Abstract
Polo-like kinase (Plk)1 is required for mitosis progression. However, although Plk1 is expressed throughout the cell cycle, its function during S-phase is unknown. Using Xenopus laevis egg extracts, we demonstrate that Plx1, the Xenopus orthologue of Plk1, is required for DNA replication in the presence of stalled replication forks induced by aphidicolin, etoposide or reduced levels of DNA-bound Mcm complexes. Plx1 binds to chromatin and suppresses the ATM/ATR-dependent intra-S-phase checkpoint that inhibits origin firing. This allows Cdc45 loading and derepression of DNA replication initiation. Checkpoint activation increases Plx1 binding to the Mcm complex through its Polo box domain. Plx1 recruitment to chromatin is independent of checkpoint mediators Tipin and Claspin. Instead, ATR-dependent phosphorylation of serine 92 of Mcm2 is required for the recruitment of Plx1 to chromatin and for the recovery of DNA replication under stress. Depletion of Plx1 leads to accumulation of chromosomal breakage that is prevented by the addition of recombinant Plx1. These data suggest that Plx1 promotes genome stability by regulating DNA replication under stressful conditions.
Keywords: checkpoint, DNA replication, Plx1, stress, Xenopus laevis
Introduction
Polo-like kinase (Plk)1 is essential for cell cycle progression (Golsteyn et al, 1995). A role for Plk1 during mitosis has been established by many studies showing that the absence of Plk1 impairs mitotic progression (Golsteyn et al, 1996). From these studies, unchallenged chromosomal DNA replication does not appear to be affected by the absence of Plk1. Plx1, the Xenopus orthologue of Plk1 (Kumagai and Dunphy, 1996; Liu and Maller, 2005), has been shown to regulate the adaptation to the ATR- and Chk1-dependent checkpoint that inhibits mitosis onset in the presence of DNA damage and unreplicated DNA (Yoo et al, 2004a). Following a prolonged ATR activation, Plx1-mediated phosphorylation of Claspin induces attenuation of ATR- and Chk1-dependent signalling allowing mitosis onset (Yoo et al, 2004a). Owing to its role in controlling the DNA damage response, Plx1 might have a role in DNA replication under stressful conditions.
In eukaryotes, DNA replication is achieved through the coordinated activation of multiple replication origins distributed throughout the DNA. The origin recognition complex 1–6 is bound to DNA and promotes the assembly of the mini-chromosome maintenance (Mcm)2–7 protein complex onto the DNA to form the pre-replicative complex (pre-RC) in cooperation with Cdc6 and Cdt1. Replication initiation requires Cdk2- and Cdc7-mediated activation of the pre-RC to unwind the DNA and to start DNA polymerization (Diffley, 1994).
The timing of replication origin firing and the activation of Mcm2–7 complexes at replication origins are regulated by a caffeine-sensitive, ATM- and ATR-dependent checkpoint (Shechter et al, 2004a). This checkpoint modulates Cdk2 and Cdc7 kinase activities regulating replication initiation during unperturbed cell cycle and halting DNA replication in the presence of DNA damage or stalled replication forks (Shechter et al, 2004a). Mcm2–7 complexes are bound to DNA in excess and some of these complexes remain inactive (Edwards et al, 2002). An important mechanism to ensure genome stability during DNA replication under stressful conditions relies on the activation of dormant Mcm2–7 complexes bound to chromatin whose reduction was shown to decrease replication efficiency in the presence of DNA damage or replication inhibitors (Woodward et al, 2006; Ge et al, 2007). However, the caffeine-sensitive checkpoint activated by replication stress strongly suppresses origin firing and Mcm2–7 activation (Marheineke and Hyrien, 2004; Shechter et al, 2004a; Woodward et al, 2006). Therefore, cells must circumvent this checkpoint to ensure efficient and full chromosomal replication under stressful conditions. As Plx1 suppresses the checkpoint at mitosis onset, it could also regulate the checkpoint that prevents the firing of dormant origins. Intriguingly, members of the Mcm complex are also phosphorylated by ATM/ATR, suggesting a direct coordination of their activity in the presence of DNA lesions activating the checkpoint (Cortez et al, 2004; Yoo et al, 2004b). Here, we have investigated the role of Plx1 in chromosomal DNA replication under stress in Xenopus laevis egg extracts. We show that Plx1 is required for DNA replication in the presence of stalled replication forks. Plx1 binds to chromatin interacting with the Mcm complex through its polo box domain and its binding is increased in the presence of stalled replication forks. Plx1 bound to the Mcm complex on chromatin promotes DNA replication under stress by suppressing the intra-S-phase checkpoint. This is achieved through regulation of Chk1 phosphorylation. Plx1-mediated downregulation of the checkpoint requires ATM/ATR-mediated phosphorylation of serine 92 of Mcm2. Importantly, the absence of Plx1 leads to accumulation of chromosomal breakage during DNA replication, suggesting that Plx1 function is critical for preventing gross chromosomal abnormalities.
Results
Effects of Plx1 depletion on DNA replication
To test whether Plx1 is required for DNA replication, we raised antibodies against recombinant Plx1. Using these antibodies, we were able to deplete Plx1 from egg extracts (Figure 1A). Normal levels of Plx1 were reconstituted by supplementing egg extracts with active recombinant Plx1 (Figure 1A and Supplementary Figure 1). Genomic DNA replication was monitored in Plx1-depleted extracts and in Plx1-depleted extracts supplemented with recombinant Plx1 in the presence of sperm nuclei and [α-32P]dATP. After 120 min, DNA was extracted and separated on an agarose gel (Figure 1B). In interphase egg extracts, Plx1 depletion did not affect DNA replication (Figure 1B). To rule out additional effects of Plx1 on basic replication processes, we monitored nuclear formation and DNA elongation in the absence of Plx1. Plx1 depletion did not affect nuclear formation (Supplementary Figure 2). In addition, replication of single-stranded DNA of M13 phage was normal in Plx1-depleted extracts, indicating that DNA elongation was not affected by Plx1 deficiency (Supplementary Figure 3). To exclude the possibility that Plx1 was impairing the kinetics of DNA replication, we performed a pulse-labelling experiment monitoring DNA replication progression. In these experiments, we showed that even in the absence of Plx1, DNA replication was initiated and terminated with similar kinetics compared to the mock-depleted extract (Supplementary Figure 4).
Figure 1.
Effects of Plx1 depletion on DNA replication. (A) Immunodepletion of Plx1. Western blot analysis with anti-Plx1 antibodies of control (C) mock-depleted (M), Plx1-depleted (ΔPlx1) and Plx1-depleted extracts reconstituted with 6 ng/μl recombinant Plx1 (+Plx1). (B) Replication assay in unchallenged extracts. Extracts supplemented with 2000 nuclei/μl were untreated (CTRL), mock depleted (Mock), Plx1 depleted (ΔPlx1) and Plx1 depleted and supplemented with recombinant Plx1 (+Plx1). DNA was replicated in the presence of [α-32P]dATP for 120 min, extracted and separated on an agarose gel. The gel shown represents a typical result. (C) Chromatin binding of Mcm7. Sperm nuclei were replicated in untreated extracts (CTRL) and extracts supplemented with a low concentration (40 nM) of geminin (GEM). Chromatin was isolated at the indicated time points and separated on SDS–PAGE. Western blots were performed with antibodies against human Mcm7. (D) Effects of geminin on DNA replication. Extracts were supplemented with increasing concentrations of geminin (40, 60 and 120 nM). (E) Phosphorylation of Chk1 serine 345 (p-CHK1) in extracts supplemented with 7000 nuclei/μl. Extracts were untreated (−) or supplemented (+) with 40 nM geminin (GEM). Western blot was performed with anti-phospho-serine 345 of human Chk1 (p-Chk1) and anti-Chk1. Chk1 phosphorylation induced by geminin was detected with ECL plus reagent (see Materials and methods). (F) Replication assay in extracts supplemented with 40 nM geminin (GEM) in the presence of 7000 nuclei/μl. Extracts were untreated (CTRL), mock depleted (Mock), Plx1 depleted (ΔPlx1) and Plx1 depleted and supplemented with recombinant Plx1 (Plx1-dep+Plx1). The gel shown represents a typical result.
Supplementary origins and low inter-origin distance ensure faster and more efficient replication in embryonic systems, facilitating the bypass of eventual obstacles to replication progression (Woodward et al, 2006). To verify whether Plx1 is required for DNA replication in more stressful conditions, we studied the role of Plx1 in DNA replication in the presence of a limited number of replication origins and increased inter-origin distance. To reduce the number of replication origins, we used geminin, an inhibitor of chromosomal DNA replication known to prevent the loading of Mcm complexes onto DNA (McGarry and Kirschner, 1998). The increased inter-origin distance was ensured by the use of a high number of nuclei (Walter and Newport, 1997). The level of Mcm complexes loaded onto DNA was reduced by geminin (Figure 1C). As reported by Blow and co-workers (Woodward et al, 2006), DNA replication levels were unaffected in the presence of reduced numbers of DNA-bound Mcm complexes unless very high doses of geminin were used (Figure 1D). In extracts supplemented with a high number of nuclei and geminin, the checkpoint protein Chk1 was phosphorylated (Figure 1E). This was likely due to the presence of replication stress induced by spontaneous stalled or damaged replication forks activating the ATM/ATR-dependent checkpoint. However, although sufficiently high to be detected, the levels of Chk1 phosphorylation were not sufficient to inhibit DNA replication (Figure 1F). Instead, in these conditions, Plx1 depletion abolished DNA replication (Figure 1F). In the absence of geminin, Plx1 deficiency did not affect replication of a high number of nuclei (Supplementary Figure 4). DNA replication in Plx1-depleted extracts could be fully restored when supplemented with recombinant Plx1 (Figure 1F). The complete rescue by recombinant Plx1 indicates the presence of a sufficient number of Mcm complexes left on chromatin after geminin treatment to support full DNA replication. Taken together, these results suggest that Plx1 is required for DNA replication under stressful conditions.
Plx1 binds to chromatin and promotes DNA replication in the presence of stalled replication forks
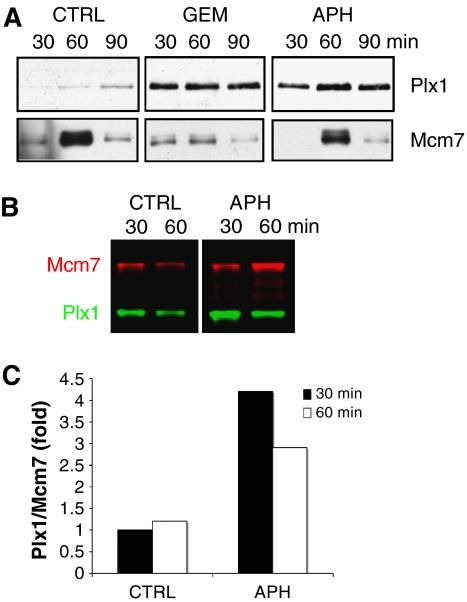
To demonstrate a direct role of Plx1 in regulating DNA replication, we monitored Plx1 binding to chromatin. We could reproducibly detect low levels of Plx1 binding to chromatin (Figure 2A). Significantly, we observed a large increase in Plx1 binding to chromatin during DNA replication in the presence of low levels of DNA-bound Mcm complexes. A similar effect was observed with low amounts of aphidicolin, a replication inhibitor known to induce stalled replication forks (Jenkins et al, 1992) (Figure 2A). The Mcm binding was not significantly affected by aphidicolin treatment (Figure 2A), suggesting that induction of Plx1 binding was due to the presence of stalled forks. To quantify the induction of Plx1 binding to chromatin, we performed a western blot using secondary antibodies labelled with infrared dyes, which have better signal-to-noise ratio and high detection sensitivity. Compared to chemiluminescence, the detection with infrared dye is linear over a very wide range and amounts from 1 pg up to 10 ng can be detected more reliably (Chen et al, 2005). Using this sensitive technique and comparing the amount of chromatin-bound Plx1 and Mcm7, we could quantify Plx1 binding in the presence of aphidicolin during the early stages of replication (Figure 2B). Compared to the levels of Mcm7 loaded onto chromatin, the binding of Plx1 increased up to four-fold (Figure 2C). These experiments demonstrate that Plx1 is recruited to chromatin in the presence of stalled replication forks.
Figure 2.
Plx1 binding to chromatin is enhanced in the presence of stalled replication forks. (A) Chromatin binding of Plx1 and Mcm7 to chromatin 30, 60 and 90 min after nuclei addition to untreated extracts (CTRL) and extracts treated with geminin (GEM) or 5 μM aphidicolin (APH). (B) Chromatin binding of Plx1 after aphidicolin treatment. Extracts were untreated (CTRL) or treated with 5 μM aphidicolin (APH) before the addition of sperm nuclei. Chromatin-bound proteins were separated on SDS–PAGE, transferred to nitrocellulose and probed for Mcm7 and Plx1, which were visualized with infrared dye-labelled secondary antibodies. (C) Quantification of chromatin-bound Plx1 at 30 and 60 min after nuclei addition to untreated (CTRL) or aphidicolin (APH)-treated extracts. Chromatin-bound proteins were quantified with an infrared imager and the ratio of Plx1 to Mcm7 was plotted. The ratio Plx1 to Mcm7 in the untreated sample (CTRL) was considered as the reference value.
Plx1 regulates replication origin firing under stressful conditions by modulating Chk1 phosphorylation
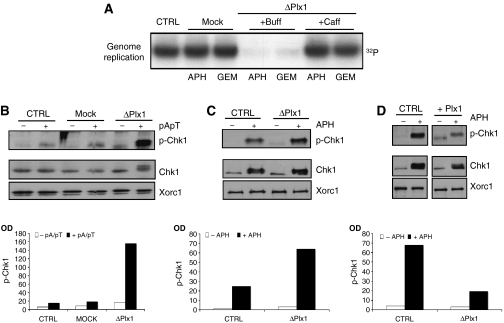
To test whether the increased Plx1 binding to chromatin was required for promoting DNA replication in the presence of stalled replication forks, we measured DNA replication in Plx1-depleted extracts treated with low amounts of aphidicolin. Similar to geminin, the supplementation of extracts with low doses of aphidicolin inhibited DNA replication in the absence of Plx1 and not in mock-depleted extracts (Figure 3A and Supplementary Figure 5). We then tested whether the effects of Plx1 on DNA replication were mediated by the caffeine-sensitive intra-S-phase checkpoint that controls origin firing and the recruitment of supplementary Mcm complexes (Shechter et al, 2004b; Woodward et al, 2006). We had previously shown that caffeine releases the inhibition of DNA replication induced by ATM and ATR in Xenopus egg extracts (Costanzo et al, 2000; Shechter et al, 2004a). Significantly, caffeine could restore DNA replication impaired by the absence of Plx1 in the presence of aphidicolin or geminin (Figure 3A), suggesting that Plx1 was suppressing the intra-S-phase checkpoint. Inhibition of topoisomerase II activity mediated by etoposide induces DNA lesions activating ATR (Costanzo and Gautier, 2003; Costanzo et al, 2003). Consistent with the results obtained with low amounts of aphidicolin, DNA replication in the presence of low amounts of etoposide was impaired in Plx1-depleted extracts (Supplementary Figure 6). This indicates that Plx1 is required for bypassing a wide range of DNA lesions that impair replication progression and activate ATR. To investigate directly the effects of Plx1 on checkpoint signaling, we monitored the activation of Chk1, a target of the caffeine-sensitive intra-S-phase checkpoint (Shechter et al, 2004b). We triggered Chk1 activation by supplementing egg extracts with annealed oligomers of poly-deoxy-(A)70 and poly-deoxy-(T)70 (pApT). These templates mimic incompletely replicated DNA and induce Chk1 phosphorylation (Figure 3B), as described previously (Yoo et al, 2004a). Strikingly, phosphorylation of Chk1 was dramatically enhanced in Plx1-depleted extracts supplemented with pApT (Figure 3B). We then monitored the activation status of Chk1 in isolated nuclei treated with aphidicolin. Again, Plx1 depletion led to a significant increase in Chk1 phosphorylation (Figure 3C). Supplementation of control extracts with recombinant Plx1 strongly suppressed Chk1 phosphorylation (Figure 3D). These results indicate that Plx1 suppresses the caffeine-sensitive intra-S-phase checkpoint that regulates origin firing.
Figure 3.
Plx1 suppresses the caffeine-sensitive intra-S-phase checkpoint. (A) The effects of caffeine on DNA replication under stress in the absence of Plx1. Sperm nuclei (7000 nuclei/μl) were replicated in extracts that were untreated (CTRL), mock depleted (Mock) or Plx1 depleted (ΔPlx1) in the presence of 2.5 μM aphidicolin (APH) or 40 nM geminin (GEM). Extracts were supplemented with buffer (Buff) or 5 mM caffeine (Caff). (B) Phosphorylation of Chk1 in untreated (CTRL), mock-depleted (Mock) and Plx1-depleted (ΔPlx1) extracts that were untreated (−) or supplemented with 50 ng/μl pApT (+). Western blot was performed with anti-phospho-Chk1 (p-Chk1), anti-Chk1 and anti-Xorc1 antibodies. Quantification was made by measuring optical density (OD) and is presented as a bar graph. (C) Chk1 phosphorylation in nuclei. Sperm nuclei were incubated with untreated extracts (CTRL) or with Plx1-depleted extracts (ΔPlx1). All extracts were untreated (−) or supplemented with 50 μM aphidicolin (+). The nuclei were isolated and the nuclear proteins were separated by SDS–PAGE. Western blot was performed with anti-phospho-Chk1 (p-Chk1), anti-Chk1 and anti-Xorc1 antibodies. (D) Sperm nuclei were incubated with untreated extracts (CTRL) or with extracts supplemented with 6 ng/μl Plx1 (Plx1) and processed as described above. All extracts were untreated (−) or supplemented with 50 μM aphidicolin (+). As described above, quantification of western blot signals in panels C and D was made by measuring OD and is presented as bar graphs.
Molecular mechanisms underlying Plx1-dependent control of origin firing
To understand the molecular mechanism of replication arrest, we monitored chromatin binding of the Mcm complex and Cdc45. In extracts treated with aphidicolin, the binding of the Mcm complex to chromatin was not affected by Plx1 depletion, as shown by normal levels of Mcm7 detected on chromatin (Figure 4A). We instead observed a strong inhibition of Cdc45 binding to chromatin in the absence of Plx1, which could be restored by supplementing extracts with recombinant Plx1 (Figure 4A). Plx1 depletion did not impair Cdc45 binding in untreated extracts (Supplementary Figure 7). As Cdc45 is loaded in the context of origin firing (Masuda et al, 2003), DNA replication arrest in the absence of Plx1 is due to suppression of origin firing. Cdc45 loading requires Cdk2 activity (Masuda et al, 2003), which can be downregulated by the activation of the checkpoint (Costanzo et al, 2000; Shechter et al, 2004a). To verify whether impaired Cdc45 loading was due to the downregulation of Cdk2 activity, we measured nuclear Cdk2 activity in the presence and absence of Plx1 in conditions in which DNA replication was impaired. Cdk2 activity was reduced in the presence of an active intra-S-phase checkpoint (Figure 4B), as expected. Significantly, the reduction of Cdk2 activity was more pronounced in the absence of Plx1. Treatment with caffeine restored Cdk2 activity to control levels in normal and Plx1-depleted extracts (Figure 4B), suggesting that downregulation of Cdk2 was mediated by hyperactivation of the checkpoint induced by the absence of Plx1.
Figure 4.
Plx1 deficiency affects Cdc45 loading and Cdk2 activity in the presence of stalled replication forks. (A) Cdc45 and Mcm7 binding to chromatin isolated from extracts that were untreated (CTRL), Plx1 depleted (ΔPlx1) or Plx1 depleted and reconstituted with recombinant Plx1 (+Plx1). All extracts were supplemented with aphidicolin. (B) Nuclear Cdk2 activity measured by monitoring 32P incorporation in histone H1 after immunoprecipitating nuclear Cdk2 with anti-Xenopus Cdk2 antibodies from extracts that were untreated (CTRL), mock depleted (Mock) or Plx1 depleted (ΔPlx1) and treated with and without 50 μM aphidicolin (APH) or 5 mM caffeine (Caff), as indicated.
Mcm-dependent recruitment of Plx1 to chromatin
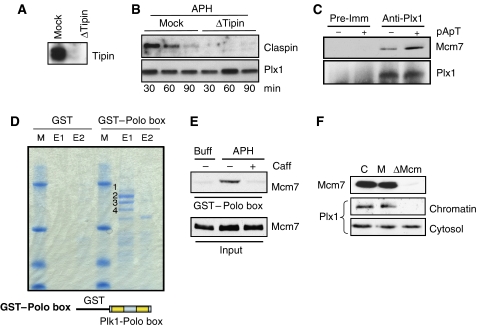
Plx1 has been shown to interact with Claspin, a mediator of Chk1 activation. Prolonged and sustained activation of the checkpoint promotes ATR-dependent recruitment of Plx1 to Claspin. Plx1 in turn phosphorylates Claspin, inducing its release from chromatin and promoting adaptation to the checkpoint that delays mitosis onset (Yoo et al, 2004a). We tested whether Claspin was also mediating the recruitment of Plx1 to chromatin at the onset of DNA replication under stressful conditions. We have recently shown that Claspin loading onto chromatin requires the action of Tipin (Errico et al, 2007). We confirmed that Tipin depletion (Figure 5A) suppresses Claspin binding to chromatin (Figure 5B). However, accumulation of Plx1 on chromatin induced by stalled forks was not affected by the absence of Claspin (Figure 5B). These results suggest that differently from adaptation, which occurs 3–4 h after persistent checkpoint activation, the rapid effects mediated by Plx1 at the onset of DNA replication are not dependent on Claspin. Adaptation to persistent checkpoint signalling also requires Plx1-dependent release of Claspin from chromatin. However, increasing the amount of Plx1 by the addition of recombinant Plx1 resulted in downregulation of Chk1 activation (Figure 3D) but did not promote Claspin release from chromatin during early S-phase (Supplementary Figure 8). Overall, these data indicate that the mechanism responsible for Plx1-mediated suppression of the checkpoint during origin firing is distinct from Plx1-mediated adaptation to the checkpoint at the G2/M transition. To identify the mechanism of Plx1 recruitment to chromatin, we tested the interaction of Plx1 with the Mcm complex. We found that Plx1 co-precipitated with Mcm7 and that its binding was enhanced by activation of the checkpoint induced by pApT (Figure 5C). To identify the region of Plx1 that binds to the Mcm complex, we used truncated proteins fused to GST in pull-down experiments. One of the truncated proteins contained the protein interaction domain known as the Polo box, which has been shown to interact with phosphorylated proteins (Elia et al, 2003). The GST–Polo box-containing fragment derived from Plk1, which is highly similar to the Polo box of Plx1, was incubated with egg extracts. Proteins bound to the Polo box domain were then eluted using a phosphorylated peptide with a strong affinity for the Polo box, and subsequently subjected to matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) to identify the sequence of the interacting proteins. As shown in Figure 5D, the GST–Polo box was able to pull down four major proteins, Mcm2, Mcm4, Mcm6 and Mcm7, as revealed by MALDI-TOF analysis. As the Polo box interacts with phosphorylated residues and the Mcm proteins could be eluted using a phosphorylated peptide, it is likely that the interaction is mediated by phosphorylated serine/threonine residues present on endogenous Mcm proteins. The binding of the Polo box to endogenous Mcm proteins was consistent with the ability of full-length Plx1 to interact with chromatin and Mcm7. We then tested whether the Polo box binding to Mcm proteins was enhanced by the activation of checkpoint. In line with the results obtained with full-length Plx1, Polo box binding to the Mcm complex was enhanced by checkpoint activation and was inhibited by caffeine (Figure 5E). Plx1 was unable to bind to chromatin in extracts depleted of the Mcm complex (Figure 5F), suggesting that the Mcm complex is the major binding site for Plx1 on chromatin during S-phase.
Figure 5.
Plx1 interacts with the Mcm complex through its Polo box domain. (A) Immunodepletion of Tipin. Extracts were mock depleted (Mock) or Tipin depleted using anti-Tipin antibodies (ΔTipin). The cytosol was then subjected to western blot with anti-Tipin antibodies. (B) Plx1 and Claspin binding in Tipin-depleted extracts. Sperm nuclei were incubated in mock-depleted (Mock) or Tipin-depleted (ΔTipin) extracts in the presence of 2.5 μM aphidicolin (APH). Chromatin was isolated at the indicated time points and probed with anti-Plx1 and anti-Claspin antibodies. (C) Co-immunoprecipitation of Plx1 and Mcm7. Extracts that were untreated (−) or treated with 50 ng/μl pA/pT (+) were immunoprecipitated with pre-immune (Pre-Imm) or anti-Plx1 (Anti-Plx1) antibodies. Western blot was performed using anti-Mcm7 and anti-Plx1 antibodies. (D) Pull-down with GST–Polo box and MALDI-TOF analysis. Interphase extracts were incubated with GST–agarose beads or agarose beads with GST fused to the Polo box derived from human Plk1. After pull-down, proteins were consecutively eluted using 3 mg/ml of phosphopeptide solution (first elution=E1, second elution=E2, see Supplementary data for the sequence of the peptide and the Polo box) and subjected to MALDI-TOF analysis. Band 1 corresponds to Mcm2, band 2 to Mcm6, band 3 to Mcm4 and band 4 to Mcm7. The GST fusion protein containing the Polo box from Plk1 is shown at the bottom. (E) GST–Polo box pull-down of nuclear proteins probed with anti-Mcm7 antibodies (GST–Polo box). The extract used for the pull-down was supplemented with nuclei and buffer (Buff), 2.5 μM aphidicolin (APH) or 2.5 μM aphidicolin and 5 mM caffeine (Caff), as indicated. The total amount of unbound Mcm7 extracted from nuclei was loaded as a control (Input). (F) Effects of Mcm complex depletion on Plx1 binding to chromatin. The extract was untreated (C), mock depleted (M) or Mcm depleted (ΔMCM) using anti-Mcm3 antibodies and incubated with sperm nuclei for 60 min in the presence of 2.5 μM APH. Cytosol and chromatin were then subjected to western blot with anti-Mcm7 and anti-Plx1 antibodies.
Phosphorylation of serine 92 of Mcm2 is required for Plx1 function
The fact that Plx1 binding to the Mcm complex is enhanced by checkpoint activation suggests that post-translational modifications of the Mcm proteins induced by the activation of the DNA damage response could increase Plx1 affinity for the Mcm complex. Mcm2 is strongly phosphorylated on serine 92 by ATM/ATR (Yoo et al, 2004b), and its phosphorylation can be induced by pA/pT, aphidicolin or etoposide (Supplementary Figure 9A). To test whether phosphorylation of serine 92 of Mcm2 has a role in Plx1 recruitment to chromatin and in Plx1-mediated suppression of the checkpoint, we produced recombinant wild-type Mcm2 (Mcm2-WT) and Mcm2 in which serine 92 had been substituted with alanine (Mcm2-S92A) rendering it resistant to ATR-mediated phosphorylation (Supplementary Figure 9B). We supplemented egg extracts with an excess of Mcm2-WT or Mcm2-S92A proteins. This procedure leads to the formation of a significant amount of endogenous Mcm complexes containing the exogenous Mcm2-WT or Mcm2-S92A proteins, which are able to assemble onto chromatin and support DNA replication, as shown previously (Yoo et al, 2004b). We confirmed that equivalent amounts of recombinant Mcm2-WT and Mcm2-S92A proteins could be loaded onto chromatin (Figure 6A). Importantly, the presence of Mcm2-S92A impaired Plx1 recruitment to chromatin following replication stress induced by etoposide, which activates ATR in this system (Costanzo et al, 2003). Plx1 loading onto chromatin was instead unaffected in the presence of Mcm2-WT protein. This suggests that Plx1 binding to the Mcm complex loaded onto chromatin requires ATR-mediated phosphorylation of serine 92. We then tested the ability of recombinant Plx1 to restore DNA replication under stressful conditions in the presence of Mcm2-WT or Mcm2-S92A. Strikingly, Plx1 could not restore DNA replication under stressful conditions induced by etoposide in Plx1-depleted extracts supplemented with recombinant Mcm2-S92A (Figure 6B). The ability of Plx1 to rescue replication was instead preserved in the presence of Mcm2-WT (Figure 6B). These results suggest that during DNA replication under stressful conditions, ATR-mediated phosphorylation of Mcm2 promotes chromatin recruitment of Plx1, which subsequently releases the block on origin firing imposed by the intra-S-phase checkpoint.
Figure 6.
Mcm2 serine 92 phosphorylation is required for Plx1 function. (A) Chromatin binding of Plx1 in the presence of Mcm2-WT and Mcm2-S92A proteins. Sperm nuclei were incubated for 60 min in extracts supplemented with 200 ng/μl recombinant histidine-tagged Mcm2-WT and Mcm2-S92A proteins in the presence of 5 μM etoposide. Chromatin was isolated and subjected to western blot analysis with anti-Plx1 (Plx1) and anti-histidine (His-Mcm2) antibodies. (B) DNA replication under stress in Plx1-depleted extracts in the presence of Mcm2-WT and Mcm2-S92A proteins. Sperm nuclei (7000 nuclei/μl) were replicated in extracts that were untreated (CTRL), mock depleted (Mock) or Plx1 depleted (ΔPlx1) in the presence of 5 μM etoposide. Extracts were supplemented with buffer (Buff), 200 ng/μl Mcm2-WT or 200 ng/μl Mcm-S92A proteins. Plx1-depleted extracts were also supplemented with 6 ng/μl recombinant Plx1, as indicated.
The absence of Plx1 leads to accumulation of DNA double-strand breaks during DNA replication
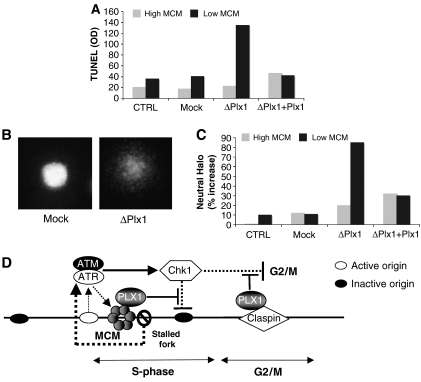
Efficient and complete replication of the genome is essential for genome stability. As Plx1 is required for efficient replication under stress, we asked whether the absence of Plx1 leads to loss of genome integrity. Unstable genomes accumulate double-strand breaks (DSBs) during DNA replication (Trenz et al, 2006). To detect the presence of chromosomal breaks occurring during DNA replication, we used a TUNEL-based assay that we previously developed (Costanzo et al, 2001; Trenz et al, 2006). Low numbers of sperm nuclei were used to allow DNA replication in the absence of Plx1. The nuclei were replicated in mock-depleted, Plx1-depleted or Plx1-depleted extracts supplemented with recombinant Plx1. The chromatin was then isolated and DNA breaks were radioactively labelled with terminal deoxynucleotidyl transferase. We measured a significant increase in chromosomal breaks in the absence of Plx1 when the levels of DNA-bound Mcm complexes were reduced (Figure 7A). The occurrence of chromosomal breaks was prevented by supplementing egg extracts with recombinant Plx1 (Figure 7A). To demonstrate that the breaks occurring during DNA replication were double stranded, we performed the neutral comet assay, which is able to specifically detect DSBs (Trenz et al, 2006). A significant number of nuclei replicated in Plx1-depleted extracts showed the presence of the characteristic neutral comet or halo, which indicates the presence of DSBs (Figure 7B). To quantify the amount of DSBs, we used the head length of the neutral halo corrected for the diameter of untreated nuclei as the main parameter. Using this parameter, we observed a significant accumulation of DSBs in the absence of Plx1 (Figure 7C). These results indicate that Plx1 prevents accumulation of DSBs during chromosomal DNA replication under stressful conditions.
Figure 7.
Plx1 prevents chromosomal breakage during DNA replication. (A) DNA breaks detected with the TUNEL assay. Egg extract was left untreated (CTRL), mock depleted (Mock), Plx1 depleted (ΔPlx1) or Plx1 depleted and supplemented with recombinant Plx1 (ΔPlx1 dep+Plx1). DNA replication was obtained in the absence (grey bar) or presence of 40 nM geminin (black bar). Sperm nuclei were replicated for 90 min, chromatin was isolated and TUNEL assay was performed in the presence of [α-32P]dATP. The DNA was isolated, precipitated and separated on an agarose gel. Incorporated 32P was quantified with the PhosphoImager and optical density (OD) was plotted. (B) Typical image of a nucleus processed with the neutral comet assay from mock-depleted extract (Mock) and from Plx1-depleted extract (ΔPlx1). (C) Quantification of DSBs. Untreated extracts (grey) and extracts supplemented with geminin (black) were left untreated (CTRL), mock depleted (Mock), Plx1 depleted (ΔPlx1) or Plx1 depleted and supplemented with recombinant Plx1 (ΔPlx1+Plx1). The percentage increase in the head length of the comets subtracted from the average nuclear diameter of undepleted samples with high Mcm was used as a parameter to measure DSBs. (D) Model representing the function of Plx1 in DNA replication under stressful conditions. In the presence of stalled replication forks, the ATR/ATM-dependent checkpoint activation promotes Plx1 binding to the Mcm complex close to stalled forks and attenuates the inhibitory activity of the checkpoint towards unfired origins. Activation of the supplementary origins overcomes the stalled replication forks. Claspin-dependent recruitment of Plx1 suppresses the checkpoint that controls mitosis onset in the presence of unreplicated DNA.
Discussion
Plks are fundamental regulators of the cell cycle. Plx1 controls the assembly of the mitotic spindle apparatus and the activation–inactivation cycles of CDK/cyclin complexes during M-phase (Liu and Maller, 2005). In mammalian cells, Plk1 kinase activity increases in M-phase (Golsteyn et al, 1995). However, Plk1 function outside mitosis has remained unknown so far. Here, we show that Plx1 is required for DNA replication in the presence of stalled replication forks induced by different treatments such as the addition of aphidicolin and etoposide or by reducing the level of Mcm complex loaded onto chromatin using low amounts of recombinant geminin. These findings reveal an unanticipated role for Plx1 in the regulation of S-phase progression under stressful conditions. We established that Plx1 is dispensable for unchallenged chromosomal DNA replication. This is consistent with the observed phenotype of Plk1 inactivation in metazoan organisms and it rules out a role for Plx1 in the basic mechanisms of DNA replication such as nuclear formation, establishment of replication complexes and DNA elongation. However, DNA is continuously subjected to damage even under physiological conditions. Therefore, it is likely that the DNA replication machinery frequently encounters damaged templates giving rise to stalled replication forks. During embryonic replication in vertebrate organisms, the number of Mcm complexes loaded onto chromatin exceeds the number of active replication origins (Edwards et al, 2002). Origin firing from the loaded Mcm complexes is dynamically regulated throughout replication by an ATM- and ATR-dependent intra-S-phase checkpoint (Marheineke and Hyrien, 2004; Shechter et al, 2004a). Transient inactivation of this intra-S-phase checkpoint could lead to the firing of supplementary origins that would not fire under normal conditions. This mechanism would ensure stalled fork bypass by recruitment of supplementary origins to complete genome replication under stress (Woodward et al, 2006). However, the mechanism of checkpoint inactivation under stress is unclear, as DNA damage or replication inhibition activates the same intra-S-phase checkpoint pathway that keeps the replication origins inactive. Plx1 has recently been shown to regulate adaptation to the ATR-mediated mitosis onset delay induced by unreplicated DNA (Yoo et al, 2004a). In unicellular organisms, adaptation to DNA damage signalling requires the Plx1 orthologue Cdc5, suggesting that this mechanism is conserved (Toczyski et al, 1997). Our data indicate that Plx1 binds to chromatin in the presence of stalled replication forks and induces suppression of the intra-S-phase checkpoint. Similar to mammalian organisms, Plx1 binds to the Mcm complex in Xenopus through its Polo box domain and its binding is enhanced by checkpoint activation (Tsvetkov and Stern, 2005; Lowery et al, 2007; Stuermer et al, 2007). However, the function of this interaction was not understood at functional level. Our data reveal a direct role for Plx1 in the control of origin firing through the interaction with the Mcm complex. Mcm7 and Mcm2 are phosphorylated by ATM/ATR in the presence of DNA damage (Cortez et al, 2004; Yoo et al, 2004b). As the interaction between Plx1 and the Mcm complex is mediated by the Polo box, which is a phospho-binding domain (Elia et al, 2003), it is plausible that direct phosphorylation of Mcm7 and/or Mcm2 by ATM/ATR could favour Plx1 interaction with the Mcm complex on chromatin. Indeed, we found that the interaction of Plx1 with Mcm proteins is increased by the induction of the checkpoint. Significantly, we show that Plx1 binding to the Mcm complex loaded onto chromatin requires Mcm2 serine 92, which is phosphorylated by ATM/ATR following DNA damage (Yoo et al, 2004b). The Mcm complex containing Mcm2-S92A mutant subunit is unable to support Plx1-mediated recovery of DNA replication under stressful conditions. These observations reveal an unanticipated function for the ATM/ATR-dependent phosphorylation of serine 92 of Mcm2, which appears to promote DNA replication under stress. It is likely that Plx1-mediated suppression of the checkpoint following its binding to the Mcm complex promotes the derepression of supplementary dormant origins. This mechanism probably operates locally to promote activation of origins that otherwise would be kept suppressed by the checkpoint.
After prolonged checkpoint activation, Plx1 interacts with Claspin following ATR-dependent phosphorylation of threonine 906 (Yoo et al, 2004a). This interaction promotes Claspin inactivation and release from chromatin. Claspin inactivation by protein degradation restrains Chk1 activity and promotes recovery from the DNA replication checkpoint in mammalian organisms (Mailand et al, 2006; Peschiaroli et al, 2006). We have recently shown that Tipin, a factor critical for replication fork stabilization (Chou and Elledge, 2006), is required for Claspin binding to chromatin and for efficient Chk1 activation (Errico et al, 2007). However, Tipin depletion does not affect Plx1 binding to chromatin, confirming that Plx1 recruitment to chromatin at the onset of DNA replication takes place through a mechanism distinct from that of Claspin-mediated adaptation to the checkpoint. Plx1-mediated inactivation of Chk1 at the onset of S-phase might happen at the level of replication origins following Mcm binding and might be due to Plx1-mediated phosphorylation of factors required for efficient Chk1 phosphorylation, such as Tipin (Errico et al, 2007). This event precedes the suppression of Chk1 phosphorylation at the transition into mitosis, which requires Plx1 binding to Claspin and probably a more robust signalling from Plx1 whose activity increases at mitosis onset in Xenopus (Kelm et al, 2002). Consistent with this interpretation, we show that, differently from mitosis onset, Plx1 does not promote Claspin release from chromatin and does not affect Claspin stability during early S-phase. These data suggest that Plx1-mediated inactivation of Claspin after persistent checkpoint activation regulates the transition from S-phase into mitosis whereas Mcm-mediated recruitment of Plx1 promotes replication progression in the presence of stalled replication forks (Figure 7D).
Checkpoint activation in Xenopus leads to downregulation of S-phase kinase activities (Costanzo et al, 2000, 2003). We have shown that Plx1-mediated suppression of the checkpoint affects Cdk2 activity. The absence of Plx1 induces a strong suppression of Cdk2 levels that can be restored by caffeine treatment. This suggests that the effects on Cdk2 activity are mediated by the hyperactivation of the checkpoint induced by Plx1 deficiency. However, we cannot exclude the existence of an alternative mechanism in the suppression of Cdk2 activity. During mitosis, Plx1 promotes Cdk1 activation through direct phosphorylation and maintenance of Cdc25 phosphatase activity, which dephosphorylates the inhibitory tyrosine 15 of Cdk1 (Kumagai and Dunphy, 1996; Liu and Maller, 2005). The checkpoint-induced downregulation of Cdk2 activity in egg extracts is mediated by the inactivating phosphorylation on tyrosine 15 of Cdk2/Cyclin E complex, which is under the control of Cdc25 (Costanzo et al, 2000). In the light of these observations, we cannot exclude that suppression of Cdk2 activity is also due to the direct role of Plx1 in controlling the activity of one of the Cdc25 phosphatases present in egg extracts.
Overall, our data suggest a model in which Plx1 is required for modulating the intra-S-phase checkpoint that regulates DNA replication progression in an embryonic model system (Figure 7D). However, Plx1 activity could also be important for somatic DNA replication. Plk1 is overexpressed in many tumours (Ahr et al, 2001). Downregulation of Plk1 expression leads to cell death in transformed cells (Liu and Erikson, 2003a, 2003b), suggesting that Plk1 is required for the cell cycle of tumour cells. The genome of tumour cells is highly unstable and has many DNA lesions that would normally interfere with replication progression. In this case, overexpression of factors such as Plk1 might drive DNA replication forward, overcoming the effects of replication stress. We show that Plx1 is required for preventing accumulation of DSBs in the presence of a restricted number of replication origins induced by geminin. This situation might actually mimic DNA replication in tumour cells where geminin is frequently deregulated (Montanari et al, 2006). Consistent with our results, replication in the absence of Plk1 in transformed human cells leads to accumulation of DSBs, as shown by the accumulation of phosphorylated histone H2AX foci (Liu et al, 2006). In addition, we show that Plx1 is required for DNA replication in the presence of etoposide, a widely used anti-cancer drug. This result could be relevant to understand the molecular mechanisms underlying the response of cancer cells to chemotherapeutic strategies based on the use of this drug. Overall, our study provides new insights into the role of Plx1, indicating that in addition to mitotic functions this enzyme preserves genome integrity by promoting DNA replication under stressful conditions.
Materials and methods
Additional Materials and methods can be found in Supplementary data.
X. laevis egg extracts
Mature X. laevis females were primed about 1 week in advance with 50 U of pregnant mare serum gonadotropin per animal. To induce ovulation, 400 U of human chorionic gonadotropin per animal was used. The eggs were collected in 90 mM NaCl. To prepare interphase extracts, the eggs were dejelled in a buffer (20 mM Tris, pH 8.5, 110 mM NaCl, 5 mM DTT) for 5 min, washed with ¼ Marc's modified Ringer (MMR) (5 × MMR: 100 mM HEPES, pH 7.5, 2 M NaCl, 10 mM KCl, 5 mM MgSO4, 10 mM CaCl2, 0.5 mM EDTA) and activated with 1 μg/ml calcium ionophore A23187 in MMR for 5 min. The activated eggs were washed with ¼ MMR and then washed three times with ice-cold S-buffer (0.25 M sucrose, 50 mM KCl, 2.5 mM MgCl2, 2 mM β-mercaptoethanol, 15 μg/ml leupeptin and 50 mM HEPES–KOH at pH 7.5). The eggs were packed by spinning and then crushed at 13 000 r.p.m. for 15 min. The cytoplasmic fraction between lipid cap and pellet was collected, supplemented with cytochalasin B (40 μg/ml final concentration) and centrifuged at 70 000 r.p.m. for 15 min to remove residual debris. The cytosolic and membrane fractions were collected and supplemented with 30 mM creatine phosphate and 150 mg/ml creatine phosphokinase. Extracts were then snap-frozen with 3% glycerol in beads of 20 μl. Frozen beads were thawed and used immediately.
Antibodies
Rabbit polyclonal antibodies against Plx1 were raised against recombinant histidine-tagged Plx1 expressed in baculovirus (Harlan). Mouse monoclonal antibodies against Mcm7 were purchased from Santa Cruz (SC-9966). To detect Chk1 phosphorylation, a phospho-specific antibody (Cell Signalling Technology, cat. no. 2341) raised against human Chk1 phosphorylated at S345 (S344 in Xenopus) was used. For detection of total Chk1 levels, an antibody from Santa Cruz Biotechnology was used (SC-8408). Antibodies against Xenopus Cdk2 and Orc1 were a gift from Julian Gannon. Antibodies against Xenopus Claspin were a gift from Howard Lindsay. Xenopus Tipin and Mcm3 antibodies have been described previously (Costanzo et al, 2000; Errico et al, 2007). Anti-phospho-human Mcm2 serine 108 antibodies (crossreacting with Xenopus Mcm2 serine 92) were obtained from Bethyl and anti-histidine antibodies were from Sigma. Infrared dye-labelled secondary antibodies were obtained from Li-COR Bioscience (goat anti-mouse IR Dye 680 for the detection of anti-Mcm7; goat anti-rabbit IR Dye 800CW for the detection of anti-Plx1). The western blot procedures were performed using ECL or ECL plus (Amersham).
Plx1, Mcm3 and Tipin depletion
Interphase extracts (500 μl) were incubated three times for 40 min with 100 μl of protein A-Sepharose beads, 4B fast flow (Sigma) coupled to 200 μl of anti-Plx1 serum, 100 μl of anti-Mcm3 or 200 μl of anti-Tipin serum at 4°C for 2 h in the presence of 1 mM AEBSF (4-(2-aminoethyl)benzenesulphonyl fluoride hydrochloride). Mock depletions were performed using beads coupled to 200 μl of pre-immune serum. The coupling to the beads was performed in EB buffer (100 mM KCl, 50 mM HEPES, pH 7.5, 2.5 mM MgCl2)/0.2% NP-40. After the coupling reaction, the beads were first washed extensively with EB buffer/0.2% NP-40 and then with EB buffer.
Recombinant proteins
6xHistidine-tagged geminin (a gift from M Micheal) was expressed in BL21(DES) cells from a pET28 plasmid and purified on a nickel-NTA column according to standard protocols. Baculoviruses encoding 6xHistidine-tagged Plx1 (a gift from J Gautier), Mcm2-WT and Mcm2-S92A (a gift from W Dunphy) were used to infect Sf9 cells, which were expanded in large flasks using complete GRACE medium according to standard protocols (Invitrogen). 6xHistidine-tagged Plx1, Mcm2-WT or Mcm2-S92A proteins were purified on a nickel-NTA column (Qiagen) according to the manufacturer's protocol.
Replication assay
DNA replication was performed as described previously (Costanzo et al, 2001). Briefly, sperm nuclei at the concentration of 2000–7000 nuclei/μl, as indicated in the figure legend, were added to interphase extracts and replicated in the presence of [α-32P]dATP at 23°C. The amounts of geminin, aphidicolin and etoposide (Sigma) were titrated for each extract due to the different checkpoint activation thresholds of each batch. Usually 2.5–5 μM aphidicolin, 20–60 nM geminin or 5–10 μM etoposide was used. EB buffer was used to compensate for volume differences after reagents addition. The experiments were repeated three times.
If not otherwise indicated the replication reactions were stopped at 120 min with Stop buffer (1% SDS, 80 mM Tris, pH 8, 8 mM EDTA). For pulse labelling, aliquots of the replication reaction were incubated for 30 min in the presence of [α-32P]dATP. The samples were digested with 1 mg/ml proteinase K for 45 min, extracted with phenol/chloroform/isoamylalcohol, ethanol-precipitated and separated on 0.8% agarose gel. The gel was fixed in 30% TCA for 20 min, dried and exposed for autoradiography. For quantification of DNA replication, the gel was exposed to a phosphoscreen, the signal was obtained with a PhosphoImager and quantified by ImageQuant software. Replication of single-stranded DNA of M13 phage was performed in a similar manner using 12.5 ng/μl of DNA.
Chromatin binding
Chromatin was isolated as described previously (Costanzo et al, 2003) with slight modifications. Chromatin was assembled in 50 μl of interphase extracts. The extracts were untreated, mock depleted, Plx1 depleted or Plx1 depleted and supplemented with 6 ng/μl Plx1 in the presence of 5 μM aphidicolin (Sigma) or a low concentration of geminin (40 nM). For the experiment in Figure 6A, the extract was supplemented with 5 μM etoposide in the presence of 200 ng/μl of Mcm2-WT or 200 ng/μl of Mcm2-SA92 proteins. For all chromatin binding experiments, 4000 sperm nuclei/μl were incubated for 30, 60 or 90 min. To isolate the chromatin, the extracts were diluted in 500 μl of EB buffer (100 mM KCl, 50 mM HEPES, pH 7.5, 2.5 mM MgCl2)/0.25% NP-40 and layered onto 100 μl of a 30% sucrose cushion made of the same buffer. The chromatin was spun at 10 000 g for 5 min at 4°C, using a swing-out bucket rotor, washed with 500 μl EB buffer and spun again at 10 000 g for 5 min, in a fixed-angle rotor. The pellet was resuspended in Laemmli loading buffer, loaded onto a 10% SDS–PAGE and analysed by western blotting with specific antibodies, as indicated in the figure legend. To quantify the amount of Plx1 loaded onto the chromatin in the presence of aphidicolin, secondary antibodies labelled with infrared dye were used. The anti-mouse secondary antibody, detecting Mcm7, had an emission at 680 nm (displayed in red in Figure 2A) and the anti-rabbit secondary antibody detecting Plx1 had an emission at 800 nm (displayed in green in Figure 2A). The amount of chromatin-bound protein was quantified with an LI-COR Infrared Imager (Biosciences).
Neutral comet assay
Sperm nuclei (2000 nuclei/μl) were replicated in 50 μl of interphase extract at 23°C. Interphase extract was used without any treatment, mock depleted, Plx1 depleted or Plx1 depleted and supplemented with recombinant Plx1 (6 ng/μl). After 120 min, the samples were diluted in 1 ml PBS, the nuclei were spun down at 2000 r.p.m. for 5 min and the supernatant was removed. Nuclei were resuspended and processed for the comet assay. The neutral version (pH 8.3) of the comet assay was performed according to Trenz et al (2006). Aliquots of 6 μl of sperm nuclei suspension were mixed with 60 μl of low-melting agarose (0.5% in PBS) and added to microscope slides (Trevigen) pre-coated with a bottom layer of 1.5% agarose. Nuclei were lysed for 1 h at 4°C in a lysis solution at pH 9.5 (2.5 M NaCl, 100 mM EDTA, 10 mM Tris base, 1% Triton X-100 and 10% dimethyl sulphoxide). The slides were washed three times in the electrophoresis buffer (100 mM Tris, 300 mM sodium acetate, adjusted to pH 8.3) and then equilibrated for 60 min in the same buffer. Slides were transferred to an electrophoresis chamber, covered with fresh buffer and electrophoresed for 60 min at 4°C at 14 V (0.48 V/cm) and 120 mA. Slides were then washed three times with alkaline buffer (300 mM NaOH, 1 mM EDTA) followed by three brief washes with 0.4 M Tris pH 7.5. The slides were dehydrated in 70% ethanol and then air-dried. Images of 100 randomly selected ethidium bromide-stained cells were analysed from each coded slide at × 400 magnification. To measure the extent of DNA damage, image analysis software (Comet IV, Perceptive Instruments) was used. DNA damage was assessed by monitoring comet head length analysing 100 cells for each slide.
Supplementary Material
Supplementary Figures
Supplementary Data
Acknowledgments
We thank T Hunt, members of Genome Stability Unit and Clare Hall Laboratories for critical discussion. We are grateful to W Dunphy for the MCM2-WT and MCM2-S92A baculoviruses. This work was supported by Cancer Research UK, the Lister Institute for Preventive Medicine and the EMBO Young Investigator program awarded to VC.
References
- Ahr A, Holtrich U, Solbach C, Scharl A, Strebhardt K, Karn T, Kaufmann M (2001) Molecular classification of breast cancer patients by gene expression profiling. J Pathol 195: 312–320 [DOI] [PubMed] [Google Scholar]
- Chen H, Kovar J, Sissons S, Cox K, Matter W, Chadwell F, Luan P, Vlahos CJ, Schutz-Geschwender A, Olive DM (2005) A cell-based immunocytochemical assay for monitoring kinase signaling pathways and drug efficacy. Anal Biochem 338: 136–142 [DOI] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ (2006) Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci USA 103: 18143–18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Glick G, Elledge SJ (2004) Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA 101: 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Gautier J (2003) Single-strand DNA gaps trigger an ATR- and Cdc7-dependent checkpoint. Cell Cycle 2: 17. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J (2001) Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Ying CY, Kim E, Avvedimento E, Gottesman M, Grieco D, Gautier J (2000) Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol Cell 6: 649–659 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J (2003) An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 11: 203–213 [DOI] [PubMed] [Google Scholar]
- Diffley JF (1994) Eukaryotic DNA replication. Curr Opin Cell Biol 6: 368–372 [DOI] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC (2002) MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem 277: 33049–33057 [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB (2003) The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115: 83–95 [DOI] [PubMed] [Google Scholar]
- Errico A, Costanzo V, Hunt T (2007) Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci USA 104: 14929–14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ (2007) Dormant origins licensed by excess Mcm2 7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn RM, Lane HA, Mundt KE, Arnaud L, Nigg EA (1996) The family of polo-like kinases. Prog Cell Cycle Res 2: 107–114 [DOI] [PubMed] [Google Scholar]
- Golsteyn RM, Mundt KE, Fry AM, Nigg EA (1995) Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol 129: 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Saxena JK, Kumar A, Wilson SH, Ackerman EJ (1992) DNA polymerase beta and DNA synthesis in Xenopus oocytes and in a nuclear extract. Science 258: 475–478 [DOI] [PubMed] [Google Scholar]
- Kelm O, Wind M, Lehmann WD, Nigg EA (2002) Cell cycle-regulated phosphorylation of the Xenopus polo-like kinase Plx1. J Biol Chem 277: 25247–25256 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (1996) Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Liu J, Maller JL (2005) Xenopus Polo-like kinase Plx1: a multifunctional mitotic kinase. Oncogene 24: 238–247 [DOI] [PubMed] [Google Scholar]
- Liu X, Erikson RL (2003a) Polo-like kinase 1 in the life and death of cancer cells. Cell Cycle 2: 424–425 [PubMed] [Google Scholar]
- Liu X, Erikson RL (2003b) Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci USA 100: 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lei M, Erikson RL (2006) Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol 26: 2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB (2007) Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J 26: 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Bartek J, Lukas J (2006) Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell 23: 307–318 [DOI] [PubMed] [Google Scholar]
- Marheineke K, Hyrien O (2004) Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J Biol Chem 279: 28071–28081 [DOI] [PubMed] [Google Scholar]
- Masuda T, Mimura S, Takisawa H (2003) CDK- and Cdc45-dependent priming of the MCM complex on chromatin during S-phase in Xenopus egg extracts: possible activation of MCM helicase by association with Cdc45. Genes Cells 8: 145–161 [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Montanari M, Macaluso M, Cittadini A, Giordano A (2006) Role of geminin: from normal control of DNA replication to cancer formation and progression? Cell Death Differ 13: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M (2006) SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell 23: 319–329 [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004a) ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 6: 648–655 [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004b) Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 3: 901–908 [DOI] [PubMed] [Google Scholar]
- Stuermer A, Hoehn K, Faul T, Auth T, Brand N, Kneissl M, Putter V, Grummt F (2007) Mouse pre-replicative complex proteins colocalise and interact with the centrosome. Eur J Cell Biol 86: 37–50 [DOI] [PubMed] [Google Scholar]
- Toczyski DP, Galgoczy DJ, Hartwell LH (1997) CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90: 1097–1106 [DOI] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov L, Stern DF (2005) Interaction of chromatin-associated Plk1 and Mcm7. J Biol Chem 280: 11943–11947 [DOI] [PubMed] [Google Scholar]
- Walter J, Newport JW (1997) Regulation of replicon size in Xenopus egg extracts. Science 275: 993–995 [DOI] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ (2006) Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2004a) Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 117: 575–588 [DOI] [PubMed] [Google Scholar]
- Yoo HY, Shevchenko A, Dunphy WG (2004b) Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem 279: 53353–53364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Data