Abstract
The GTP-binding protein Rap1 regulates integrin-mediated and other cell adhesion processes. Unlike most other Ras-related proteins, it contains a threonine in switch II instead of a glutamine (Gln61 in Ras), a residue crucial for the GTPase reaction of most G proteins. Furthermore, unlike most other GTPase-activating proteins (GAPs) for small G proteins, which supply a catalytically important Arg-finger, no arginine residue of RapGAP makes a significant contribution to the GTPase reaction of Rap1. For a detailed understanding of the reaction mechanism, we have solved the structure of Rap1 in complex with Rap1GAP. It shows that the Thr61 of Rap is away from the active site and that an invariant asparagine of RapGAPs, the Asn-thumb, takes over the role of the cis-glutamine of Ras, Rho or Ran. The structure and biochemical data allow to further explain the mechanism and to define the important role of a conserved tyrosine. The structure and biochemical data furthermore show that the RapGAP homologous region of the tumour suppressor Tuberin is sufficient for catalysis on Rheb.
Keywords: GAP, G proteins, GTPase, Rap
Introduction
The Rap1 protein has an essential function in integrin-mediated cell–cell adhesion and other cell adhesion processes (Bos et al, 2001). It has also been reported to activate extracellular signal-regulated kinases (Hattori and Minato, 2003). Rap1 belongs to the superfamily of Ras-like guanine-nucleotide-binding proteins (GNBPs, G proteins) sharing more than 50% sequence identity with Ras. Of the five human Rap isoforms, Rap1A/B, Rap2A/B/C, Rap1A and Rap1B share more than 90% sequence identity and show no apparent difference in cellular function. G proteins cycle between an inactive, GDP-bound and an active, GTP-bound state. The active conformation allows interaction with effector proteins activating different signalling cascades. Regulation of this cycle is achieved by specific sets of guanine-nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (Vetter and Wittinghofer, 2001; Bos et al, 2007). GEFs catalyse the nucleotide-releasing step. An excess of GTP over GDP in the cell subsequently results in binding of GTP and reactivation of the GNBP. In contrast, GAPs stimulate the inefficient intrinsic GTP-hydrolysis by orders of magnitude. GEFs bind to G protein–nucleotide complexes and use a variety of ways to decrease the affinity of the nucleotide. GAPs stimulate hydrolysis by complementing and/or stabilising the G protein active site.
GTPase-activation in Ras, Rho and Ran is based on a correct positioning of the nucleophilic water by a crucial glutamine residue from the G protein, while Sar1 and most likely elongation factors such EF1 use an His for the same purpose (Bi et al, 2002; Daviter et al, 2003). In case of Rabs, the glutamine is supplied in trans by the RabGAP TBC domain (Pan et al, 2006). Furthermore, GAPs for Ras, Rho and Rab supply an arginine residue into the active site whose positive charge neutralises developing negative charge in the hydrolysis step and thus lowers the activating energy of the chemical step. In Rap proteins, the crucial glutamine residue (Gln61 in Ras) is replaced by threonine (Thr61), and this residue has been shown to be required for binding rather than catalysis (Chakrabarti et al, 2007). Sequence analysis has identified a number of different RapGAPs containing seven conserved arginines. Mutational analysis has shown that they have, if anything, only a minor effect on catalysis, which led us to conclude that RapGAP-mediated GTP hydrolysis on Rap follows an alternative mechanism (Brinkmann et al, 2002; Kraemer et al, 2002). Biochemical studies and the X-ray structure of Rap1GAP (Daumke et al, 2004) led us to propose that an invariant asparagine, Asn290 in Rap1GAP, is a crucial residue in the catalytic mechanism of Rap1GAP, and that the ‘Asn-thumb' may insert into the active site of Rap1.
Tuberin (Tsc2) together with Hamartin (TSC1) forms the tuberous sclerosis complex (TSC). Mutations in TSC1 or TSC2 lead to the formation of hamartomas in a wide range of tissues. Tuberin has been shown to inactivate Rheb, a protein involved in the mTOR signalling pathway (Garami et al, 2003). The C-terminal end of Tuberin shares significant sequence similarity with the catalytic, but not the dimerisation domain of Rap1GAP (Supplementary Figure S1). Due to this high similarity, the presence of the ‘Asn-thumb' in Tuberin and its mutation in TSC patients, it has been assumed that Rheb-inactivation by Tuberin follows the same mechanism as in Rap–Rap1GAP (Inoki et al, 2003; Li et al, 2004).
Structures of G protein–GAP complexes have been determined for Ras, Rho, Ran, Sar1 and Rab; however, no structural data for the Rap–Rap1GAP complex is available. Here, we present the complex structure of Rap–Rap1GAP in the GDP-BeFx-bound state at a resolution of 3.4 Å, showing that the ‘Asn-thumb' occupies the position of the catalytic glutamine residue in those other systems, and that Thr61 of Rap has no role in catalysis. Based on its structure, we performed mutational studies to analyse complex formation and hydrolysis. The knowledge gained from Rap–Rap1GAP was then applied to the Rheb–Tuberin reaction, which suggests that the RapGAP homology domain of Tuberin is sufficient for the chemistry of the Rheb GTPase reaction.
Results
Complex formation and crystallisation
To isolate the complex between Rap1B and Rap1GAP, several phosphate mimics and nucleotide analogues have been tested. Using GDP in combination with BeFx, a mimic of the ground state of GTP, or AlFx, mimicking the transition state of hydrolysis, a tight complex (see also Chakrabarti et al, 2007) could be isolated by gel filtration and showed equimolar stoichiometry based on band intensities in SDS–PAGE. Alternatively, GTP analogues such as GppNHp also resulted in a complex of Rap1B–Rap1GAP. Although all three forms could be crystallised under similar conditions, only crystals of the BeFx-bound complex exhibited improved diffraction quality compared with crystals obtained with GDP-AlFx or GppNHp. This complex crystallised in space group P3(1)21 (Table I), containing three molecules of Rap1GAP (molecules A,B,C) and one molecule of Rap (molecule D). Two of these Rap1GAP molecules (molecules B and C) form a dimer within the asymmetric unit corresponding to the Rap1GAP homodimer that has already been described by Daumke et al (2004), one of which is bound to Rap1B. The third Rap1GAP (molecule A) forms a dimer with its symmetry related molecule A* (Supplementary Figure S2). Although complex purification resulted in an apparently equimolar complex, crystal packing and the low affinity of the complex apparently favoured the incorporation of only one Rap1B–Rap1GAP complex and two additional Rap1GAP molecules.
Table 1.
Data collection and refinement statistics
| Rap–GDP·BeF3−-Rap1GAP(Q204A) | |
|---|---|
| Data collection | |
| X-ray source | SLS X10SA PXII |
| Space group | P3(1)21 |
| Cell parameters | a=b=209.73 Å, c=108.22 Å |
| Resolution (Å) | 20.0–3.4 (3.5–3.4) |
| Wavelength (Å) | 0.97916 |
| Completeness (%) | 98.3 (94.9) |
| Unique reflections | 37 307 (2979) |
| Redundancy | 6.6 (3.4) |
| Rsym (%) | 8.7 (32.4) |
| I/σI | 15.6 (3.7) |
| Refinement | |
| PDB code | 3BRW |
| Rwork (%)a | 23.4 |
| Rfree (%)b | 28.0 |
| Reflections (work/free) | 35 759 (33 881/1878) |
| RMSD/av. (sigma) | |
| Bond length (Å) | 0.008/0.022 |
| Bond angle (deg) | 1.128/1.962 |
| Ramachandran plot region most favoured/additional allowed/generously allowed/ disallowed (%) | 82.9/14.8/1.2/1.1 |
| No. of atoms (protein/water/ ligand) | 9481 (9436/12/33) |
| Avg B-factor (Å2) | 97.48 |
| Values in parentheses are for last-resolution shell. | |
| aRwork=∑h∣Fo–Fc∣/∑h∣Fo∣, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h. | |
| bRfree is the same as Rwork, but calculated on the reflections set aside from refinement. | |
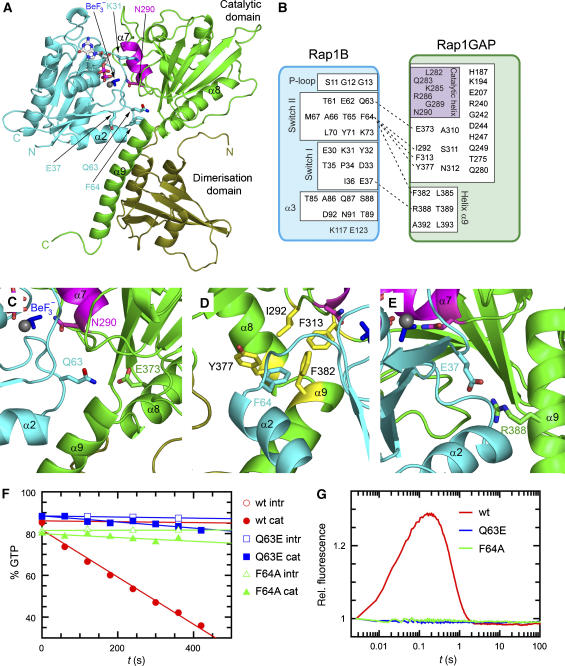
The contact between Rap1B and Rap1GAP involves, as expected, the nucleotide-binding site and both switch regions on Rap1B. Previously, we had identified Rap1GAP to consist of two subdomains. If we consider helix α9 as part of the catalytic domain, which is reasonable considering its higher conservation compared with the dimerisation domain, the major contact on Rap1GAP is via the catalytic domain (green, Figure 1a and b). The interface buries a total surface of approximately 2300 Å2 (calculated using CNS with probe radius of 1.5 Å; Brunger et al, 1998). A detailed analysis of the heterodimer interface (Figure 1b) shows that most residues of switch I and II are located in the interface and involved in the interaction, explaining the specificity for the triphosphate-bound state. In switch I, all residues from Glu30 to Glu37 are involved, including Lys31, which is the residue-determining specificity for the interaction of Ras and Rap with effectors such as Raf and RalGDS (Nassar et al, 1996). Similarly, many residues of switch II appear crucial for binding, including Gln63 and Phe64 (see below) that are different between Ras and Rap and, together with Lys31, are likely determinants of specificity of the GAP reaction. Apart from the switch regions and the P-loop, helix α3 additionally contributes to complex formation.
Figure 1.
Rap–Rap1GAP complex and interface analysis. (A) Ribbon representation of the Rap–GDP·BeF3−-Rap1GAP complex with Rap1B in cyan and Rap1GAP in green (catalytic domain green; dimerisation domain olive green). The catalytic helix containing the Asn-thumb (Asn290) is shown in magenta, GDP-BeF3− as ball-and-stick. (B) Schematic representation of interacting residues. Interactions shown in detail in (C–E) are depicted with a dashed line. (C–E) Structural details of interactions between Rap1B and Rap1GAP, with colours as in (A). (F) HPLC-based analysis of the Rap-stimulated GTPase reaction, with 200 μM wt and mutant Rap and 100 nM Rap1GAP. (G) Stopped-flow analysis of the interaction between 2 μM Aedans-labelled wt and mutant Rap and 50 μM Rap1GAP; reaction was followed by monitoring fluorescence through a 408 nm cutoff filter. Wt and mutant Rap contain the A86C mutation, which has been shown to behave as wild type, as described earlier (Kraemer et al, 2002; Chakrabarti et al, 2007).
Based on previous mutagenesis studies, helix α7 (shown in magenta) had been assigned the catalytic centre of Rap1GAP. The structure shows indeed that α7, the most highly conserved part of the molecule, is close to the active site of Rap1. All residues of α7 are involved in numerous contacts to either the rest of the RapGAP molecule or to Rap1. The residues on the exposed side of the helix, Leu282G, Lys285G, Arg286G, Gly289, Asn290G (R and G superscript denote residues on Rap1B and Rap1GAP, respectively) are all involved in forming the protein–protein interface. The interface with Rap1 involves hydrophobic contacts towards Gly12R, Tyr32R and Pro34R and polar contacts with Ser88R (Supplementary Figure S3). The position of helix α7 is extensively stabilised by residues within the catalytic domain of Rap1GAP. We have previously shown that the positioning of this helix on the surface of Rap1GAP is apparently crucial for its function, as mutations destabilising its location have a dramatic effect on catalytic efficiency without having an effect on kcat. Helix α7 is close to the P-loop as well as to switch I in Rap1B. The close proximity of α7 and particularly Asn290, the Asn-thumb, to the γ-phosphate of RapGTP (here BeF3− mimicking the γ-phosphate) supports its presumed role in the stimulation of GTP hydrolysis (see below).
Affinity mutants
The crystal structure of the complex allowed us to learn more about the requirements of the interaction and to find mutants suitable to function as constitutive GTPase-negative Rap variants, as we have previously shown that the G12V mutation of Rap1 is still responsive to GAP-mediated GTP hydrolysis. Gln63R from switch II forms a contact to the conserved Glu373G as shown in Figure 1c. Mutation of Gln63R to glutamate, as found in Ras, was expected to result in charge repulsion and inhibition of complex formation. Phe64R points into a hydrophobic cavity on Rap1GAP, which is formed by Ile292G, Phe313G, Tyr377G and Phe382G (Figure 1d). Residue Glu37R in switch I interacts with the invariant Arg388G on helix α9 (Figure 1e). Both the Q63ER and F64AR mutants were analysed for the GAP-stimulated GTPase reaction using HPLC. The respective initial rates for the intrinsic and Rap1GAP-catalysed reactions were determined by linear regression using one standard set of concentrations (200 μM Rap1; 100 nM Rap1GAP). Under the conditions used, the activity is reduced 7.7 and 12.5 fold for Q63ER (0.030 μM/s) and F64AR (0.018 μM/s), respectively, as compared with wild-type Rap1B (0.224 μM/s), such that the reaction is almost nonstimulated as compared with the intrinsic wild-type reaction (0.004 μM/s; Figure 1f). For a more mechanistic insight, we used a previously developed stopped-flow system (Kraemer et al, 2002), whereby fluorescently labelled Rap1B is rapidly mixed with Rap1GAP. The wild-type Rap1B shows a rapid increase and subsequent decrease of fluorescence indicating the association and post-hydrolysis dissociation reaction, whereas no fluorescence increase is observed for the Rap mutants, indicating that loss of GTPase activity is due to the inability to form a Rap–RapGAP complex (Figure 1g). The R388AG mutation, which, as derived from the complex structure, disrupts the interaction with E37R, was previously shown (Chakrabarti et al, 2007) to drastically reduce the affinity to Rap without affecting the catalytic step. This was measured by FTIR at high protein concentrations to overcome the affinity and complex-formation problem. Based on those previous results and the lack of association signals in the stopped-flow experiment (Figure 1g), the three Rap1 mutants are well suited as mutants blocking the interaction between Rap1 and Rap1GAP. Mutation of E37R, a switch I residue located in and close to the interface of complexes of Ras and Rap with Ras association (RA) or Ras-binding (RB) domains, is expected to disturb the interaction with effector proteins. We would, however, expect that mutation of either Q63R or F64R does not interfere with effector binding, if it involves switch I residues and the RA/RB domains. These two mutations could thus be useful as GAP-insensitive mutants for further in vitro and in vivo studies, assuming that effector-binding capacity is not perturbed.
The Asparagine thumb
Rho, Rab, Ran and Ras subfamily proteins use a glutamine residue to position the nucleophilic water relative to the γ-phosphate (Vetter and Wittinghofer, 2001; Bos et al, 2007). GAPs specific for Rho, Rab and Ras supply an Arg residue to stabilise the position of the catalytic Gln and to neutralise negative charge (Rittinger et al, 1997; Scheffzek et al, 1997; Seewald et al, 2002; Pan et al, 2006). As the arginine is inserted into the active site, it has been called the arginine finger (Scheffzek et al, 1997). The Rap–RapGAP system lacks both the intrinsic Gln from Rap and the Arg-finger in trans (Brinkmann et al, 2002).
Previous biochemical analysis of the Rap–Rap1GAP system by Daumke et al (2004) already pointed to the crucial role of Asn290, mutation of which resulted in a drastically reduced activity without any negative effect on complex formation.
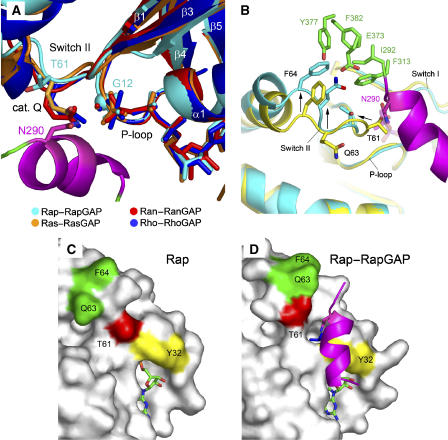
The structure of the Rap–RapGAP complex now allows the role of Asn290 to be analysed. It is situated on helix α7 and points indeed into the active site, justifying it being called the Asn-thumb in analogy to the arginine finger. It is close to BeFx, which is modelled as tetrahedrally coordinated (Figure 1c and e; Supplementary Figure S4a). BeF3− is expected to mimic the stereochemistry of the γ-phosphate in the ground state and would be different from AlFx complexes where AlFx forms a flat tri- or tetragonal base with oxygens from the GDP β-phosphate and the nucleophile occupying the apical positions of the bipyramidal arrangement. Figure 2a shows an overlay of the Rap1B–Rap1GAP complex with previously published complex structures of small G proteins and their respective GAPs. Overall the secondary structure elements (α1, α2, β1, β2) and the P-loop around the active sites of the four structures look very similar. The catalytic (cis) Gln residues of Ran (Gln69), Ras (Gln61) and Rho (Gln63) in the cognate GAP complexes overlay well and are in position to stabilise (and/or polarise) the water molecule for the cleavage step. Rap instead carries a threonine (Thr61) at this position that points away from the active site, whereas in the GAP-free structure, it is in a position to block access of the Asn-thumb to the active site (Figure 2b–d). Upon complex formation, switch II is rearranged to release blockade by reorienting Thr61. This process involves the residues Q63 and F64, both of which pull switch II into the new conformation upon interaction with Rap1GAP (see also Supplementary Figure S4b). This allows the Asn-thumb Asn290 to be introduced into the active site of Rap in trans. Asn290 approaches the active site from a totally different angle, but overlays with the glutamines such that the carboxamide side chains are in very similar positions. Although the nucleophilic water is not visible in this structure due to the lower resolution, it can be assumed that the role of the Asn is analogous to that of Gln. Mutation of the cis-Gln in Ras, Rho and Ran, the trans-Gln in RabGAP or the trans-Asn in RapGAPs leads to a total loss of GAP-stimulated GTPase activity, stressing the importance of a properly located carboxamide side chain for catalysis of small G proteins.
Figure 2.
The active site. (A) Active site of Rap–Rap1GAP, shown as superimposition with Ras, Ran and Rho in complex with their cognate GAPs. Asn290 in Rap1GAP occupies the position of the catalytic Gln in Ras (Gln61), Ran (Gln69) and Rho (Gln63). The G12 position in Rap1B is marked with a sphere. (B) Superimposition of uncomplexed Rap (yellow) and the Rap–Rap1GAP (cyan/green) complex. Interaction of Gln63 and Phe64 with residues on Rap1GAP (green) forces switch II into an alternative conformation (arrows) to release blockade by Thr61 thereby allowing Asn290 to enter the active site. (C, D) Surface representation of uncomplexed Rap (C) and Rap in complex with Rap1GAP (D). The switch II residues T61 (red) and Q63/F64 (green) undergo a drastic conformational change upon complex formation with Rap1GAP to allow access for the Asn-thumb to the active site.
The role of Tyr32
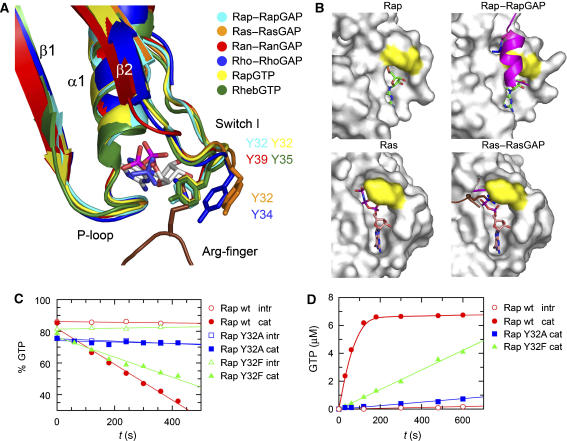
As mentioned in the introduction, GAPs for Rho and Ras introduce an arginine finger into the active site to facilitate the GTP hydrolysis, whereas in GAPs for Rap and Ran this catalytic residue is not present. To further analyse and compare small G proteins that use and do not use an arginine finger, we analysed the structural differences between these two groups of proteins. An overlay in the switch I region indeed shows a different localisation of a highly conserved tyrosine in the two systems (Figure 3a; Supplementary Figure S4c). Tyr32 in Ras and Tyr34 in Rho are in a different position compared with Tyr32 of Rap and Tyr39 of Ran. The tyrosines in Ras-RasGAP and Rho-RhoGAP complexes are in an open conformation, which allows the catalytic arginine to insert into the active site, while in the case of the Ran–RanGAP and Rap–RapGAP complexes, it is in a closed conformation where it would clash with a potential catalytic arginine (Figure 3a). Incidentally, both RanGAP and RapGAP have an arginine close to the active site: Arg286 in the case of RapGAP, which, however, do not have an appreciable effect on catalysis (Brinkmann et al, 2002; Seewald et al, 2002). In the structure of Rap2-GTP (Cherfils et al, 1997), as well as in the structure of Rheb-GTP or GppNHp (Yu et al, 2005), Tyr32 was found in a position identical to that in the presence of RapGAP (Figure 3a), indicating that it is already positioned for GAP binding and catalysis. It has also been speculated that the hydrogen bond between the phenolic OH and the γ-phosphate is important for catalysis of Rap proteins (Cherfils et al, 1997).
Figure 3.
Role of Tyr32. (A) Superimposition of active sites from various structures as indicated, with an emphasis on the conformation of Tyr32. Systems using an Arg-finger (RasGAP/RhoGAP) show Tyr32 in a more open versus a more closed conformation for Ran and Rap. (B) Surface representation of uncomplexed Rap/Ras or in complex with their respective GAPs, with residue Tyr32 labelled in yellow. The Rap or Ras structures are shown in slightly different orientations. Catalytic elements from the GAP, the catalytic helix with the Asn-thumb and the Arg-finger (magenta and brown, respectively) are shown as ribbon. (C) HPLC-based analysis of the GTPase stimulation by Rap1GAP for Rap wt and Y32-mutants (described in Figure 1F). (D) GTPase stimulation of Rap wt and Y32 mutants analysed by radioactive charcoal assay. Rap (10 μM) and Rap1GAP (50 nM) were incubated as described before (Kupzig et al, 2006). The concentration of released 32P-labelled Pi (correscponding to hydrolysed GTP) was plotted against the reaction time and initial rates were determined by linear regression fitting.
Tyr32 in Rap was mutated to Phe or Ala and the mutants were tested for the influence on GAP-stimulated GTPase activity. HPLC-based analysis of the GAP-catalysed reaction (Figure 3c) showed a 25-fold reduced (0.009 μM/s) activity for Y32AR in comparison with the wild-type protein (0.224 μM/s) under the conditions used. In the case of Y32FR, the initial rate was reduced only less than twofold (0.126 μM/s). These measurements were confirmed using a radioactive charcoal assay measuring the release of 32P-labelled Pi upon GTP hydrolysis. The Y32AR mutation has a drastically reduced GAP-mediated activity, whereas the effect of Y32FR is less pronounced with initial rates of 0.07, 0.007 and 0.001 μM/s for wt, Y32F and Y32A, respectively (Figure 3d). In previous studies, Y32WR was found to be inactive, with steric hindrance by the bulkier Trp side chain being the proposed explanation. We conclude that an aromatic side chain is necessary for efficient catalysis, and that, in contrast to previous conclusions (Brinkmann et al, 2002), the phenolic OH group further supports catalysis.
In the surface representations of Rap and Ras in complex with their respective GAPs (Figure 3b), the role of Tyr32 becomes apparent. In the Rap–Rap1GAP system, the closed conformation of Tyr32 shields the active site from bulk solvent and allows easy access to the catalytic helix of Rap1GAP. Access of the catalytic Asn to the pre-assembled active site would be blocked by an open conformation of Tyr32. This function can also be assumed by Phe, but not by the less bulky Ala or the bulkier Trp side chain. The phenolic hydroxyl group stabilises the conformation due to its interaction with the γ-phosphate. In contrast, Tyr32 in the Ras active site is in an open conformation and rather flexible, thus offering enough space for insertion of an arginine and allowing RasGAP to assemble the active site.
Implications for the Rheb–Tuberin interaction
A high sequence similarity has been discovered between Rap1GAP and a C-terminal part of TSC2/Tuberin (Supplementary Figure S1), and genetic and biochemical experiments suggested that Tuberin, which acts as a negative regulator of the mTOR pathway (Tee et al, 2003; Nobukini and Thomas, 2004), acts as a GAP towards the Ras subfamily protein Rheb. As Tuberin forms a complex with TSC1/Hamartin, it has been argued that complex formation between Tuberin and Hamartin is necessary for Rheb-GAP activity. This was supported by the finding that Tuberin is only homologous to the catalytic, but not the dimerisation, domain of RapGAP, suggesting that the other domain of RapGAP might be supplied by Hamartin. As shown by Inoki et al (2003) full-length Tuberin, overexpressed in HEK293 cells and isolated by immunoprecipitation, can indeed stimulate GTP hydrolysis in Rheb, whereas fragments of Tuberin (containing the GAP-domain) expressed in Escherichia coli did not show activity against Rheb-GTP. Zhang et al (2003) expressed a fragment of Tuberin (1384–1847) as GST-fusion protein encompassing not only the GAP domain but also the N- and C-terminal flanking elements as well. By using this construct, an activity against Rheb-GTP had been detected.
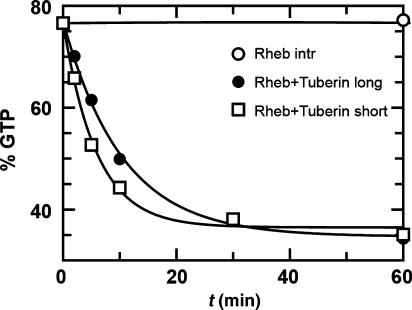
The structure of the Rap–RapGAP complex shows that the Tuberin fragment homologous to the catalytic domain of RapGAP should, in principle, be sufficient to act as GAP for Rheb. We used two recombinant Tuberin fragments encompassing the GAP-domain alone (Tubshort: 1538–1729; Tublong: 1532–1760) expressed in E. coli to investigate whether the fragment homologous to the catalytic domain of RapGAP is sufficient for catalysis. In our study, both fragments can inactivate Rheb efficiently, with Tubshort being slightly more active (Figure 4). Tublong, in addition to the short construct, contains the long helix (helix α9 in Rap1GAP) with the conserved Arg1743 (R388 in Rap1GAP), which in the Rap–Rap1GAP system contributes to complex association (see above). Here, we see no increased activity for the construct containing Arg1743. To efficiently stimulate GTP hydrolysis in Rheb, we had to use much higher concentrations of Rheb and Tuberin (80 and 100 μM, respectively), as compared with the Rap–Rap1GAP reaction (100 nM Rap1GAP with 200 μM Rap). Previous experiments were using smaller concentrations similar to those used for Rap–Rap1GAP, which might have led to the erroneous conclusion that Tuberin does not contain the machinery for GTPase stimulation.
Figure 4.
Stimulation of Rheb GTP hydrolysis by Tuberin. HPLC-based analysis of intrinsic and Tuberin-stimulated GTPase of Rheb, with two different constructs of the catalytic domain of Tuberin, using 80 μM Rheb and 100 μM Tuberin (Tuberinlong: 1532–1760; Tuberinshort: 1538–1729).
Discussion
GTP-binding proteins regulate signal transduction and transport processes as binary switches. The switch-OFF GTPase reaction is intrinsically very slow and stimulated via a variety of mechanisms. These involve either heterodimer formation between the G protein and GAPs, as found for most members of the Ras superfamily, or by formation of homodimers or higher oligomers as in the case of dynamin and the dynamin-like protein hGBP1 (Ghosh et al, 2006), or HypB (Gasper et al, 2006) and MnmE (Scrima and Wittinghofer, 2006). In most cases, the active site is rearranged or complemented by complex formation to allow an efficient phosphoryl transfer reaction. X-ray crystallographic and biochemical analyses have identified an unexpectedly large variety of activation mechanisms.
After a number of structures of complexes of Ras-like proteins and their respective GAPs have been solved, a common principle emerges whereby the correct positioning and/or activation of the nucleophilic water for an in-line attack on the γ-phosphate is the most important aspect of catalysis. In the case of Ras, Rho and Ran, this is accomplished by an intrinsic glutamine on switch II. In Sar1, a member of the Arf subfamily, the function of Gln is replaced by a His, and in the Rab–RabGAP complex, Gln is supplied in trans, although Rab proteins do contain a conserved intrinsic Gln. Here, we have shown that the missing intrinsic Gln in Rap is replaced by an Asn in trans called the Asn-thumb, which is inserted into the active site and occupies exactly the same position as Gln in the other G protein-GAP systems. Conversely, it seems that the arginine finger, that is, the positively charged arginine side chain of RasGAP, RhoGAP and RabGAPs has a less important role in catalysis, as its mutation often has less dramatic consequences than the Gln/Asn one, which completely eliminates activity. Mutation of Arg in RasGAP and RhoGAP reduces the activity by 2000 and 50- to 240–fold, respectively, compared with an overall 105-fold stimulation (Ahmadian et al, 1997; Nassar et al, 1998; Graham et al, 1999). As the catalytic Gln in most GAP-free structures is highly mobile and becomes properly oriented and immobile by complex formation with its cognate GAP, it appears that even part of the arginine effect could well be due to the stabilisation of the carboxamide in the active site. The fact that systems such as that of Ran and Rap show efficient catalysis in the absence of an arginine finger seems to indicate a mechanistic difference between the systems. It has been proposed that a positively charged residue contacting the transferred phosphate makes an important contribution to an associative, but is anticatalytic for a dissociative transition state (Maegley et al, 1996). Therefore, we would argue that in the case of RapGAP-mediated GTPase reaction on Rap, and in the Ran–RanGAP system, we are observing a more dissociative transition state with a metaphosphate-like configuration, in contrast to the reaction of Ras, Rab and Rho with a penta-coordinated γ-phosphate as the transition state of a more associative mechanism.
The complex structure not only explains the mechanistic basis for GTPase stimulation in Rap, but also allows drawing conclusions concerning the Rheb inactivation by Tuberin. Even though Hamartin and Tuberin (TSC1 and TSC2) exist and are active as a complex in the regulation of the mTOR pathway, our data show that the RapGAP homologous domain in TSC2 apparently contains the full catalytic machinery. It may, however, require TSC1 or other domains of TSC2 for supporting function, just as the dimerisation domain of RapGAP supports the catalytic domain. We can, in fact, show that the catalytic domain of RapGAP is much less active than the full-length protein, as it contributes to affinity. Helix α9 is partially sequence conserved between RapGAP and Tuberin. We speculate that the position of helix α9 is highly mobile in the absence of the second domain and renders the catalytic domain less active, which also seems to be the case in Tuberin. Even the presence of Arg1743 in Tublong does not increase the GAP efficiency, and, therefore, possibly needs Hamartin as binding partner to stabilise helix α9 in Tuberin. Alternatively, helix α9 could be already stabilised in full-length Tuberin, as Inoki et al (2003) could only show efficient inactivation of Rheb with full-length Tuberin, but not with fragments thereof.
Rheb, although sharing high homology with Rap1, contains a glutamine (Gln64) corresponding to Gln61 in Ras and Thr61 in Rap. In the P-loop position corresponding to Gly12 in Rap, Rheb carries an Arg (Arg15). As reported by Li et al (2004), Rheb Q64L is still sensitive to TSC1/2-catalysed inactivation as well as Rheb R15V (which in Ras (G12V) leads to oncogenic transformation and is resistant to GAP inactivation). Mutation of Arg15 to Gly rendered the protein more resistant to TSC1/2-induced inactivation. In our study (using Tubshort in an HPLC-based GTP hydrolysis assay as in Figure 4), Rheb Q64A and R15G show a similarly reduced sensitivity towards Tubshort (data not shown). With Q64A, we observed a stimulated GTP hydrolysis, which is, however, weaker as compared with wt, whereas R15G only showed a very weak inactivation by Tubshort. Previously, we have shown by FTIR using high protein concentrations that mutations of Thr61 in Rap drastically reduce catalysis, and that this is solely due to a loss of affinity. As Gln64 would be in a similar position in the interface of the Rheb–Tuberin complex, it is also likely involved in binding rather than in catalysis.
Based on these results and as the catalytic Asn (Asn1643) is conserved in Tuberin and mutated in TSC patients, we assume that Gln64 is not involved in catalysis and that Rheb downregulation by Tuberin is achieved by a mechanism that is identical to the Rap–RapGAP reaction.
Materials and methods
Protein expression and purification
Rap1B (1–167) and Rap1GAP Q204A (75–415) were expressed and purified as described previously (Brinkmann et al, 2002). Complex was purified by incubation of 5 mg Rap1B and 5 mg Rap1GAP (twofold molar excess of Rap1B) in the presence of 1 mM BeCl2, 10 mM NaF and 1 mM GDP for 30 min on ice in 50 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2 and 5 mM DTE. The stoichiometrically formed complex was afterwards isolated by size-exclusion chromatography on a Superdex S200 10/30 column in the buffer described above supplied with 1 mM BeCl2 and 10 mM NaF. Complexes with GppNHp or GDP+AlFx were purified accordingly.
Rheb (1–170) and Tuberin constructs were expressed as GST-fusion proteins in TB-media. At OD600≈0.6, expression was induced by addition of 100 μM IPTG; the proteins were expressed overnight at 25°C. Cells were collected and lysed in 20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2, 5 mM DTE, 1 mM ATP and 0.1 mM PMSF. Soluble protein supernatant was applied onto a GSH-column for affinity purification, according to the manufacturer's manual. Lysis and washing buffers contain ATP for removal of chaperone contaminations bound to the fusion protein. Cleavage of the fusion protein was carried out on the column by addition of Thrombin and circulation for at least 4 h. Subsequently, proteins were separated by size-exclusion chromatography in 20 mM HEPES 7.5, 100 mM NaCl, 5 mM MgCl2 and 5 mM DTE. Finally, the target protein was concentrated, flash-frozen in liquid nitrogen and stored at −80°C.
Mutants were generated using the QuikChange protocol (Stratagene).
Crystallography and structure solution
Rap-GDP·BeF3−-Rap1GAP(Q204A) was crystallised at 20°C by the hanging drop vapour diffusion method. A 1 μl portion of protein at a concentration of 60 g/l was mixed with 1 μl of reservoir solution containing 8–11% PEG2000 MME, 100 mM MES pH 6.5. Crystals were cryoprotected for data collection by slowly adding ethyleneglycol to the mother liquor to a final concentration of 20%.
Collected data were processed with XDS (Kabsch, 1993). Molecular replacement was performed with Molrep from the CCP4-package (Vagin and Teplyakov, 1997) using the Rap1GAP structure as searching model (1SRG, Daumke et al, 2004). The additional electron density was clearly identified as Rap1B and the model was completed by building using XtalView/Xfit (McRee, 1999). Refinement was carried out with REFMAC5 (Murshudov et al, 1997). Figures were generated using Pymol (DeLano Scientific LLC).
Atomic coordinates and structural factors have been deposited within the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) under the accession code 3BRW.
Biochemistry
For all biochemical experiments (Stopped-flow, HPLC and charcoal assay), Rap1GAP Q204A was used as ‘wild type', a mutant showing higher protein stability, but retaining properties of the nonmutated form. For Rap1B, all measurements were made with A86C (used for fluorescence labelling, see main text and Figure legends) as ‘wild type'.
The single-turnover fluorescence-based assay (stopped-flow) was performed as described in Kraemer et al (2002). Rap1B-AedansGTP (2 μM) was mixed with 50 μM Rap1GAP in a stopped-flow apparatus (SM-17; Applied Photophysics). The Aedans-fluorophor was excited at 350 nm and change in fluorescence was monitored through a 408 nm cutoff filter. Experiments were carried out at 10°C in 50 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2 and 5 mM DTE. Data were processed with GraFit 3.0 (Erithacus Software Limited).
For HPLC-based multiple-turnover measurements, 100 nM Rap1GAP were incubated with 200 μM Rap1B-GTP or 80 μM Rheb-GTP with 100 μM Tuberin (at 20°C in 50 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2 and 5 mM DTE). Aliquots were flash-frozen in liquid N2 at the given time points. Aliquots were incubated at 95°C for 2 min, denatured protein was removed by centrifugation (1 min, 13000 r.p.m.) and the supernatant was applied to the HPLC (Beckman-Coulter, System Gold). Nucleotides were separated on a hydrophobic C18-column (Beckman-Coulter) with 100 mM potassium-phosphate pH 6.5, 10 mM tert-butyl-ammonium-bromide and 7.5% acetonitril as polar, mobile phase. The concentration of nonhydrolysed GTP was plotted against reaction time. Initial reaction rates were determined by linear regression.
The radioactive charcoal assay was performed as described previously (Brinkmann et al, 2002; Kupzig et al, 2006). Briefly, 10 μM of radioactively labelled Rap-[γ-32P]GTP were added to 50 nM of Rap1GAP and the release of 32P-labelled Pi was plotted against the reaction time. Initial rates were determined by linear regression fitting.
PDB codes of structures used for figures
Ras-RasGAP (1WQ1), Rho-RhoGAP (1TX4), Ran–RanGAP (1K5D), RapGTP (3RAP), Rheb-GTP (1XTS), Ras-GppNHp (5P21).
Supplementary Material
Supplementary Figures S1–S4
Acknowledgments
We thank Michael Weyand, Wulf Blankenfeldt, Neelakshi Gohain, Ilme Schlichting, Anton Meinhard, Thomas Barends and Bernhard Loll for data collection. The data sets were collected at the Swiss Light Source, beamline X10SA, Paul Scherrer Institut, Villigen, Switzerland. We thank Michael Weyand and Ingrid R Vetter for crystallographic assistance and Dorothee Kühlmann for technical assistance.
References
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 [DOI] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419: 271–277 [DOI] [PubMed] [Google Scholar]
- Bos JL, de Rooij J, Reedquist KA (2001) Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol 2: 369–377 [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129: 865–877 [DOI] [PubMed] [Google Scholar]
- Brinkmann T, Daumke O, Herbrand U, Kühlmann D, Stege P, Ahmadian MR, Wittinghofer A (2002) Rap-specific GTPase activating protein follows an alternative mechanism. J Biol Chem 277: 12525–12531 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chakrabarti PP, Daumke O, Suveyzdis Y, Kötting C, Gerwert K, Wittinghofer A (2007) Insight into catalysis of a unique GTPase reaction by a combined biochemical and FTIR approach. J Mol Biol 367: 983–995 [DOI] [PubMed] [Google Scholar]
- Cherfils J, Menetrey J, Le Bras G, Janoueix-Lerosey I, de Gunzburg J, Garel JR, Auzat I (1997) Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS. EMBO J 16: 5582–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A (2004) The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429: 197–201 [DOI] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV (2003) Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 332: 689–699 [DOI] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Rocciom M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G (2003) Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466 [DOI] [PubMed] [Google Scholar]
- Gasper R, Scrima A, Wittinghofer A (2006) Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem 281: 27492–27502 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C (2006) How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440: 101–104 [DOI] [PubMed] [Google Scholar]
- Graham DL, Eccleston JF, Lowe PN (1999) The conserved arginine in rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with Rho.GDP and aluminium fluoride. Biochemistry 38: 985–991 [DOI] [PubMed] [Google Scholar]
- Hattori M, Minato N (2003) Rap1 GTPase: functions, regulation, and malignancy. J Biochem (Tokyo) 134: 479–484 [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL (2003) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17: 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26: 795–800 [Google Scholar]
- Kraemer A, Brinkmann T, Plettner I, Goody R, Wittinghofer A (2002) Fluorescently labelled guanine nucleotide binding proteins to analyse elementary steps of GAP-catalysed reactions. J Mol Biol 324: 763–774 [DOI] [PubMed] [Google Scholar]
- Kupzig S, Deaconescu D, Bouyoucef D, Walker SA, Liu Q, Polte CL, Daumke O, Ishizaki T, Lockyer PJ, Wittinghofer A, Cullen PJ (2006) GAP1 family members constitute bifunctional Ras and Rap GTPase-activating proteins. J Biol Chem 281: 9891–9900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Inoki K, Guan KL (2004) Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol 24: 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegley KA, Admiraal SJ, Herschlag D (1996) Ras-catalyzed hydrolysis of GTP: a new perspective from model studies. Proc Natl Acad Sci USA 93: 8160–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA (1998) Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol 5: 1047–1052 [DOI] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Block C, Janknecht R, Wittinghofer A (1996) Ras/Rap effector specificity determined by charge reversal. Nat Struct Biol 3: 723–729 [DOI] [PubMed] [Google Scholar]
- Nobukini T, Thomas G (2004) The mTOR/S6K signalling pathway: the role of the TSC1/2 tumour suppressor complex and the proto-oncogene Rheb. Novartis Found Symp 262: 148–154 [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442: 303–306 [DOI] [PubMed] [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ (1997) Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature 389: 758–762 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277: 333–338 [DOI] [PubMed] [Google Scholar]
- Scrima A, Wittinghofer A (2006) Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J 25: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seewald MJ, Körner C, Wittinghofer A, Vetter IR (2002) RanGAP mediates GTP hydrolysis without an arginine finger. Nature 415: 662–666 [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (2003) Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13: 1259–1268 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Yu Y, Li S, Xu X, Li Y, Guan K, Arnold E, Ding J (2005) Structural basis for the unique biological function of small GTPase RHEB. J Biol Chem 280: 17093–17100 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D (2003) Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5: 578–581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S4