transition (EMT) and subsequent invasion/metastasis.
Fig. 1.
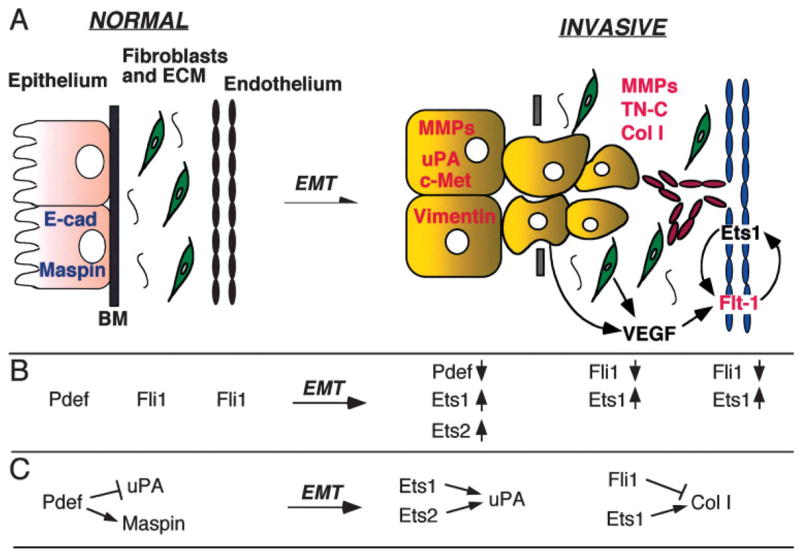
Ets conversion in EMT. Selected examples of Ets factors and target genes are used to illustrate the Ets conversion model during EMT. A: The epithelial and stromal (fibroblasts, ECM, and endothelium) compartments are shown for normal and invasive tissues. Ets targets that are expressed at high levels in normal epithelium are shown in blue. They are down-regulated in the invasive tumor cells. In the invasive phase, several up-regulated Ets target genes are shown in red and in their respective compartments. Tumor-induced neo-vasculature is shown in red. VEGF signaling, emanating from both tumor cells and fibroblasts, is provided as an example for tumor–stroma interaction and feed-back regulatory loop mediated by Ets. B: Examples of different Ets factors that are involved in the Ets conversion program during EMT within different cellular compartments. Arrow direction indicates up-regulation (
 ) or down-regulation (
) or down-regulation (
 ) in invasive cancer. C: Examples of target genes differentially regulated by the indicated Ets factors as a result of Ets conversion in different cellular compartments. Abbreviations: BM, basement membrane; Col I, collagen I; E-cad, E-cadherin; ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; MMP, matrix metalloproteinase; TN-C, tenascin C; uPA, urokinase-like plasminogen activator.
) in invasive cancer. C: Examples of target genes differentially regulated by the indicated Ets factors as a result of Ets conversion in different cellular compartments. Abbreviations: BM, basement membrane; Col I, collagen I; E-cad, E-cadherin; ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; MMP, matrix metalloproteinase; TN-C, tenascin C; uPA, urokinase-like plasminogen activator.
Ets Involvement in Migration, EMT, and Cancer Metastasis
Common to most cancers is the acquisition of new functional capacities that allow: (1) independence from mitogenic/growth signals; (2) loss of sensitivity to “anti-growth” signals; (3) evasion of apoptosis; (4) release from senescence (limitless proliferation); (5) angiogenic conversion; (6) invasiveness and metastasis [Hanahan and Weinberg, 2000]. The last two, sustained angiogenesis and tissue invasion and metastasis, are common features of the most aggressive and lethal tumors. These events require that cells lose contacts with their neighbors, become motile, and invade surrounding areas where they proliferate and undergo further invasive behavior. Thus, while abnormal epithelial growth and differentiation contribute to initial development of cancer, epithelial cells must undergo multiple molecular changes by which they acquire the ability to migrate prior to progression to aggressive cancer. At these later stages of tumor progression, epithelial tumor cells transdifferentiate into mesenchyme, a phenomenon termed EMT. In contrast to epithelia, mesenchymal cells have the ability to invade and migrate through the extracellular matrix (ECM). Thus, EMT is a critical event leading to dissemination and formation of micrometastatic tumor cells, predisposing the conversion from carcinoma in situ (CIS) to invasive and metastatic carcinoma [Thiery, 2002].
Another critical property of the metastatic cells is that they must acquire proliferative capacity at the metastatic site. This means that they need to proliferate while facing different combinations and levels of growth factors as well as new growth substrates.
The involvement of Ets genes in cancer was first demonstrated by the presence of Ets sequence in the oncogenic virus E26. Their importance in carcinogenesis is supported by the observations that human Ets genes are located at translocation breakpoints, are chromosomally deleted or have altered expression patterns in leukemia and solid tumors [Watson and Seth, 2000; Watson et al., 2001; Dittmer, 2003; Oikawa and Yamada, 2003]. Notably, translocation of the Fli1 gene sequence to the EWS locus in Ewing sarcoma results in the transforming EWS-Fli1 chimeric protein, which is a stronger transcription factor than Fli1 and may regulate ectopic target genes. In addition, individual Ets factors are overexpressed or down-regulated in cancers. For example, we and others have shown that Ets2 is over-expressed in prostate and breast cancer and that overexpressed Ets2 is necessary for transformed properties of the cancer cells. Ets1 expression is correlated with more malignant carcinomas and is a negative prognostic indicator [Oikawa and Yamada, 2003]. Conversely, PDEF expression is lost in many epithelial cancers [Feldman et al., 2003].
Among the multiple Ets target genes that are important for cancer progression are those that function in control of cell proliferation (cyclins and cdks), adhesion [cadherins, integrins, cell adhesion molecules (CAMs)], motility/migration (hepatocyte growth factor receptor c-Met, vimentin), cell survival (Bcl-2), invasion (uPA & uPAR, PAI, MMPs, TIMPs, heparanase), extravasation (MMPs, integrins), micro-metastasis [osteopontin, parathyroid hormone-related peptide (PTHrP), chemokines/chemokine receptors (RANTES, MIP-3α), CD44], and establishment and maintenance of distant site metastasis and angiogenesis (integrin β3, VEGF, Flt-1/KDR, Tie2) [Sementchenko and Watson, 2000; Oikawa and Yamada, 2003].
Epithelial–Stromal Cell Interactions and the Tumor Micro-Environment
Tumor metastases are the result of a complex network of interactions between tumor and stromal cells. Tumor- and fibroblast-derived angiogenic factors, such as vascular endothelial growth factor (VEGF), stimulate endothelial cell proliferation and invasion by inducing VPF/VEGF receptors, as well as stimulating production of ECM proteins and proteolytic enzymes from the endothelium. Other tumor-derived factors, such as connective tissue growth factor (CTGF), activate stromal fibroblasts to produce increased amounts of collagen type I, fibronectin and other ECM proteins. Correlation between increased collagen synthesis and poor prognosis was observed for multiple cancers. Activated fibroblasts also contribute to production of growth factors such as TGFβ and matrix-degrading enzymes [Bissell and Radisky, 2001; Mareel and Leroy, 2003]. Therefore, the reactive stroma can be induced and sustained by both tumor and stromal cells. A recent gene profiling analysis [Kang et al., 2003] comparing non-metastatic breast cancer cells and their metastatic derivatives identified CTGF and FGF5 as among only a handful of metastatic signature genes. CTGF is known to have pro-angiogenic as well as profibrogenic properties [Moussad and Brigstock, 2000], and FGF5 was originally isolated also as a factor that sustained NIH3T3 fibroblast growth in the absence of serum. Furthermore, FGF5 knockout mice showed abnormal hair growth [Hebert et al., 1994], again implicating this growth factor as a stromal regulator. Thus, it appears that a significant part of the overall “metastasis program” is targeted at the stroma. Interestingly, among those prominent stroma-modifying factors, many have known linkage to Ets factors.
A particularly significant aspect of the Ets program is that different Ets factors can function in different compartments and mediate epithelial–stromal interactions. For example, Ets1 is a downstream effector of the EMT-promoting hepatocyte growth factor (HGF), emanating from the stroma, while in tumor cells Ets1 and PEA3 can induce the expression of EMT markers such as vimentin and MMPs. Ets1 is also an activator of the HGF receptor c-Met, thus forming a positive feed-back loop. Ets can also mediate similar communication across different tumor and stroma compartments. VEGF, produced by tumor cells and fibroblasts, can induce Ets1 expression in endothelial cells [Lavenburg et al., 2003]. Concomitantly, Ets1, in cooperation with Hif-2a, activates the transcription of VEGF receptor 2 [Elvert et al., 2002]. Both Ets1 and Fli1 are downstream effectors of and are differentially regulated by TGFβ (see below) and these two factors have divergent functions in both fibroblasts and endothelial cells (Fig. 1). CTGF is induced by TGFβ [Moussad and Brigstock, 2000], emanating either from fibroblasts or immune cells within the stroma, although whether this functional correlation is mediated by Ets factors remains to be elucidated. FGF5-induced signaling has not been specifically linked to Ets factors; however, this model is likely as FGF receptor-mediated Ras-MAPK signaling is the prototype Ets modulator.
Significantly, our recent studies have identified two novel Ets targets that implicate Ets functions in stroma remodeling: tenascin-C and collagen type I [Shirasaki et al., 1999; Czuwara-Ladykowska et al., 2001, 2002; Jinnin et al., in press]. Interestingly, the activities of Fli1 and Ets1 toward the expression of these target genes are modulated by acetylation in a TGFβ-dependent manner. These data suggest that Ets1 and Fli1 are the effectors of the TGFβ signaling pathway through novel, previously undescribed regulatory mechanisms. However, while Ets1, in a combinatorial action with Smad 3 and Sp1, activates tenascin-C [Jinnin et al., in press], Fli-1 is a potent repressor of collagen 1 expression. It is therefore likely that the balance between these two Ets factors within the fibroblast population, regulated at least by TGFβ, is critical for ECM homeostasis.
Thus, coordinated functions of Ets factors in epithelial and stromal compartments provide critical control of interplay between these different cell types to modulate tissue home-ostasis (Fig. 1).
PERSPECTIVES
Much of our current understanding of Ets function is based upon in vitro 2-D culture, complemented by knockout and transgenic in vivo models. While these areas of investigation have yielded much information, we now have the opportunity to more directly assess Ets function in specific cellular compartments through two recently developed experimental approaches: in vitro 3-D culture systems and the use of conditional gain-of-function and loss-of-function models.
The use of 3D culture—growing cells in or on gels—has revealed cellular behavior that has never been observed in conventional culture systems [Bissell et al., 2002], such as integrin-mediated cell death (anoikis), tubule formation, matrix-induced cell shape change, etc., that are more relevant to EMT and metastasis. Conceivably, the target gene repertory of Ets factors obtained from gene profiling of cells grown in 3-D culture would be quite different when compared with the existing sets obtained from cells grown in conventional culture conditions. The 3-D system can also be adopted for co-culture studies, enabling assessment of tumor–stromal interactions in vitro. As reagents (e.g., collagens and Matrigel™) and methodology become more accessible, 3-D culture should become an important complement to the standard cell culture system.
For in vivo studies, bitransgenic mice will allow tissue-, tissue compartment- and temporal-specific analyses of gain (e.g., rtTA) or loss of function (loxP/Cre and loxP/CreER). These systems will permit direct assessment of the role of specific Ets factors in epithelial versus stromal compartments. An impressive battery of new strains of mice is constantly being generated and characterized for such studies. These can also be combined with existing or developing models of human cancer to test specific hypotheses in vivo.
Furthermore, genetic screens for modifiers of Ets function will further enhance our understanding of the mechanisms that control the expression of Ets target genes. One important approach is to use genetically accessible model systems. As microarray analyses can identify only Ets downstream genes, genetic modifiers screen can potentially isolate all components relevant to the Ets functions. Particularly, the availability of the complete sequence of the Drosophila genome and the use of Drosophila genetics [Rebay et al., 2000] provide an approach to identify upstream regulators of the Ets activities as well as downstream targets. Once identified, these genes may serve as probes for the identification of relevant mammalian target genes and modulators of Ets transcription factors.
While previous studies have focused on single Ets factors in the context of specific promoters, future studies should consider the functional impact of multiple Ets proteins present within a specific cell type. Multiple Ets factors may be able to control the same genes, albeit at different magnitude or direction of regulation. Inaddition, functional antagonism between different Ets factors and between Ets and other transcription factors has been observed and it is likely to contribute to the final transcriptome. Additional parameters that need to be carefully examined include the magnitude and kinetics (constitutive vs. transient expression) of Ets expression, which have already been shown experimentally to affect hematopoietic lineage selection. Complete assessment of the regulatory network of Ets factors—the Ets conversion program—will also require knowledge of their upstream and downstream effectors. Such a “network” approach should represent an improved biomarker set for tumorigenesis and cancer progression. It will therefore be important to determine whether the composition of Ets factors as well as the collection of upstream effector molecules allows one to predict target gene expression profiles. In turn, future studies may allow prediction of biological response based on Ets factor composition and target gene expression. Through such approaches, upstream and downstream effectors of Ets functions critical for metastasis will be identified, many of which represent novel therapeutic targets.
Acknowledgments
We apologize to the many researchers whose studies could not be cited because of space limitations or was only cited indirectly by referring to reviews or more recent articles.
Grant sponsor: National Institutes of Health; Grant numbers: P01 CA78582, R01 CA102297, RO1 GM057843, R01 AR42334.
References
- Aplin AE, Stewart SA, Assoian RK, Juliano RL. Integrin-mediated adhesion regulates ERK nuclear translocation and phosphorylation of Elk-1. J Cell Biol. 2001;153:273–282. doi: 10.1083/jcb.153.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: Microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio PM, Boccaccio C, Trusolino L. Interactions between growth factor receptors and adhesion molecules: Breaking the rules. Curr Opin Cell Biol. 2003;15:565–571. doi: 10.1016/s0955-0674(03)00096-6. [DOI] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson DK, Trojanowska M. Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. J Biol Chem. 2001;276:20839–20848. doi: 10.1074/jbc.M010133200. [DOI] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Sementchenko VI, Watson DK, Trojanowska M. Ets1 is an effector of the transforming growth factor beta (TGF-beta) signaling pathway and an antagonist of the profibrotic effects of TGF-beta. J Biol Chem. 2002;277:20399–20408. doi: 10.1074/jbc.M200206200. [DOI] [PubMed] [Google Scholar]
- Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29–49. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvert G, Kappel A, Heidenreich R, Englmeier U, Lanz S, Acker T, Rauter M, Plate K, Sieweke M, Breier G, Flamme I. Cooperative interaction of hypoxia inducible factor (HIF)-2a and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J Biol Chem. 2002;278:7520–7530. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003;63:4626–4631. [PubMed] [Google Scholar]
- Goel A, Janknecht R. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 2003;23:6243–6254. doi: 10.1128/MCB.23.17.6243-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, Favier R, Bernstein A. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Rosenquist T, Gotz J, Martin GR. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Jinnin M, Ihn H, Asano Y, Yamane K, Trojanowska M, Tamaki K. Tenascin-C upregulation by TGF-β in human dermal fibroblasts involves Smad3, Sp1 and Ets1. Oncogene. doi: 10.1038/sj.onc.1207064. (in press) [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kawada H, Ito T, Pharr PN, Spyropoulos DD, Watson DK, Ogawa M. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int J Hematol. 2001;73:463–468. doi: 10.1007/BF02994008. [DOI] [PubMed] [Google Scholar]
- Klappacher GW, Lunyak VV, Sykes DB, Sawka-Verhelle D, Sage J, Brard G, Ngo SD, Gangadharan D, Jacks T, Kamps MP, Rose DW, Rosenfeld MG, Glass CK. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell. 2002;109:169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- Kubo M, Czuwara-Ladykowska J, Moussa O, Markiewicz M, Smith E, Silver RM, Jablonska S, Blaszczyk M, Watson DK, Trojanowska M. Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol. 2003;163:571–581. doi: 10.1016/S0002-9440(10)63685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenburg KR, Ivey J, Hsu T, Muise-Helmericks RC. Coordinated functions of Akt/PKB and ETS1 in tubule formation. Faseb J. 2003;17:2278–2280. doi: 10.1096/fj.03-0040fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei H, Watson DK. Regulation of Ets function by protein–protein interactions. Oncogene. 2000a;19:6514–6523. doi: 10.1038/sj.onc.1204035. [DOI] [PubMed] [Google Scholar]
- Li R, Pei H, Watson DK, Papas TS. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000b;19:745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. The microenvironment of the tumour–host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- Moussad EE, Brigstock DR. Connective tissue growth factor: What’s in a name? Mol Genet Metab. 2000;71:276–292. doi: 10.1006/mgme.2000.3059. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK. EAPII interacts with Ets1 and modulates its transcriptional function. Oncogene. 2003;22:2699–2709. doi: 10.1038/sj.onc.1206374. [DOI] [PubMed] [Google Scholar]
- Rebay I, Chen F, Hsiao F, Kolodziej PA, Kuang BH, Laverty T, Suh C, Voas M, Williams A, Rubin GM. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif- containing protein. Genetics. 2000;154:695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sementchenko VI, Watson DK. Ets target genes: Past, present, and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- Shirasaki F, Makhulf HA, LeRoy C, Watson DK, Trojanowska M. Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter. Oncogene. 1999;18:7755–7764. doi: 10.1038/sj.onc.1203360. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Watson DK, Seth A. Ets gene family. In: Jenkins J, Reddy EP, editors. Oncogene reviews. London: Nature Publishing Group; 2000. pp. 6393–6548. [Google Scholar]
- Watson DK, Li R, Sementchenko VI, Mavrothalassitis G, Seth A. The ETS genes. In: Bertino JR, editor. Encyclopedia of cancer. San Diego: Academic Press; 2001. pp. 189–196. [Google Scholar]
- Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]


