Abstract
The general secretory, Sec, system translocates precursor polypeptides from the cytosol across the cytoplasmic membrane in Escherichia coli. SecB, a small cytosolic chaperone, captures the precursor polypeptides before they fold and delivers them to the membrane translocon through interactions with SecA. Both SecB and SecA display twofold symmetry and yet the complex between the two is stabilized by contacts that are distributed asymmetrically. Two distinct regions of interaction have been defined previously and here we identify a third. Calorimetric studies of complexes stabilized by different subsets of these interactions were carried out to determine the binding affinities and the thermodynamic parameters that underlie them. We show here that there is no change in affinity when either one of two contact areas out of the three is lacking. This fact and the asymmetry of the binding contacts may be important to the function of the complex in protein export.
Keywords: SecA, SecB, export, protein interactions, calorimetry
The gram negative bacterium Escherichia coli utilizes two systems to translocate proteins either into or completely across the cytoplasmic membrane. One system, the TAT system, transports proteins that have acquired their final folded structure, whereas the other, the general secretory, or Sec, system can transfer proteins only if they are devoid of stably folded structure. Thus, in addition to a pathway through the membrane, provided by the heterotrimeric SecYEG translocon, the Sec system must capture polypeptides before they fold. SecB, a homotetramer of molecular weight of 69,000, is one of a group of small cytoplasmic chaperones that serves this function. SecB delivers its bound polypeptide ligand to the translocon through specific interaction with SecA, a homodimer of molecular weight of 204,000. SecA functions as a peripheral component of the translocon to provide the energy for translocation through a cycle of binding and hydrolysis of ATP. Understanding the details of the interactions among the proteins involved in the export pathway should help us elucidate the mechanism of the transfer of the polypeptide ligand from SecB to SecA and finally through the translocon. Studies of the complex formed between SecA and SecB led to the unexpected conclusion that, even though each protein displays twofold symmetry, the contacts that stabilize the complex are asymmetric (Fig. 1; Randall et al. 2005). Stabilizing interactions involving two distinct regions of each protein have been defined previously: One set of contacts is between a zinc-containing domain at the extreme C terminus of the SecA protomer and the negatively charged surface of the flat β-sheet formed by each dimer of the SecB tetramer (Fig. 1; Fekkes et al. 1999; Randall et al. 2004). A second interaction involves the extreme C-terminal region of the SecB protomers and the interfacial region of the SecA dimer (Fig. 1; Randall et al. 2004, 2005). The wild-type complex contains a dimer of SecA bound to a tetramer of SecB. If the contact between the zinc domain of SecA and the side of SecB cannot form, then one protomer of SecA dissociates. The inability to make the contact between the zinc domain and side of SecB can arise from mutational changes in SecA or in SecB, or in the case of the wild-type pair can result from competition for the site on SecB by a zinc-containing peptide with the sequence of the last 21 residues of SecA (Randall et al. 2005). The resulting complex has a monomer of SecA bound to a tetramer of SecB (Randall et al. 2005). The striking difference in stoichiometry of these complexes made it of interest to determine the binding constants and the thermodynamic parameters that underlie them.
Figure 1.
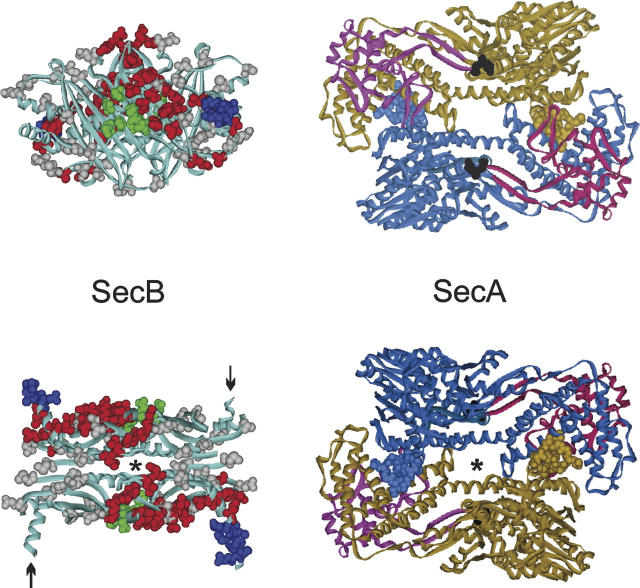
Docking between SecB and SecA. (Left) SecB X-ray crystal structure. The structure in the lower panel is related to the view in the upper panel by a rotation of 90° toward the viewer around the horizontal axis. The residues shown on the structure as CPK models are those analyzed by site-directed spin labeling or shown to be contacts by X-ray crystallography as described in the text. The positions that showed changes in spectral line shape in the spin label study when SecA was added are displayed in red or blue; those that showed no change, in gray. The contacts defined by X-ray crystallography are shown in green. In the lower panel, the C-terminal α-helices shown in blue CPK models emerge from the upper face, while those indicated by arrows emerge from the lower face. The structure is that of Haemophilus influenzae because the C-terminal residues are resolved (Xu et al. 2000). (Right) SecA X-ray crystal structure from Bacillus subtilis (Hunt et al. 2002). The structure in the upper panel has the PPXD (red and magenta) on the upper surface. The structure in the lower panel is related to the upper panel by a rotation of 180° around the horizontal axis and has the PPXD on the lower surface. The C-terminal zinc-domain of SecA was not resolved in the crystal structure but would emerge from the residues displayed as black CPK models. Amino acid residues 1–11 are shown for each monomer as blue and brown CPK models. The axes of symmetry for both SecB and SecA are indicated (*). The views in the lower panel are positioned to facilitate envisioning docking. The SecA would slide over the top of SecB with the symbol indicating the axes of symmetry aligned. The PDB codes are 1M74, SecA; 1FX3, SecB.
Results and Discussion
We used isothermal titration calorimetry in a previous study of complex formation between SecA and SecB to provide evidence for the two distinct regions of interaction (Randall et al. 2004). Two species of SecB, SecBL75Q and SecBD20A, which have mutational changes that prohibit interaction with the C-terminal zinc-containing region of SecA, were shown to bind SecA with an affinity twofold lower than that of wild-type SecB. These complexes, which lacked the interaction with the β-sheet region of SecB, contained a monomer of SecA bound to tetrameric SecB and were stabilized by contacts that involved the C-terminal 13 residues of SecB. A second study (Randall et al. 2005) showed that those C-terminal regions of SecB bind in the interfacial region of the SecA dimer. In addition, even though the wild-type complex has a stoichiometry of one SecA dimer bound to one SecB tetramer, the binding occurs in an asymmetric manner.
Complexes of different stoichiometry
Here we have extended the calorimetric analyses of the complexes that display different stoichiometries. In this work the complex of monomeric SecA bound to tetrameric SecB was generated not by use of SecB variants as in the previous work but by using altered forms of SecA as follows: SecAN880, a truncated version lacking the zinc-containing domain; SecAC4, a full-length version that lacks zinc because the coordinating cysteines are replaced by serines; and SecAdN10, a form of SecA rendered monomeric by deletion of 10 residues from the N-terminal region.
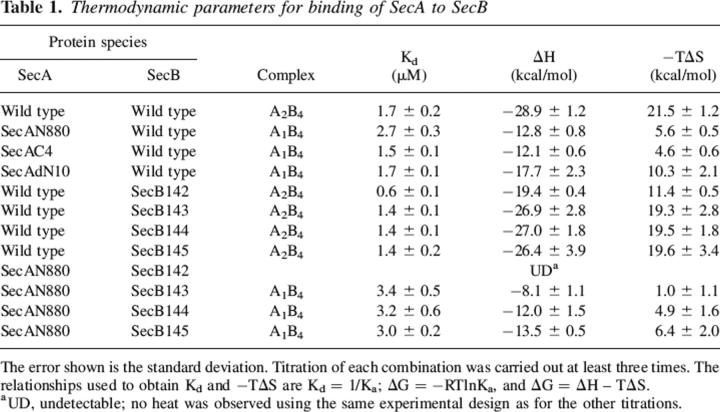
For all combinations of SecA and SecB, titration was performed by successive injections of SecB into a solution of 5 μM SecA dimer held at 8°C in the reaction cell. Each injection resulted in an exothermic heat effect until the SecA in the cell was saturated (Fig. 2, upper panel; SecB into wild-type SecA shown as an example). The reaction heat for each injection was obtained by integration of the deflection from baseline and corrected by subtraction of the integrated heat of dilution. The heats, normalized to the moles of SecB injected, were plotted as shown in Figure 2, lower panel, and the values for the dissociation constant (Kd), change in enthalpy (ΔH), and change in entropy (ΔS) were obtained from the best fit of the data. Table 1 gives a summary of the binding parameters. The complexes have essentially the same dissociation constants whether one or two protomers of SecA are present. In the case of the truncated SecA and the zinc-free SecA, the ability to form side contacts on SecB is lacking, whereas for SecAdN10 those contacts can be made but the interfacial region of SecA is missing and thus the C-terminal region of SecB cannot bind (Randall et al. 2005). In all cases the loss of bonds results in a decrease in favorable enthalpy that is compensated by an increase in favorable entropy such that the free energy of binding does not change significantly.
Figure 2.
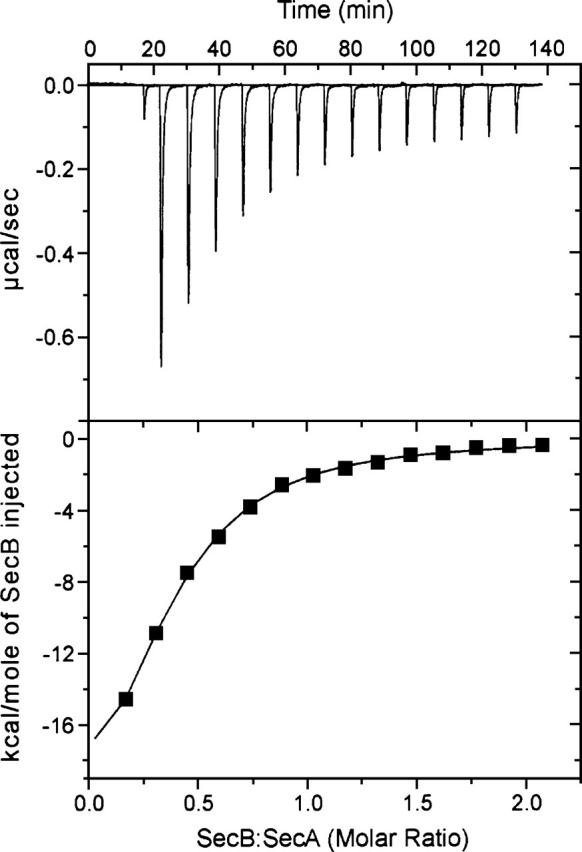
Binding of SecB to SecA. Wild-type SecA was loaded into the cell at a concentration of 5 μM dimer in 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2, 2 mM TCEP (pH 7.6). Wild-type SecB (held in the syringe at 200 μM tetramer) in the same buffer was added in a sequence of 14 injections of 10 μL each after a first injection of 2 μL, which is included to expel any air that might be in the tip of the syringe. The heat of the first injection is not included in the analysis. (Upper) Raw data. (Lower) Integrated area of heat as a function of the molar ratio of the reactants. The points are the experimental data and the solid line is the calculated best fit.
Table 1.
Thermodynamic parameters for binding of SecA to SecB
Contacts with the C-terminal region of SecB
We concluded previously that when all contacts are present within a complex the interactions between the C-terminal region of SecB and the interface of the SecA dimer provide a negative contribution to the overall binding energy. This conclusion was based on the observation that wild-type SecA bound a truncated version of SecB, SecB142 lacking the final 13 residues, more tightly than it bound full-length SecB (Table 1; Randall et al. 2005). Since in the free state the C-terminal regions of SecB are disordered (Volkert et al. 1999; Crane et al. 2005), it is reasonable to postulate that the negative contribution to the binding energy arises from a loss of disorder when the flexible region of SecB is immobilized by interaction with SecA. To determine what portion of the C-terminal region was necessary to result in weakened binding, a set of truncated variants of SecB was generated by incrementally lengthening SecB142 to create SecB143, SecB144, and SecB145. The values for binding enthalpy and entropy for these variants binding to wild-type SecA approached wild-type levels with the addition of a single amino acid residue, Gln143. The dissociation constant for the complex with SecB143 as well as with the longer SecB variants was 1.4 μM, which is very close to that of a complex with full-length SecB (Kd 1.7 μM) as opposed to 0.6 μM for SecB142. The abrupt decrease in affinity is the result of a large increase in unfavorable entropy that is not completely compensated by the increase in favorable enthalpy.
The presence of the C-terminal region of SecB weakens binding to wild-type SecA; however, if the side contact is not formed as in the complex between SecAN880 and SecB, the C termini can provide sufficient energy of binding to form a stable complex. In contrast, SecB that has the C termini removed by truncation at residue 142, SecB142, cannot form a stable complex with SecAN880. No heat was detected by calorimetry (see Table 1), and no complexes were observed by column chromatography or analytical centrifugation (Randall et al. 2005).
Addition of a single residue, Gln143, restored binding to the truncated SecA (see SecAN880 and SecB143 in Table 1). The dissociation constant of this complex was essentially the same as that of the complex with full-length SecB and the longer variants, SecB144 and SecB145. This constant value of the binding-free energy is maintained through enthalpy/entropy compensation. The favorable binding enthalpy increases incrementally from −8.1 kcal/mol to −13.5 kcal/mol as amino acid residues are added, presumably due to an increase in contact surface area. As the enthalpy becomes more favorable, the entropy becomes less favorable, going from a value for −TΔS of 1.0 kcal/mol for SecB143 to 6.4 kcal/mol for SecB145, which is essentially the same as full-length SecB (6.2 kcal/mol).
Enthalpy of binding
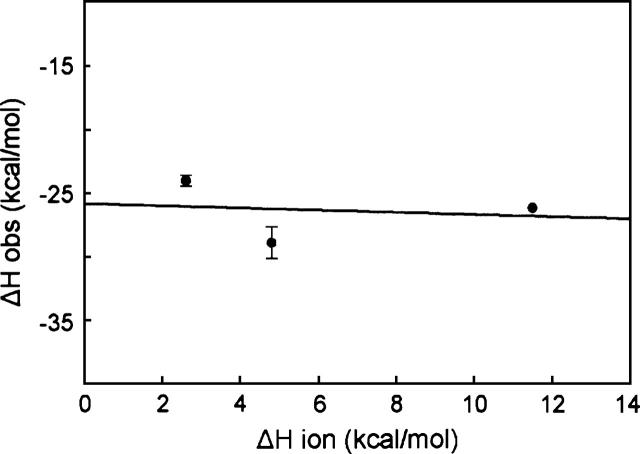
The change in enthalpy measured in these experiments is the total change in heat of the system. After correction for heats of dilution the observed enthalpy could include heat from at least three sources: formation of direct contacts, reorganization of water when the proteins interact, and protonation events coupled to binding. We carried out titrations in buffers that have different heats of ionization to determine whether the observed enthalpy change included protonation events. Figure 3 shows only a slight dependence of the observed ΔH on the buffer used, and thus the heat of ionization does not contribute significantly to the enthalpy change.
Figure 3.
Dependence of observed change in enthalpy on heat of ionization of buffers. The values for ΔHobs were determined in buffers at 8°C (pH 7.6). The buffers used and their corresponding ΔHion at 8°C were PIPES, 2.6 kcal/mol; HEPES, 4.8 kcal/mol; and Tris, 11.5 kcal/mol. The solid line is the linear regression analysis that gives a slope of −0.09 (the number of protons released to solvent per mole of complex formed) and an intercept of −25.8 kcal/mol (the corrected ΔH of binding). Titrations in PIPES and Tris were carried out twice and in HEPES four times. The standard deviation is shown. Note that the standard deviation is contained within the data point for Tris.
Definition of a third area of interaction
Two regions of association between SecA and SecB have been identified by previous studies (Fekkes et al. 1999; Randall et al. 2004, 2005). Evidence for a third area of interaction comes from the observation reported here that addition of a single amino acid residue to SecB142 restored the ability to form a complex between truncated SecB and SecAN880, a pair that lacks both previously defined regions of interaction. We postulate that SecAN880 and SecB142 are in contact with each other but that the surface area of interaction is too small to maintain a stable association. The minimal area of contact required for a stable complex has been estimated to be 1200 Å2, that is, 600 Å2 on each of the binding partners (Chothia and Janin 1975; Jones and Thornton 1995). Addition of the glutamine residue to SecB142 would raise the contact surface area over the 1200 Å2 threshold. That same contact surface would be present in the complex between SecAdN10 and SecB.
The notion that the size of this common area of contact is poised near the threshold for stability is supported by the observation that the alteration of a single amino acid, D20A, on SecB in the site that binds the zinc domain of SecA results in the inability to stably bind SecAdN10 (Fig. 4). Size-exclusion chromatography coupled with static light scatter was used to determine whether complexes were present in mixtures of SecAdN10 and SecB. The position of elution of a protein species depends on hydrodynamic properties that arise from both mass and shape. Therefore, the absolute masses of the eluted species were determined by passing the eluent through a detector for determination of concentration in series with a multiangle static light scatter detector. The intensity of static light scatter is proportional to the product of the concentration and the weight average molar mass of the particles in solution that are scattering the light. For these studies, concentrations were determined using a refractive index detector, since the change in refractive index as a function of protein concentration (dn/dc) is constant for all proteins, whereas the extinction coefficients for SecA and SecB are different. When applied to the column separately, SecAdN10 (Fig. 4, blue) and SecB, either wild type (not shown) or SecBD20A (Fig. 4, pink), each eluted at the same position even though the determined molar masses, 70 kDa for the SecB species and 108 kDa for SecAdN10, were in excellent agreement with the expected masses of 68,610 for SecB (determined by mass spectrometry; Smith et al. 1996) and 101 kDa for SecAdN10 (calculated from the sequence). This observation illustrates the danger of taking position of elution as a measure of mass.
Figure 4.
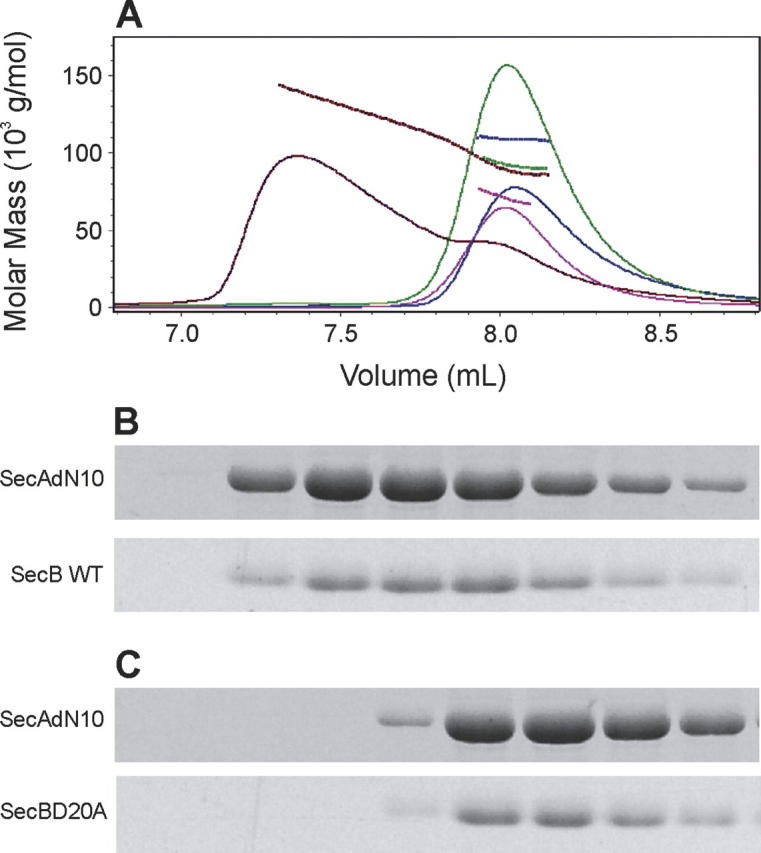
Complexes between SecA and SecB. (A) Protein mixtures were subjected to size-exclusion chromatography and the eluent was monitored to determine protein concentration by change in refractive index (solid traces) and molar mass by light scatter (dashed lines). The samples contained SecAdN10 and SecB wild type (red), SecAdN10 and SecBD20A (green), SecBD20A only (pink), or SecAdN10 only (blue). In each case SecAdN10 was applied at 12 μM monomer and SecBD20A or SecB wild type at 12 μM tetramer. The protein content of each fraction was analyzed by SDS gel electrophoresis. Fractions shown in B and C correspond to the elution profile. (B) SecAdN10 and SecB wild type, red trace in A. (C) SecAdN10 and SecBD20A, green trace in A. The positions of migration of SecAdN10 and SecBD20A or SecB wild type are indicated.
Analysis of a mixture of SecAdN10 and wild-type SecB showed a complex eluting with a molar mass of 143 kDa at the point of the highest concentration where the complex would be most populated. Examination of the protein content of each of the fractions from the chromatograph showed that each contained both SecAdN10 and SecB. No complex was detected when a mixture of SecAdN10 and SecBD20A was analyzed. All protein eluted at the position of the uncomplexed species. The mass determined, 92 kDa, is the weighted average of the masses of the two free species.
Concluding remarks
We are able to assign the third area of contact to a particular region on the surface of SecB by examination of the model of docking between SecA and SecB that is based on a study of the pair using site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy. The proposed docking interface is displayed in Figure 1. It is a compilation of data from the EPR study and from an X-ray structure of SecB in complex with a zinc-containing peptide having the sequence of the C-terminal 27 amino acids of SecA (Zhou and Xu 2003). The residues shown in the crystal structure to form bonds with the zinc domain are displayed as CPK models in green. The residues examined in the EPR study are displayed in gray, blue, and red. Those displayed in red and blue demonstrated restricted movement of the side chain in the complex relative to the mobility of the same residue in free SecB and are considered to be sites of contact. The gray residues are those that showed no change in spectral line shape. Taken together the green, red, and blue residues define the docking interface. The blue residues are those that are removed in the truncated SecB142 and are involved in binding into the interface of SecA. The residues colored red are those that are candidates for the third binding surface. The lower panel of Figure 1 has SecB and SecA positioned to facilitate envisioning how docking might occur. One should envision sliding SecA over the top of SecB so that the twofold axes of symmetry overlay. In this position the blue residues on SecB are aligned under SecA at the proper distance apart so that they could insert into the dimer interface (residues 2 through 10 on SecA shown in blue and brown CPK models). It is important to note that, although this region is α-helical in the X-ray crystal structure, it is disordered in solution as shown by NMR (Volkert et al. 1999) and EPR (Crane et al. 2005). The zinc domains that interact with the green residues on the sides of SecB were not resolved in the structure but would emerge from the lower face of SecA down into the plane of the page and thus could make contact with the sides as SecA straddles SecB. The upper panel of Figure 1 shows SecA rotated 180° so that the position of the zinc domain is clear (indicated by black CPK models). It should be noted that we have not yet determined which regions bind asymmetrically, but not all contacts will be made in any one complex. In addition, the observation of restricted motion of residues by EPR, although indicating that the residues lie on the contact surface, does not give any information about binding energy, which might be derived from those contacts.
The large change in enthalpy observed when SecA binds SecB might arise not only from direct contacts but also from structural rearrangements. Comparison of crystal structures of SecA shows that the protomer of SecA within a dimer has a different conformation from that of a monomeric form of SecA (Hunt et al. 2002; Osborne et al. 2004). A domain (PPXD, indicated in red and magenta in Fig. 1) shown to bind the precursors of exported polypeptides (Kimura et al. 1991; Kourtz and Oliver 2000; Baud et al. 2002; Papanikou et al. 2005) is closely packed onto the main body of the protein in the dimer, but in the monomeric form it is rotated 60°, thereby opening a groove that is large enough to accommodate a precursor polypeptide. We have proposed that, in the complex between SecA and SecB, binding of the C-terminal regions of SecB disrupts the interface of the SecA dimer and allows SecA to undergo a similar conformational change. The opening of the groove would provide a pathway for transfer of the precursor ligand from SecB to SecA during active export. Such a change in conformation could be accompanied by changes in enthalpy as a consequence of structural rearrangement of solvent (Chervenak and Toone 1994). It has been shown that reactions involving solvent reorganization, particularly in hydrogen-bonding solvents, have a propensity toward enthalpy/entropy compensation (Grunwald and Steel 1995; Liu and Guo 2001; Sola et al. 2005). The complete enthalpy/entropy compensation observed upon interaction of the series of incrementally lengthened SecB species with SecAN880 supports the interpretation that the change in enthalpy results from solvent reorganization. Chervenak and Toone showed that enthalpy changes that result from solvent reorganization are strongly correlated with the change in heat capacity (ΔCp) of the system. Unfortunately we cannot determine the ΔCp for SecB binding to SecA because SecA is thermally unstable.
Although the heat of binding for SecA and SecB is large, it is counteracted by a large unfavorable entropy, and thus the binding affinity for the complexes is modest. The dissociation constants are in the micromolar range. Such modest binding affinity is necessary for proper physiological function of SecA and SecB. During the process of export, SecB undergoes a cycle of binding and release of its binding partners. SecB displays two types of binding. In addition to the specific protein interaction with SecA described here, SecB acts as a chaperone and binds promiscuously to many precursor polypeptides. The ternary complex of SecB:precursor:SecA engages the membrane-associated translocon SecYEG via the affinity of SecA for SecY. During translocation the precursor must be passed from SecB to SecA and then through the channel formed by SecYEG. The interactions of the proteins must not be so tight that the efficiency of the cycle is compromised. Although the affinity of SecB as a chaperone for unstructured polypeptides is high (Kd in the range of 10–50 nM, Randall et al. 1998), the binding is characterized by high on and off rates (Hardy and Randall 1991; Fekkes et al. 1995; Randall and Hardy 1995). The precursor samples the free state but is rebound before it folds into a stable structure. In the ternary complex, transfer between SecB and SecA could be readily achieved if the binding groove in SecA opened to accept the polypeptide at the time of dissociation from SecB.
The unusual properties of interaction between SecA and SecB, that the binding contacts are asymmetric and that the loss of either one of two contact areas out of the three described here results in no decrease in affinity, may play a role during transfer of the precursor polypeptide from SecB to SecA. Perhaps transfer is achieved in a stepwise manner by making and breaking contacts in different regions without complete dissociation. As we proposed earlier (Randall et al. 2005) the two protomers of SecA in the dimer may act sequentially. SecB might first insert its C-terminal region into one of the two symmetrically related contacts between the residues that make interfacial contacts in the SecA dimer (Fig. 1, blue and brown CPK models) and the opposite surface to allow one protomer to open a binding cleft for a precursor polypeptide. Subsequently, the symmetrically related interfacial region might be opened. The transfer presents a topological challenge since the precursor wraps around the surface of the chaperone and when SecA docks it straddles the groove formed at the interface of the dimers of SecB, which has been proposed to be the binding site for the precursor. Previous studies have come to conflicting conclusions as to whether SecA functions as a monomer (Or et al. 2002, 2005) or as a dimer (de Keyzer et al. 2005; Jilaveanu et al. 2005; Randall et al. 2005; Jilaveanu and Oliver 2006). Perhaps the difference lies in whether or not transfer from SecB is involved.
The small chaperone SecB (69,000 MW) must make use of much of its surface to bind its ligands. It has been shown for three precursor ligands that a stretch of ∼150 amino acids of the polypeptides are in contact with the chaperone (Topping and Randall 1994; Khisty et al. 1995; Smith et al. 1997). Such binding would impose asymmetry on the ligand-occupied SecB. In this state it may be that only one of the two binding sites for the zinc domain of SecA would be accessible. Even so, because of the properties of the interaction, the affinity of SecA for SecB would not be compromised. Deciphering the details of this dynamic interaction will require further research.
Materials and methods
Protein purification
The proteins were purified using published procedures as described (Randall et al. 2005). The construction of the strains harboring plasmids for the SecA variants has been described: SecAN880 (Woodbury et al. 2000); SecAdN10, SecAC4 (Ramamurthy and Oliver 1997). Concentrations of SecA species were determined spectrophotometrically at 280 nm using coefficients of extinction (for monomer) as follows: 78,900 M−1 cm−1 for SecA and SecAdN10; 78,937 M−1 cm−1 for SecA lacking the cysteines; 77,200 M−1 cm−1 for SecAN880. The zinc content of purified SecA, determined spectrophotometrically as described (Zhou et al. 1999), was >85% for wild-type SecA and SecAdN10. SecAN880 and the SecA lacking the zinc-coordinating cysteines contained insignificant levels of zinc. Concentrations of SecB species were determined spectrophotometrically at 280 nm using 47,600 M−1 cm−1 as the coefficient of extinction for SecB tetramer. Matrix-assisted laser desorption/ionization mass spectrometry indicated that all purified proteins were quantitatively intact with the exception of wild-type SecA, which was ∼85% intact with 15% missing 7–9 amino acids from the N terminus.
Titration calorimetry
All titrations were carried out using the VP-ITC titration calorimeter (Microcal, Inc.) and the Origin software supplied with the instrument. The “one set of sites” curve-fitting model supplied with the software was used to determine Kd, ΔH, and ΔS. In all cases SecA was held at 8°C in the cell at 5 μM dimer and was titrated with SecB held in the syringe at 200 μM tetramer. SecB was added in a sequence of 14 injections of 10 μL each, spaced at 500-sec intervals. Determination of the heat of dilution for each species of SecB was carried out identically except that SecA was omitted from the reaction cell. The standard buffer used was 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2, 2 mM Tris-(2-carboxyethyl) phosphine hydrochloride (TCEP) (pH 7.6). In experiments to determine heat of protonation the solutions contained the same salts and TCEP buffered at pH 7.6 with PIPES, Tris, or HEPES. The values used for the change in enthalpy of ionization (ΔHion) at 8°C were calculated using values for changes in enthalpy and heat capacity for deprotonation of the buffers taken from the literature (Morin and Freire 1991; Fukada and Takahashi 1998).
Size-exclusion chromatography and molar mass determination
High performance liquid chromatography was performed on a TSK G3000SW (TosoHaas) size-exclusion column (7.5 mm inner diameter × 60 cm) equilibrated in 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2 (pH 7.6), at 7°C. The absolute molar mass of proteins was determined directly using static light scatter by passing the eluent through a multi-angle laser light scatter detector followed by a differential refractometer (DAWN-EOS and Optilab, respectively; Wyatt Technology Corp.). The molar mass was determined using a specific refractive index increment (dn/dc) of 0.182 mL/gm and the Debye plotting formalism of the Astra software supplied with the instrument. The relationship between the weight average molar mass (Mw) and the excess Rayleigh ratio R(θ) at the low protein concentrations used here is given by:
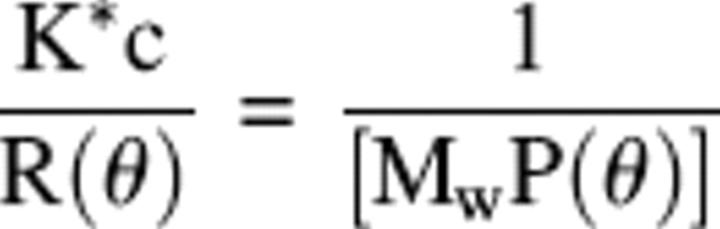 |
where R(θ) is the light scattered by the solution at angle θ in excess of that scattered by pure solvent divided by the incident light intensity, c is the concentration of protein, P(θ) is the form factor that describes the angular dependence of the scatter, and K* is a constant dependent on the parameters of the system used in the study. Further details are given in Woodbury et al. (2002). Fractions were collected and a portion of each (4%) was subjected to SDS-polyacrylamide electrophoresis using 14% polyacrylamide gels as described (Randall et al. 1998). The gels were stained with Coomassie brilliant blue.
Acknowledgments
We thank Gseping Liu for help in purifying SecA, Angela A. Lilly for construction of the SecB variants, Hilary C. Vasel for the analysis by SDS gel electrophoresis, and Beverly DaGue of The Proteomics Center at the University of Missouri for MALDI mass spectrometry. SecAC4 is the kind gift of Donald B. Oliver. We are grateful to Jennine M. Crane for preparing and critically reading the manuscript. We thank Elizabeth Howell for drawing our attention to the work of Chervenak and Toone. This work was supported in part by NIH research grant GM29798 (L.L.R.) and an endowment from the Hugo Wurdack Trust at the University of Missouri (L.L.R.).
Footnotes
Reprint requests to: Linda L. Randall, Department of Biochemistry, 117 Schweitzer Hall, University of Missouri, Columbia, MO 65211, USA; e-mail: craneje@missouri.edu; fax: (573) 882-5635.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062141006.
References
- Baud C., Karamanou S., Sianidis G., Vrontou E., Politou A.S., Economou A. 2002. Allosteric communication between signal peptides and the SecA protein DEAD motor ATPase domain J. Biol. Chem. 277 13724–13731. [DOI] [PubMed] [Google Scholar]
- Chervenak M.C. and Toone E.J. 1994. A direct measure of the contribution of solvent reorganization to the enthalpy of ligand binding J. Am. Chem. Soc. 116 10533–10539. [Google Scholar]
- Chothia C. and Janin J. 1975. Principles of protein–protein recognition Nature 256 705–708. [DOI] [PubMed] [Google Scholar]
- Crane J.M., Mao C., Lilly A.A., Smith V.F., Suo Y., Hubbell W.L., Randall L.L. 2005. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: Insight into the mechanism of ligand transfer during protein export J. Mol. Biol. 353 295–307. [DOI] [PubMed] [Google Scholar]
- de Keyzer J., van der Sluis E.O., Spelbrink R.E., Nijstad N., de Kruijff B., Nouwen N., van der Does C., Driessen A.J. 2005. Covalently dimerized SecA is functional in protein translocation J. Biol. Chem. 280 35255–35260. [DOI] [PubMed] [Google Scholar]
- Fekkes P., den Blaauwen T., Driessen A.J. 1995. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB Biochemistry 34 10078–10085. [DOI] [PubMed] [Google Scholar]
- Fekkes P., de Wit J.G., Boorsma A., Friesen R.H., Driessen A.J. 1999. Zinc stabilizes the SecB binding site of SecA Biochemistry 38 5111–5116. [DOI] [PubMed] [Google Scholar]
- Fukada H. and Takahashi K. 1998. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride Proteins 33 159–166. [PubMed] [Google Scholar]
- Grunwald E. and Steel C. 1995. Solvent reorganization and thermodynamic enthalpy-entropy compensation J. Am. Chem. Soc. 117 5687–5692. [Google Scholar]
- Hardy S.J.S. and Randall L.L. 1991. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB Science 251 439–443. [DOI] [PubMed] [Google Scholar]
- Hunt J.F., Weinkauf S., Henry L., Fak J.J., McNicholas P., Oliver D.B., Deisenhofer J. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA Science 297 2018–2026. [DOI] [PubMed] [Google Scholar]
- Jilaveanu L.B. and Oliver D. 2006. SecA dimer cross-linked at its subunit interface is functional for protein translocation J. Bacteriol. 188 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilaveanu L.B., Zito C.R., Oliver D. 2005. Dimeric SecA is essential for protein translocation Proc. Natl. Acad. Sci. 102 7511–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. and Thornton J.M. 1995. Protein–protein interactions: A review of protein dimer structures Prog. Biophys. Mol. Biol. 63 31–59. [DOI] [PubMed] [Google Scholar]
- Khisty V.J., Munske G.R., Randall L.L. 1995. Mapping of the binding frame for the chaperone SecB within a natural ligand, galactose-binding protein J. Biol. Chem. 270 25920–25927. [DOI] [PubMed] [Google Scholar]
- Kimura E., Akita M., Matsuyama S., Mizushima S. 1991. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli J. Biol. Chem. 266 6600–6606. [PubMed] [Google Scholar]
- Kourtz L. and Oliver D. 2000. Tyr-326 plays a critical role in controlling SecA-preprotein interaction Mol. Microbiol. 37 1342–1356. [DOI] [PubMed] [Google Scholar]
- Liu L. and Guo Q.X. 2001. Isokinetic relationship, isoequilibrium relationship, and enthalpy-entropy compensation Chem. Rev. 101 673–695. [DOI] [PubMed] [Google Scholar]
- Morin P.E. and Freire E. 1991. Direct calorimetric analysis of the enzymatic activity of yeast Cytochrome C Oxidase Biochemistry 30 8494–8500. [DOI] [PubMed] [Google Scholar]
- Or E., Navon A., Rapoport T. 2002. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane EMBO J. 21 4470–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E., Boyd D., Gon S., Beckwith J., Rapoport T. 2005. The bacterial ATPase SecA functions as a monomer in protein translocation J. Biol. Chem. 280 9097–9105. [DOI] [PubMed] [Google Scholar]
- Osborne A.R., Clemons W.M. Jr., Rapoport T.A. 2004. A large conformational change of the translocation ATPase SecA Proc. Natl. Acad. Sci. 101 10937–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikou E., Karamanou S., Baud C., Frank M., Sianidis G., Keramisanou D., Kalodimos C.G., Kuhn A., Economou A. 2005. Identification of the preprotein binding domain of SecA J. Biol. Chem. 280 43209–43217. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V. and Oliver D. 1997. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding J. Biol. Chem. 272 23239–23246. [DOI] [PubMed] [Google Scholar]
- Randall L.L. and Hardy S.J.S. 1995. High selectivity with low specificity: How SecB has solved the paradox of chaperone binding Trends Biochem. Sci. 20 65–69. [DOI] [PubMed] [Google Scholar]
- Randall L.L., Topping T.B., Suciu D., Hardy S.J.S. 1998. Calorimetric analyses of the interaction between SecB and its ligands Protein Sci. 7 1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L.L., Crane J.M., Liu G., Hardy S.J.S. 2004. Sites of interaction between SecA and the chaperone SecB, two proteins involved in export Protein Sci. 13 1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L.L., Crane J.M., Lilly A.A., Liu G., Mao C., Patel C.N., Hardy S.J.S. 2005. Asymmetric binding between SecA and SecB two symmetric proteins: Implications for function in export J. Mol. Biol. 348 479–489. [DOI] [PubMed] [Google Scholar]
- Smith V.F., Schwartz B.L., Randall L.L., Smith R.D. 1996. Electrospray mass spectrometric investigation of the chaperone SecB Protein Sci. 5 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V.F., Hardy S.J.S., Randall L.L. 1997. Determination of the binding frame of the chaperone SecB within the physiological ligand oligopeptide-binding protein Protein Sci. 6 1746–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola M., Battistuzzi G., Borsari M. 2005. Modulation of the free energy of reduction in metalloproteins Chemtracts 18 73–86. [Google Scholar]
- Topping T.B. and Randall L.L. 1994. Determination of the binding frame within a physiological ligand for the chaperone SecB Protein Sci. 3 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert T.L., Baleja J.D., Kumamoto C.A. 1999. A highly mobile C-terminal tail of the Escherichia coli protein export chaperone SecB Biochem. Biophys. Res. Commun. 264 949–954. [DOI] [PubMed] [Google Scholar]
- Woodbury R.L., Topping T.B., Diamond D.L., Suciu D., Kumamoto C.A., Hardy S.J.S., Randall L.L. 2000. Complexes between protein export chaperone SecB and SecA. Evidence for separate sites on SecA providing binding energy and regulatory interactions J. Biol. Chem. 275 24191–24198. [DOI] [PubMed] [Google Scholar]
- Woodbury R.L., Hardy S.J.S., Randall L.L. 2002. Complex behavior in solution of homodimeric SecA Protein Sci. 11 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Knafels J.D., Yoshino K. 2000. Crystal structure of the bacterial protein export chaperone SecB Nat. Struct. Biol. 7 1172–1177. [DOI] [PubMed] [Google Scholar]
- Zhou J. and Xu Z. 2003. Structural determinants of SecB recognition by SecA in bacterial protein translocation Nat. Struct. Biol. 10 942–947. [DOI] [PubMed] [Google Scholar]
- Zhou Z.S., Peariso K., Penner-Hahn J.E., Matthews R.G. 1999. Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli Biochemistry 38 15915–15926. [DOI] [PubMed] [Google Scholar]