Abstract
Progress toward understanding the pathogenesis of cystic fibrosis (CF) and developing effective therapies has been hampered by lack of a relevant animal model. CF mice fail to develop the lung and pancreatic disease that cause most of the morbidity and mortality in patients with CF. Pigs may be better animals than mice in which to model human genetic diseases because their anatomy, biochemistry, physiology, size, and genetics are more similar to those of humans. However, to date, gene-targeted mammalian models of human genetic disease have not been reported for any species other than mice. Here we describe the first steps toward the generation of a pig model of CF. We used recombinant adeno-associated virus (rAAV) vectors to deliver genetic constructs targeting the CF transmembrane conductance receptor (CFTR) gene to pig fetal fibroblasts. We generated cells with the CFTR gene either disrupted or containing the most common CF-associated mutation (ΔF508). These cells were used as nuclear donors for somatic cell nuclear transfer to porcine oocytes. We thereby generated heterozygote male piglets with each mutation. These pigs should be of value in producing new models of CF. In addition, because gene-modified mice often fail to replicate human diseases, this approach could be used to generate models of other human genetic diseases in species other than mice.
Introduction
Animals are an important resource in biomedical research. Mice with genetic alterations, in particular, have been responsible for many advances in our understanding of human disease pathogenesis and in the development of treatments and preventative measures (1). However, for many diseases, mice fail to replicate the human phenotype. One example is cystic fibrosis (CF). CF is an autosomal recessive disease caused by mutations in the gene encoding the CF transmembrane conductance regulator (CFTR) (2, 3). CFTR is a regulated anion channel expressed predominantly in epithelia (4). More than 11 different CF mice have been generated by targeting the mouse CFTR gene, yet none of them develop the lung or pancreatic disease that causes most of the mortality and morbidity in humans with CF (5, 6). The obvious differences in size, anatomy, biochemistry, and physiology as well as differences between the mouse and human genomes are likely responsible. The limitations of CF mice as a model have impeded understanding of disease pathogenesis and the development of novel therapeutic and preventive strategies.
Progress toward developing other gene-targeted animal models has been hindered by the inability to apply mouse embryonic stem (ES) cell technology to other species. Despite many attempts in other species, functional ES cells that can form chimeras and contribute to the germ line have been developed only for mice (1, 7). This hurdle was overcome in part with the development of somatic cell nuclear transfer (SCNT) and the production of a cloned sheep in 1997 (8). In the ensuing 10 years, numerous animal species have been cloned using similar methods (9–16).
Another limiting factor has been difficulty in genetically modifying cells. Unlike mouse ES cells, classical homologous recombination in somatic cells results in extremely low gene targeting efficiencies (17, 18). Homologous recombination strategies often employ a promoter-trap strategy that improves gene targeting efficiency by decreasing the number of random integration events. This strategy requires that the target gene be expressed in the nuclear donor cell. Because fetal fibroblasts are one of the few cell types known to be suitable for nuclear transfer, the gene of interest must be expressed in those cells for that approach to work. An additional limitation of primary somatic cells is their relatively short life span in culture; they often senesce before the intended gene modifications can be identified. Nuclear transfer efficiency also falls when donor cells undergo extensive culturing (19); to be suitable nuclear donors, it is thought that somatic cells should not exceed 30 population doublings (20).
The strategy for delivering the targeting vector also influences gene targeting efficiency. Electroporation and lipid-mediated transfection can deliver the targeting construct to a high percentage of cells, but they deliver a large quantity of targeting vector, resulting in many random, nonhomologous integrations (21, 22). As a result, it can be necessary to test a large number of antibiotic-resistant cells to find the targeted cell. Nuclear microinjection has yielded favorable gene targeting ratios, likely due to delivery of the targeting vector directly to the nucleus (23). However, it is difficult to microinject a sufficient number of cells in a timely manner.
Because of these barriers, the number of successfully targeted genes reported in mammals other than mice is small. They include disruption of the α-1,3-galactosyltransferase gene in pigs (24, 25), the prion protein gene (PRNP) in cows and goats (26, 27), and the immunoglobulin μ (IGHM) gene in cows (28). In addition, a transgene was targeted into a noncoding region of the α1 procollagen gene in sheep, but it did not prevent type I collagen expression (29). None of these attempted to reproduce a human genetic disease.
As a new model for CF, we chose the pig because it has been extensively used in biomedical research, and it offers potential advantages. Its lungs share many anatomical, histological, biochemical, and physiological features with human lungs (30–32). Porcine lungs have also been used as a model for pulmonary abnormalities that are relevant to CF, including infection and inflammation (33, 34). Additional evidence of the similarity between porcine and human organs is the major effort to develop them for xenotransplantation (35). The reproductive characteristics of swine also recommend them as a model. Their relatively fast maturation rate and the large number of offspring generated from a single sow in 1 year allow a colony to rapidly expand. Furthermore, the longevity of pigs offers advantages for investigating the pathogenesis of CF, the long-term therapeutic efficacy of pharmaceuticals and gene therapy, and adverse effects of interventions that might only be revealed with time.
We wished to generate pigs with 2 different alterations in their CFTR gene, a null allele and the ΔF508 mutation. A null allele would lack any CFTR function. It should therefore prove useful for assessing the porcine CF phenotype, for comparing the consequences of other CF-associated mutations, for exploring pathogenesis, and for evaluating many therapeutic strategies. The ΔF508 mutation deletes Phe508 and is the most common CF-associated mutation, accounting for approximately 70% of CF alleles (36). In humans, this mutation disrupts processing of the protein, so that nearly all CFTR-ΔF508 is retained in the endoplasmic reticulum (ER) and degraded, preventing maturation to the plasma membrane. In addition, this deletion reduces the activity of single CFTR channels and shortens their lifetime on the cell surface (37–39). Earlier work showed that reducing the incubation temperature and other interventions allowed some of the mutant protein to escape the ER and traffic to the cell surface, where it retained significant activity (40). These findings plus the prevalence of the ΔF508 mutation have driven efforts to correct the CFTR-ΔF508 defects (41, 42). We recently found that porcine CFTR-ΔF508 showed at least partial processing in vitro (43). A pig with the ΔF508 mutation could be of value for understanding the mechanisms responsible for the CFTR-ΔF508 biosynthetic defects in vivo and for developing pharmacological agents to correct the CFTR-ΔF508 biosynthetic defects. To begin developing these porcine models of CF, we combined gene targeting and SCNT.
Results
Fetal pig fibroblasts express little CFTR.
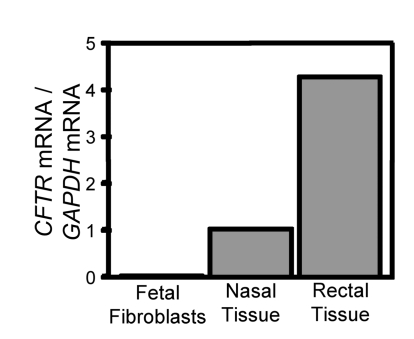
We worked with fetal fibroblasts from domestic pigs (Sus scrofa), since they have been used successfully for transgenic SCNT (44). Because a promoter-trap strategy was previously used in porcine fibroblasts (24), we asked if CFTR is expressed in fetal fibroblasts. We used quantitative RT-PCR and compared the results to transcript levels in nasal and rectal epithelia, which are known to express CFTR at low levels (45). Figure 1 shows that the primary fibroblasts produced very little CFTR mRNA. This result prevented the use of a promoter-trap strategy, as was done for the only other gene targeted in pigs (24, 25).
Figure 1. CFTR expression in pig fetal fibroblasts.
Data are quantitative RT-PCR of pig CFTR mRNA relative to GAPDH in primary pig fetal fibroblasts, nasal epithelia, and rectal epithelia. Similar results were obtained on 2 other occasions.
Developing vectors to target the pig CFTR gene.
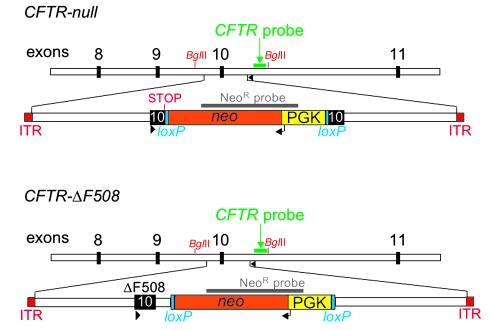
We designed a “null” targeting construct to disrupt CFTR exon 10 with a neomycin resistance cassette (NeoR) (Figure 2). Because CFTR can exhibit some alternative splicing, we chose to disrupt exon 10, which encodes a portion of nucleotide-binding domain 1; this exon is required for CFTR function. We also included an engineered stop codon at position 508. Therefore, F508X would be expected to trigger nonsense-mediated mRNA decay as well as prematurely interrupt any translation of CFTR. The ΔF508 targeting vector was designed to delete residue Phe508 (Figure 2). We also inserted a NeoR in the intron downstream of exon 10 as a positive selection marker. In this vector, NeoR was flanked by loxP sites so that it could be removed at a later time if it was found to markedly reduce the level of the CFTR-ΔF508 mRNA, a situation encountered in some attempts to make a CFTR-ΔF508 mouse (46, 47).
Figure 2. Schematic of targeting constructs for homologous recombination for CFTR-null and CFTR-ΔF508.
Exons 8–11 of pig CFTR are depicted as black boxes. NeoR contains a neomycin resistance cDNA (orange) driven by the phosphoglycerate kinase (PGK) promoter (yellow) and flanked by loxP sites (blue). The engineered stop codon is indicated in the CFTR-null targeting vector. The inverted terminal repeats (ITRs) are at both ends of the targeting construct. Position of probes for NeoR and CFTR Southern blots are indicated. PCR screen primers are depicted as arrowheads. BglII sites are indicated. Not drawn to scale.
We initially used nuclear microinjection and then electroporation to deliver the null targeting vector to fetal fibroblasts. However, we recovered no clones with homologous recombination, and we abandoned these approaches. We then investigated adeno-associated virus–mediated (AAV-mediated) gene targeting, which has been used to deliver targeting vectors to cell lines and primary cells (48–51). Using an AAV vector has the advantage that it delivers single-stranded DNA to the nucleus, the amount of DNA per cell is small, and it can infect many cell types (52). To first determine which AAV serotypes could infect pig fetal fibroblasts, we infected them with EGFP-expressing recombinant AAV1 (rAAV1), -2, and -5 (each with AAV2 inverted terminal repeats [ITRs]). Each rAAV infected the cells with at least 50%–80% efficiency, however rAAV1 appeared to infect nearly 100% of cells (data not shown). Because of rAAV genome size constraints, the total length of the targeting vectors was limited to approximately 4.5 kb. NeoR was centrally located in both vectors (Figure 2).
rAAV vectors introduced the CFTR-null and CFTR-ΔF508 alleles.
We obtained fetal fibroblasts from males so that all our clones would be male; that would allow us to more rapidly expand the number of animals. Primary cultures of pig fetal fibroblasts were infected with rAAV1, carrying the null targeting vector. After 24 h, cells were transferred to a series of 96-well plates. Approximately 2 weeks later, cells in each well of the 96-well plates were passaged by splitting among 3 plates: 96-well culture plates for cell expansion, 96-well culture plates for potential cryopreservation, and 96-well PCR plates for cell lysis. We screened cell lysates by PCR to identify wells containing gene-targeted clones (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI34773DS1) and then hybridized lysates with a NeoR-specific probe to test for inclusion of this marker (Supplemental Figure 1B). We then froze positive clones; by that time, cells had been in culture 15–17 days. We also passaged positive clones from the “cell expansion” plates to generate DNA for genotype determination. Southern blots with CFTR- and NeoR-specific probes identified clones with a targeted CFTR allele that were free of random integration (Supplemental Figure 2). On average, 75% of PCR-positive clones were also positive by Southern blot and were clonal.
We used identical procedures to introduce the CFTR-ΔF508 construct and screen for homologous recombinants. We identified numerous PCR-positive clones (Supplemental Figure 3A) that were confirmed by Southern blotting with a ΔF508 allele-specific probe (Supplemental Figure 3B). Eighteen of 25 (72%) PCR-positive clones contained the F508 deletion. The other 28% failed to contain the ΔF508 deletion, suggesting that gene targeting had occurred, but crossing over was downstream of F508 deletion. Subsequent Southern blots revealed CFTR-ΔF508–targeted clones (Supplemental Figure 4, A and B).
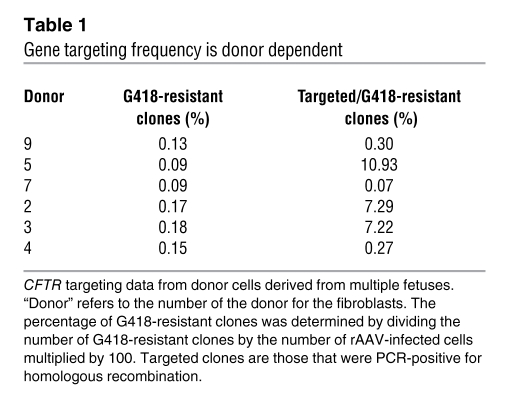
Variability in homologous recombination depended on the donor.
Over the course of these studies, we targeted the CFTR gene in fibroblasts derived from several fetuses. The fetuses were all siblings harvested from the same uterus at the same time. Yet, surprisingly, we saw a striking fetus-to-fetus variability in targeting frequency (Table 1). Even when fibroblasts from different fetuses were infected and screened at the same time, with the same reagents and by the same people, pronounced differences occurred; an example is fetus 5 versus fetus 7 in Table 1. These results suggest the difference was not due to experimental process.
Table 1 .
Gene targeting frequency is donor dependent
SCNT produced gene-targeted piglets.
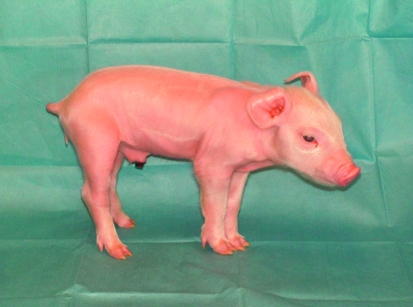
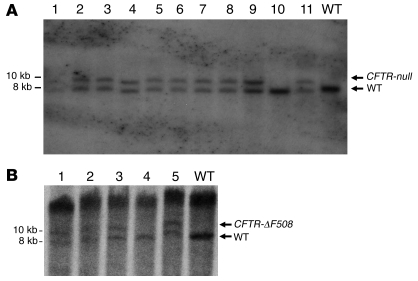
To produce heterozygote pigs, we used the CFTR-null–targeted fetal fibroblasts as nuclear donors for transfer to enucleated oocytes. Then to each of 8 surrogate females, we transferred between 94 and 144 SCNT embryos. At 117–118 days of gestation (full term), we delivered piglets by cesarean section. Five surrogates produced 10 males; 3 surrogates did not produce offspring. Figure 3 shows the first CFTR+/– piglet. Southern blots revealed that 9 of the 10 offspring were CFTR-null heterozygotes and 1 was wild type (Figure 4). The CFTR+/– males reached sexual maturity, and they sired numerous litters of heterozygote offspring, both males and females.
Figure 3. Photo of the first CFTR+/– piglet taken at 1 day of age.
Figure 4. Southern blot of genomic DNA from CFTR-targeted pigs.
BglII-digested genomic DNA was hybridized with a probe that detects pig CFTR downstream of the targeting vector boundary, shown in Figure 2. CFTR-null and CFTR-ΔF508–targeted alleles produced an approximately 9.7-kb band, and the wild-type band is approximately 7.9 kb. (A) CFTR-null. Lanes 1–11 contain DNA from individual cloned pigs. Note that pig 10 was wild type. WT well contains DNA from a wild-type control. (B) CFTR-ΔF508. Lanes 1–5 contain DNA from individual cloned pigs. Note that pig 4 was wild type. WT well contains DNA from a wild-type control.
In addition, each of 4 surrogates received 103–185 CFTR-ΔF508 SCNT embryos. Five males were recovered from 3 surrogates on days 116–117. Southern blots revealed that 4 were CFTR-ΔF508 heterozygotes and 1 was a wild type. The CFTR-ΔF508 males have not yet reached sexual maturity. All of the CFTR+/– and CFTR+/ΔF508 were phenotypically normal.
The ΔF508 allele, but not the null allele generated mRNA.
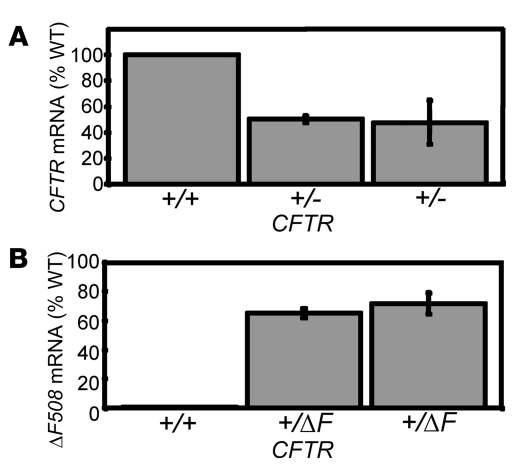
We asked whether the targeted alleles were transcriptionally active in an epithelium, in which CFTR is normally expressed. We biopsied rectal epithelia and measured CFTR mRNA using quantitative RT-PCR. In CFTR+/– animals, mRNA was present at approximately 50% of wild-type levels (Figure 5A). We cannot be certain that the remaining mRNA arose from the nontargeted allele; however, the result is consistent with disruption of 1 CFTR allele and nonsense-mediated mRNA decay.
Figure 5. CFTR mRNA expression in CFTR+/– and CFTR+/ΔF508 pigs.
(A) Quantitative RT-PCR was used to measure wild-type CFTR mRNA levels in rectal epithelial samples from CFTR+/– and wild-type pigs. (B) Quantitative RT-PCR was used to measure ΔF508-CFTR mRNA relative to wild-type mRNA levels in CFTR+/ΔF508 and wild-type pigs. Error bars represent SD.
To assess the influence of the NeoR that resides in the intron downstream of exon 10, we used probes specific for wild-type CFTR and CFTR-ΔF508. CFTR-ΔF508 mRNA was present at 65%–70% of wild-type levels (Figure 5B). This expression level suggests that the retained NeoR has only minimal effects on transcription. Moreover, this amount of transcript is likely sufficient to produce relatively normal amounts of CFTR-ΔF508 protein.
Discussion
Better animal models are needed for many human genetic diseases, including CF. SCNT opened the door for many different species to be considered, but a current limiting step has been generating genetically modified nuclear donor cells. We have developed a gene targeting approach that combines AAV and an efficient screen to successfully target the CFTR gene in pig fetal fibroblasts. Importantly, we have made both null (knockout) and ΔF508 (knockin) modifications, demonstrating the utility and repeatability of this strategy. Another article in this issue describes a similar approach to generate CFTR-null heterozygote ferrets (53). In doing so, we have overcome several obstacles that have hindered progress.
One challenge has been homologous recombination, which is generally thought to be more efficient at transcriptionally active loci (25, 54). In addition, promoter-trap strategies exploit active loci (17). However, CFTR and many other human disease genes are either not expressed or expressed at very low levels in fetal fibroblasts. By using AAV as a vector, we were able to target a nonexpressed disease gene in somatic cells. The fact that AAV delivers single-stranded targeting DNA to the nucleus in low numbers may have overcome many of the inefficient steps in homologous recombination.
Another challenge in gene targeting primary cells for use as nuclear donors is to perform all the necessary steps before they senesce or undergo so many cell doublings that they can not be reprogrammed. SCNT efficiency falls considerably after 30 cell doublings (20); this is thought to be related in part to altered DNA methylation as cells divide. With increasing numbers of cell divisions, a point is reached at which the cells can no longer be reprogrammed to an embryonic-like state. The pig fetal fibroblasts doubled about every 24–36 h. With our procedure, cells were infected, selected, screened, and frozen in 17 days, well within the safe window of operation.
We found substantial fetus-to-fetus variability in gene targeting efficiency. One potential explanation is that homologous recombination occurred efficiently with only 1 allele inherited from the parents of the fibroblast donor. This might occur if the targeting vector shared greater identity with that 1 allele. In that case, the allele that was preferentially targeted would be expected to be present in 50% of the fetal lines. Indeed, we saw good targeting rates in 50% of the lines tested. This speculation would be consistent with some observations that isogenic DNA may be targeted with greater efficiency than nonisogenic DNA (28). Another possibility is that the fetal fibroblasts express different amounts of the cellular machinery required for homologous recombination. We are currently investigating these possibilities. A better understanding of the factors that influence AAV-mediated gene targeting in primary somatic cells could further enhance the ability to specifically modify their genomes.
We did not attempt to identify a phenotype in the CFTR-null or CFTR-ΔF508 heterozygote pigs. Noninvasive procedures, such as measuring the voltage across nasal airway epithelia (55–57) or measuring sweat chloride concentrations (55, 56), do not distinguish people who are heterozygotes for a CFTR mutation from people who possess wild-type CFTR at both alleles. CFTR heterozygotes show intermediate rates of cAMP-stimulated sweat secretion relative to CF and wild type (58, 59). However, the large variability prevents interpretation from a small number of measurements.
The development of CFTR-null and CFTR-ΔF508 heterozygotes will allow new opportunities to understand CF and to develop novel therapeutic strategies. The methods we describe should also enable the production of other new animal models of human disease.
Methods
Fetal fibroblasts.
Fetal fibroblasts were isolated from day 35 fetuses as previously described (20). Cells were grown at 39°C in F10 media (Invitrogen), containing 20% FCS and 30 μg/ml gentamicin. Fetus gender was determined by PCR amplification of the Y chromosome–specific Sry gene (60).
Genomic clone construction.
Genomic DNA was isolated (Puregene; Gentra) from pig fetal fibroblasts. A 5,683-bp PCR product, including CFTR exon 10 and flanking intronic sequence, was amplified from pig fetal fibroblast genomic DNA, using primers GC1F and GC8R (for primer sequences see Supplemental Table 1) and a high fidelity polymerase (PfuUltra; Stratagene). Primers were designed based on the domestic pig genomic sequence from the NIH Intramural Sequencing Center (NISC) Comparative Vertebrate Sequencing Project (Genbank accession no. AC092478 and AC092497). This PCR product was subcloned into pCR-Blunt II-TOPO (Invitrogen), verified by sequencing (using primers GC1F–GC8R; Supplemental Table 1), and served as the template for PCR amplification of 5′ and 3′ targeting arms. This plasmid is referred to as pG16.
CFTR-null targeting vector construction.
Using PCR, the 5′ and 3′ homologous recombination arms were amplified from pG16 and sequentially subcloned upstream and downstream of the NeoR in p–phosphoglycerate kinase-Neo-I (p-PGK-Neo-I) (a generous gift from Tim Ley, Washington University in St. Louis, St. Louis, Missouri, USA; Genbank accession no. AF335419), such that the NeoR is in the opposite orientation to the CFTR sequence. The following primers were used: 5′ arm, G16-Neo5′F-XhoI and G16-Neo5′R-EcoRV; 3′ arm, G16-Neo3′F-BamHI and G16-Neo3′R-HindIII (Supplemental Table 1). The NeoR consists of a NeoR cDNA driven by the PGK promoter and is flanked by loxP sites. In the resulting construct, the NeoR disrupts CFTR exon 10 immediately after an in-frame stop codon that was introduced to follow isoleucine 507. Thymidine 1531 is effectively deleted, becoming the first nucleotide of the stop codon. This targeting construct is referred to as pG16-Neo.
CFTR-ΔF508 targeting vector construction.
The CFTR-ΔF508 targeting vector was constructed in a similar way, using the following primers: 5′ arm, dF-Neo 5′F-XhoI and dF-Neo 5′R-EcoRV; 3′ arm, dF-Neo 3′F-BamHI and dF-Neo 3′R-HindIII (Supplemental Table 1). To recreate the ΔF508 mutation seen in humans, cytidine 1521 and thymidines 1522 and 1523 were deleted from exon 10 using PCR mutagenesis. This targeting construct is referred to as pdF-Neo.
rAAV production.
The targeting vector sequences were amplified from pG16-Neo and pdF-Neo by PCR to include flanking SbfI sites and were subcloned into the rAAV2 proviral plasmid, pAV2 (ATCC 37216). Because of rAAV genome size constraints, the total length of the targeting vectors is approximately 4.5 kb with the NeoR centrally located (G16-Neo, 5′ targeting arm = 1,510 bp; 3′ targeting arm = 1,274 bp; NeoR = 1,706 bp and dF-Neo, 5′ targeting arm = 1,475 bp; 3′ targeting arm = 1,296 bp; NeoR = 1,706 bp). The only viral sequences present in the targeting vector are the ITRs, and they would only be inserted into the porcine genome at the site of targeting or in the case of random integration. We rule out the former with PCR sequencing and the latter with the Southern blots (Supplemental Figures 2 and 3). pAV2-G16-Neo was grown in SURE2 cells (Stratagene) and purified via a CsCl2 method (61). rAAV1 (with AAV2 ITRs) was prepared as previously described (62). Helper-free virus stocks were treated with nuclease and purified by high-performance liquid chromatography. Physical titers of rAAV were determined by slot-blot hybridization. These viruses are referred to as AAV-G16-Neo and AAV-dF-Neo.
Infection and selection.
Fetal fibroblasts (1.5 × 106) were thawed and plated on a 100 mm collagen-coated culture dish. Twenty-four h later, cells were infected with AAV-G16-Neo or AAV-dF-Neo (200 μl, 2.5 × 1012 particles/ml). Twenty-four h later, cells were trypsinized and transferred to forty-eight 96-well collagen-coated plates (BD Biosciences). Forty-eight h later, G418 (100 μg/ml) was added to the cell media. Ten days later, each well was trypsinized (60 μl trypsin, 0.5% EDTA) and split among 3 different vessels. For cell freezing, one-third of the cells were transferred to 96-well collagen-coated culture dishes and returned to the incubator for growth. For cell propagation, one-third of the cells were transferred to 96-well collagen-coated culture dishes and returned to the incubator for growth. For PCR screening, one-third of the cells were transferred to 96-well PCR plates.
PCR screen and PCR Southern blot.
Cells in the 96-well PCR plates were spun down and resuspended in lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-Cl, pH 8.5, 0.5% Nonidet P40, 0.5% Tween, 400 μg/ml Proteinase K) (29). Most wells (~70%) contained only dead cell debris following selection, but all wells were processed to minimize human error. The cells were lysed for 30 min at 65°C, followed by 10 min at 95°C, and 1 μl of lysate was used in a 50 μl PCR reaction. PCR conditions were as follows: 2 min at 95°C, 30 cycles of 95°C for 20 s, 56°C for 20 s, and 68°C for 4 min, and then 68°C for 5 min. Primers Ex10a5F and GC7R (Supplemental Table 1) were expected to amplify a 2.0-kb product from wild-type alleles and a 3.7-kb product from G16-Neo–targeted alleles. PCR products were electrophoresed on 1.0% E-Gel 96 gels (Invitrogen). Positive PCR reactions were also electrophoresed on standard 1.0% agarose gels and transferred to a nylon membrane. The membranes were probed with biotin-labeled NeoR-specific or ΔF508-allele–specific oligonucleotides and detected by chemiluminescence (North2South; Pierce).
Processing screen-positive cells.
Following identification of PCR-positive clones, the corresponding cells from the “freezing” plate were grown to confluence (~10,000 cells). Cells were detached with 60 μl trypsin, and 20 μl of detached cells were placed into each of 3 cryovials. Three hundred microliters freezing media was added to each cryovial, and the vials were transferred to an isopropanol cryofreezing container at –70°C. After 24 h, the vials were transferred to liquid nitrogen. The corresponding cells from the propagation plate were transferred to 24-well plates, and subsequently to 6-well and 100 mm culture dishes. The sequential transfer to increasingly larger culture dishes was carried out to achieve consistent cell growth and viability.
Southern blotting.
For CFTR-null targeting, genomic DNA was isolated from 100 mm dishes (Gentra) and 10 μg was digested with BglII overnight. For CFTR-ΔF508 targeting, genomic DNA was isolated from 24-well dishes. Ten nanograms were used for whole genome amplification (Repli-G; Qiagen), and 25 μg amplified DNA was digested with BglII overnight. Genomic digests were electrophoresed on a 0.7% agarose gel and transferred to a positively charged nylon membrane (Roche) by using an alkaline transfer procedure. Blots were prehybridized for 15 min at 65°C in Rapid-hyb buffer (Amersham). The blot was then hybridized in Rapid-hyb buffer with a 32P-labeled probe specific for a region of CFTR that is outside of the targeting vector boundaries. For Neo-specific probing, blots were either stripped or, in most cases, the BglII digest and Southern blot procedure was repeated using a 32P-labeled NeoR-specific probe.
Preparation of donor cells for SCNT.
Frozen aliquots of CFTR-targeted cells were thawed at 37°C and prewarmed in F-10 medium (Invitrogen) with 20% FCS. The cells were washed twice by centrifugation and cultured (F-10, Invitrogen; 20% FCS, Hyclone; gentamicin, 2.5 ng/ml, FGF and G418, Invitrogen) for 1–2 days in 24-well collagen-coated plates (35-4408, Biocoat cellware). Confluent cells were dispersed with 0.05% trypsin/EDTA for 3–5 min at 38.5°C and 500 μl F-10 with 20 % FBS, followed by centrifugation twice at 850 g for 5 min. The supernatant was removed, and the cells were resuspended in micromanipulation medium (25 mM HEPES TCM 199, Gibco; 0.3% BSA).
Oocyte maturation and SCNT.
Oocytes were received from BoMed Inc. approximately 24 h after placing them into maturation medium and were then transferred to a 4-well dish and cultured for a total maturation of 42–44 h at 38.5°C in a humidified atmosphere of 5% CO2 in air. After 42–44 h of in vitro maturation, oocytes were stripped of their cumulus cells by gentle vortexing in 0.5 mg/ml hyaluronidase. After removal of the cumulus cells, oocytes with good morphology and a visible polar body (metaphase II) were selected and kept in micromanipulation medium at 38.5°C until SCNT.
SCNT was performed essentially as previously described (24, 63) in micromanipulation medium supplemented with 7.5 μg/ml cytochalasin B. The metaphase II chromosomes and the polar body were aspirated by inserting a micropipette through the zona pellucida and aspirating the polar body and the adjacent cytoplasm into the pipette. Next a donor cell was aspirated into the same pipette, the pipette was inserted into the previously made hole in the zona pellucida, and the cell was deposited under the zona pellucida. The nuclear transfer complex was fused in a medium with a low Ca2+ concentration (0.3 M mannitol, 0.1 mM CaCl2.2H2O, 0.1 mM MgCl2.6H2O, and 0.5 mM HEPES), activated with 200 μM thimerosal for 10 min in the dark, and then rinsed and treated with 8 mM dithiothreitol (DTT) for 30 min. Finally, the oocytes were rinsed to remove any traces of DTT (64). Following fusion/activation, oocytes were washed 3 times with PZM3 as previously described for 30 min (65). Those that had fused were cultured for 15–21 h until surgical embryo transfer to a surrogate.
Surrogate preparation and embryo transfer.
The embryonic cleavage rate was examined before transferring the reconstructed embryos into recipients. The recipients were synchronized by administering 18–20 mg Regumate and hCG as previously described (20). Twelve surrogates on the first day of estrus (designated day 0) or the first day after standing estrus were used. Embryo transfer was performed surgically as previously described (20), and 94–185 embryos were inserted into 1 oviduct through the ovarian fimbria. Surrogates were checked for pregnancy by abdominal ultrasound examination after day 21, then checked weekly throughout gestation, and were allowed to go to term. A cesarean section was performed to recover the piglets on days 116–118. After delivery the piglets were provided medical care, fed colostrum, and initially raised on a commercial pig milk replacer until mature enough to be placed on standard pig diets.
Rectal biopsy.
Pigs were lightly anesthetized with ketamine (20 mg/kg) and acepromazine (0.2 mg/kg). A 10-cm anoscope was partially inserted in the rectum, and rectal tissue was collected using gastrointestinal biopsy forceps (2.2 mm). Tissue samples were immediately placed in RNAlater (Ambion). Recovery from anesthesia was monitored continuously until the pigs returned to normal activity (2–4 h). All animal experiments were approved by the Institutional Animal Care and Use Committees of the University of Iowa and the University of Missouri.
Quantitative RT-PCR.
Quantitative RT-PCR using TaqMan chemistry and an ABI 7500 Fast Real-Time PCR System was used to measure pig CFTR mRNA. Briefly, total RNA was isolated from fibroblasts or nasal and rectal biopsy tissue (RNeasy; Qiagen). First-strand cDNA was synthesized with random primers (SuperScript III; Invitrogen). Sequence-specific primers and probes for pig CFTR and GAPDH were designed and ordered using Assays-by-design (Applied Biosystems). For measuring total CFTR mRNA, primer/probe sets spanning exons 18 and 19 of CFTR and GAPDH were used in separate reactions. For measuring ΔF508 mRNA levels, 1 primer set and 2 probes (F508 and ΔF508) were used in separate reactions. Primer and probe sequences are included in Supplemental Table 1. TaqMan Fast Universal PCR Master Mix was used for all reactions. The reaction volume was 20 μl (10 μl of 2× Master Mix without UNG, 1 μl of 20× target primer and probe, 8 μl of nuclease-free water, and 1 μl of cDNA sample). The reaction plates were covered with optical film and centrifuged briefly. The thermocycler conditions were as follows: 20 s at 95°C, 40 cycles of 95°C for 3 s, and 60°C for 30 s. All experiments were run in triplicate. Because the efficiencies of CFTR and GAPDH amplification were not equal, the relative quantification of transcript levels was performed using the standard curve method.
Supplementary Material
Acknowledgments
We thank Philip Karp, Tom Moninger, Vlad Snitsarev, and Theresa Mayhew for excellent assistance. This work was supported by the National Heart, Lung, and Blood Institute (HL51670), the National Institute of Diabetes and Digestive and Kidney Diseases (DK54759), Food for the 21st Century, the Cystic Fibrosis Foundation, and the Howard Hughes Medical Institute. C.S. Rogers was supported by NIH Training Grant HL07638. M.J. Welsh is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Nonstandard abbreviations used: AAV, adeno-associated virus; CF, cystic fibrosis; NeoR, neomycin resistance cassette; rAAV, recombinant AAV; SCNT, somatic cell nuclear transfer.
Conflict of interest: The authors have declared that no conflict of interest exists at this time.
Citation for this article: J. Clin. Invest. 118:1571–1577 (2008). doi:10.1172/JCI34773
Christopher S. Rogers and Yanhong Hao are co–first authors.
References
- 1.Capecchi M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2. Welsh, M.J., Ramsey, B.W., Accurso, F., and Cutting, G.R. 2001. Cystic fibrosis. InThe metabolic and molecular basis of inherited disease . C.R. Scriver, et al., editors. McGraw-Hill. New York, New York, USA. 5121–5189. [Google Scholar]
- 3.Davis P.B., Drumm M.L., Konstan M.W. Cystic fibrosis. Am. J. Respir. Crit. Care Med. 1996;154:1229–1256. doi: 10.1164/ajrccm.154.5.8912731. [DOI] [PubMed] [Google Scholar]
- 4.Schwiebert E.M., Benos D.J., Egan M.E., Stutts J., Guggino W.B. CFTR is a conductance regulator as well as a chloride channel. Physiol. Rev. 1999;79:S145–S166. doi: 10.1152/physrev.1999.79.1.S145. [DOI] [PubMed] [Google Scholar]
- 5.Grubb B.R., Boucher R.C. Pathophysiology of gene-targeted mouse models for cystic fibrosis. Physiol. Rev. 1999;79:S193–S214. doi: 10.1152/physrev.1999.79.1.S193. [DOI] [PubMed] [Google Scholar]
- 6.Guilbault C., Saeed Z., Downey G.P., Radzioch D. Cystic fibrosis mouse models. Am. J. Respir. Cell Mol. Biol. 2007;36:1–7. doi: 10.1165/rcmb.2006-0184TR. [DOI] [PubMed] [Google Scholar]
- 7.Capecchi M.R. Choose your target. Nat. Genet. 2000;26:159–161. doi: 10.1038/82825. [DOI] [PubMed] [Google Scholar]
- 8.Wilmut I., Schnieke A.E., McWhir J., Kind A.J., Campbell K.H.S. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 9.Cibelli J.B., et al. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280:1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- 10.Wakayama T., Perry A.C., Zuccotti M., Johnson K.R., Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 11.Baguisi A., et al. Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 1999;17:456–461. doi: 10.1038/8632. [DOI] [PubMed] [Google Scholar]
- 12.Polejaeva I.A., et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- 13.Chesne P., et al. Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol. 2002;20:366–369. doi: 10.1038/nbt0402-366. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Q., et al. Generation of fertile cloned rats by regulating oocyte activation. Science. 2003;302:1179. doi: 10.1126/science.1088313. [DOI] [PubMed] [Google Scholar]
- 15.Lee B.C., et al. Dogs cloned from adult somatic cells. Nature. 2005;436:641. doi: 10.1038/436641a. [DOI] [PubMed] [Google Scholar]
- 16.Li Z., et al. Cloned ferrets produced by somatic cell nuclear transfer. Dev. Biol. 2006;293:439–448. doi: 10.1016/j.ydbio.2006.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sedivy J.M., Dutriaux A. Gene targeting and somatic cell genetics — a rebirth or a coming of age? Trends Genet. 1999;15:88–90. doi: 10.1016/S0168-9525(98)01689-8. [DOI] [PubMed] [Google Scholar]
- 18.Williams S.H., et al. Evaluation of gene targeting by homologous recombination in ovine somatic cells. Mol. Reprod. Dev. 2003;66:115–125. doi: 10.1002/mrd.10340. [DOI] [PubMed] [Google Scholar]
- 19.Wilmut L., et al. Somatic cell nuclear transfer. Nature. 2002;419:583–586. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 20.Lai L., Prather R.S. Production of cloned pigs by using somatic cells as donors. Cloning Stem Cells. 2003;5:233–241. doi: 10.1089/153623003772032754. [DOI] [PubMed] [Google Scholar]
- 21.Yanez R.J., Porter A.C. Influence of DNA delivery method on gene targeting frequencies in human cells. Somat. Cell Mol. Genet. 1999;25:27–31. doi: 10.1023/B:SCAM.0000007137.28557.73. [DOI] [PubMed] [Google Scholar]
- 22.Vasquez K.M., Marburger K., Intody Z., Wilson J.H. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas K.R., Folger K.R., Capecchi M.R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 24.Lai L., et al. Production of α-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295:1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- 25.Dai Y., et al. Targeted disruption of the α1,3-galactosyltransferase gene in cloned pigs. Nat. Biotechnol. 2002;20:251–255. doi: 10.1038/nbt0302-251. [DOI] [PubMed] [Google Scholar]
- 26.Yu G., et al. Functional disruption of the prion protein gene in cloned goats. J. Gen. Virol. 2006;87:1019–1027. doi: 10.1099/vir.0.81384-0. [DOI] [PubMed] [Google Scholar]
- 27.Richt J.A., et al. Production of cattle lacking prion protein. Nat. Biotechnol. 2007;25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroiwa Y., et al. Sequential targeting of the genes encoding immunoglobulin-mu and prion protein in cattle. Nat. Genet. 2004;36:775–780. doi: 10.1038/ng1373. [DOI] [PubMed] [Google Scholar]
- 29.McCreath K.J., et al. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 30.Robinson N.E. Some functional consequences of species differences in lung anatomy. Adv. Vet. Sci. Comp. Med. 1982;26:1–33. [PubMed] [Google Scholar]
- 31.McKay K.O., Wiggs B.R., Paré P.D., Kamm R.D. Zero-stress state of intra- and extraparenchymal airways from human, pig, rabbit, and sheep lung. J. Appl. Physiol. 2002;92:1261–1266. doi: 10.1152/japplphysiol.00131.2001. [DOI] [PubMed] [Google Scholar]
- 32.Maina J.N., van Gils P. Morphometric characterization of the airway and vascular systems of the lung of the domestic pig, Sus scrofa: comparisons of the airway, arterial and venous systems. . Comp. Biochem. Physiol. A Mol. Integr. Physiol. . 2001;130:781–798. doi: 10.1016/S1095-6433(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 33.Pabst R. The respiratory immune system of pigs. Vet. Immunol. Immunopathol. 1996;54:191–195. doi: 10.1016/S0165-2427(96)05700-5. [DOI] [PubMed] [Google Scholar]
- 34.Pabst R., Binns R.M. The immune system of the respiratory tract in pigs. Vet. Immunol. Immunopathol. 1994;43:151–156. doi: 10.1016/0165-2427(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 35.Cooper D.K.C., Gollackner B., Sachs D.H. Will the pig solve the transplantation backlog? Annu. Rev. Med. 2002;53:133–147. doi: 10.1146/annurev.med.53.082901.103900. [DOI] [PubMed] [Google Scholar]
- 36.Zielenski J., Tsui L.C. Cystic fibrosis: genotypic and phenotypic variations. Annu. Rev. Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]
- 37.Dalemans W., et al. Altered chloride ion channel kinetics associated with the ΔF508 cystic fibrosis mutation. Nature. 1991;354:526–528. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 38.Teem J.L., Carson M.R., Welsh M.J. Mutation of R555 in CFTR-ΔF508 enhances function and partially corrects defective processing. Receptors Channels. 1996;4:63–72. [PubMed] [Google Scholar]
- 39.Skach W.R. Defects in processing and trafficking of the cystic fibrosis transmembrane conductance regulator. Kidney Int. 2000;57:825–831. doi: 10.1046/j.1523-1755.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- 40.Denning G.M., et al. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 41.Lukacs G.L., Durie P.R. Pharmacologic approaches to correcting the basic defect in cystic fibrosis. N. Engl. J. Med. 2003;349:1401–1404. doi: 10.1056/NEJMp038113. [DOI] [PubMed] [Google Scholar]
- 42.Verkman A.S., Lukacs G.L., Galietta L.J. CFTR chloride channel drug discovery — inhibitors as antidiarrheals and activators for therapy of cystic fibrosis. Curr. Pharm. Des. 2006;12:2235–2247. doi: 10.2174/138161206777585148. [DOI] [PubMed] [Google Scholar]
- 43.Ostedgaard L.S., et al. Processing and function of CFTR-ΔF508 are species-dependent. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15370–15375. doi: 10.1073/pnas.0706974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park K.W., et al. Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Anim. Biotechnol. 2001;12:173–181. doi: 10.1081/ABIO-100108344. [DOI] [PubMed] [Google Scholar]
- 45.Trapnell B.C., et al. Expression of the cystic fibrosis transmembrane conductance regulator gene in the respiratory tract of normal individuals and individuals with cystic fibrosis. Proc. Natl. Acad. Sci. U. S. A. 1991;88:6565–6569. doi: 10.1073/pnas.88.15.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colledge W.H., et al. Generation and characterization of a ΔF508 cystic fibrosis mouse model. Nat. Genet. 1995;10:445–452. doi: 10.1038/ng0895-445. [DOI] [PubMed] [Google Scholar]
- 47.Zeiher B.G., et al. A mouse model for the ΔF508 allele of cystic fibrosis. J. Clin. Invest. 1995;96:2051–2064. doi: 10.1172/JCI118253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue N., Hirata R.K., Russell D.W. High-fidelity correction of mutations at multiple chromosomal positions by adeno-associated virus vectors. J. Virol. 1999;73:7376–7380. doi: 10.1128/jvi.73.9.7376-7380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirata R., Chamberlain J.S., Dong R., Russell D.W. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 50.Porteus M.H., Cathomen T., Weitzman M.D., Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell D.W., Hirata R.K. Human gene targeting by viral vectors. Nat. Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hendrie P.C., Russell D.W. Gene targeting with viral vectors. Mol. Ther. 2005;12:9–17. doi: 10.1016/j.ymthe.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Sun X., et al. Adeno-associated virus–targeted disruption of the CFTR gene in cloned ferrets. . J. Clin. Invest. 2008;118:1578–1583. . doi: 10.1172/JCI34599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson A.J., Marques M.M., McWhir J. Gene targeting in livestock. Reprod. Suppl. 2003;61:495–508. [PubMed] [Google Scholar]
- 55.Wilschanski M., et al. Mutations in the cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. Am. J. Respir. Crit. Care Med. 2006;174:787–794. doi: 10.1164/rccm.200509-1377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson D.C., et al. Uncertainty in the diagnosis of cystic fibrosis: possible role of in vivo nasal potential difference measurements. J. Pediatr. 1998;132:596–599. doi: 10.1016/S0022-3476(98)70345-2. [DOI] [PubMed] [Google Scholar]
- 57.Gelrud A., et al. Analysis of cystic fibrosis gener product (CFTR) function in patients with pancreas divisum and recurrent acute pancreatitis. Am. J. Gastroenterol. 2004;99:1557–1562. doi: 10.1111/j.1572-0241.2004.30834.x. [DOI] [PubMed] [Google Scholar]
- 58.Behm J.K., Hagiwara G., Lewiston N.J., Quinton P.M., Wine J.J. Hyposecretion of beta-adrenergically induced sweating in cystic fibrosis heterozygotes. Pediatr. Res. 1987;22:271–276. doi: 10.1203/00006450-198709000-00007. [DOI] [PubMed] [Google Scholar]
- 59.Sato K., Sato F. Variable reduction in β-adrenergic sweat secretion in cystic fibrosis heterozygotes. J. Lab. Clin. Med. 1988;111:511–518. [PubMed] [Google Scholar]
- 60.Pomp D., Good B.A., Geisert R.D., Corbin C.J., Conley A.J. Sex identification in mammals with polymerase chain reaction and its use to examine sex effects on diameter of day-10 or -11 pig embryos. J. Anim. Sci. 1995;73:1408–1415. doi: 10.2527/1995.7351408x. [DOI] [PubMed] [Google Scholar]
- 61. Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989.Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 999 pp. [Google Scholar]
- 62.Yan Z., et al. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai L., et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat. Biotechnol. 2006;24:435–436. doi: 10.1038/nbt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machaty Z., Wang W.H., Day B.N., Prather R.S. Complete activation of porcine oocytes induced by the sulfhydryl reagent, thimerosal. Biol. Reprod. 1997;57:1123–1127. doi: 10.1095/biolreprod57.5.1123. [DOI] [PubMed] [Google Scholar]
- 65.Im G.S., et al. In vitro development of preimplantation porcine nuclear transfer embryos cultured in different media and gas atmospheres. Theriogenology. 2004;61:1125–1135. doi: 10.1016/j.theriogenology.2003.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.