Abstract
Cells adapt to hypoxia by a cellular response, where hypoxia-inducible factor 1α (HIF-1α) becomes stabilized and directly activates transcription of downstream genes. In addition to this “canonical” response, certain aspects of the pathway require integration with Notch signaling, i.e., HIF-1α can interact with the Notch intracellular domain (ICD) to augment the Notch downstream response. In this work, we demonstrate an additional level of complexity in this cross-talk: factor-inhibiting HIF-1 (FIH-1) regulates not only HIF activity, but also the Notch signaling output and, in addition, plays a role in how Notch signaling modulates the hypoxic response. We show that FIH-1 hydroxylates Notch ICD at two residues (N1945 and N2012) that are critical for the function of Notch ICD as a transactivator within cells and during neurogenesis and myogenesis in vivo. FIH-1 negatively regulates Notch activity and accelerates myogenic differentiation. In its modulation of the hypoxic response, Notch ICD enhances recruitment of HIF-1α to its target promoters and derepresses HIF-1α function. Addition of FIH-1, which has a higher affinity for Notch ICD than for HIF-1α, abrogates the derepression, suggesting that Notch ICD sequesters FIH-1 away from HIF-1α. In conclusion, the data reveal posttranslational modification of the activated form of the Notch receptor and an intricate mode of cross-coupling between the Notch and hypoxia signaling pathways.
Keywords: gene regulation, hydroxylation, signal transduction
Reduced oxygen levels (hypoxia) lead to a set of cellular adaptations, including increased angiogenesis and erythropoiesis and a switch to glycolytic metabolism. The cellular machinery that senses hypoxia is composed of several proteins. A critical component is the transcription factor hypoxia-inducible factor 1α (HIF-1α) (1). The level and activity of HIF-1α are controlled by oxygen-dependent prolyl (PHD) and asparaginyl factor-inhibiting HIF-1α (FIH-1)] hydroxylases. PHDs hydroxylate two proline residues in the degradation domain of HIF-1α in normoxia, which makes HIF-1α a substrate for the von Hippel–Lindau E3 ubiquitin ligase and proteasomal degradation. After stabilization in hypoxia, HIF-1α interacts with aryl hydrocarbon receptor nuclear translocator (ARNT) to bind to hypoxia response elements (HREs) and activate target genes (1). FIH-1, in contrast, was originally found to be a negative regulator of HIF-1α (2) and was later shown to be an asparaginyl hydroxylase, capable of hydroxylating N803 in the C-terminal activation domain (CAD) of human HIF-1α (3, 4). This hydroxylation, which is more prevalent in normoxia, correlates with repression of HIF-1α function (5).
In addition to this “canonical” hypoxic signaling mechanism, certain aspects of the hypoxic response, for example, in the control of stem cell differentiation, require integration with the Notch pathway (6, 7). In Notch signaling, the Notch transmembrane receptor (Notch 1–Notch 4 in vertebrates) is activated by a family of membrane-bound ligands (DSL, including Delta-like and Jagged). Interaction with ligand leads to two consecutive cleavages, eventually liberating the Notch intracellular domain (ICD) from the plasma membrane. Notch ICD then enters the nucleus, where it interacts with the DNA-binding protein CSL (8). Notch ICD displaces corepressors from CSL, recruits a transcriptional activation complex, including Mastermind (MAML1) and p300/CBP, and it may also in some situations control the recruitment of CSL to its corresponding binding sites in Notch-responsive promoters (9). Notch signaling leads to transcriptional activation of immediate target genes such as Hes and Herp/Hey (8).
We observed that hypoxia promotes the undifferentiated cell state in myogenic and neural progenitors in a Notch-dependent manner. At the molecular level, HIF-1α interacts with Notch 1 ICD and is recruited to a Notch-responsive promoter under conditions of active Notch signaling in hypoxia (6). In addition to hypoxia-induced elevation of the Notch downstream response, a converse Notch-mediated potentiation of the hypoxic downstream response was noted, i.e., Notch signaling augmented hypoxia-induced expression of a bona fide HIF-1α-inducible gene, PGK1, and the recruitment of HIF-1α to its promoter (6). Because Notch ICD could not be detected at the PGK1 promoter, the action of Notch ICD may be more indirect, indicating that additional molecular mechanisms operate in the integration between Notch and hypoxia signaling. In this work, we have evaluated a possible role of FIH-1 in cross-coupling between the Notch and HIF pathways. Our data show that FIH-1 hydroxylates Notch ICD at two asparagine residues. Furthermore, Notch ICD potentiates recruitment of HIF-1α to HIF target promoters. The fact that Notch ICD has a higher affinity than HIF-1α for FIH-1 suggests that the observed Notch-mediated derepression of HIF-1α function, at least in part, is caused by ICD sequestering FIH-1 away from HIF-1α.
Results and Discussion
FIH-1-Dependent Hydroxylation of N1945 and N2012 in the Notch ICD.
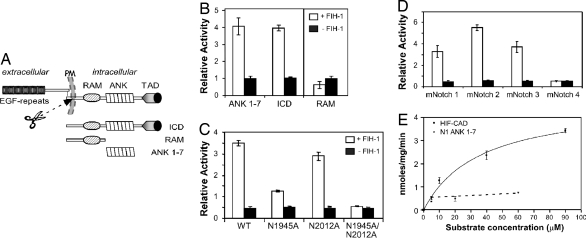
To determine whether Notch 1 is a direct target of FIH-1, a thioredoxin-histidine6 (Trx-H6)-tagged Notch ICD was tested for FIH-1-dependent hydroxylation in a 2-oxo-glutarate decarboxylation assay. Full-length mNotch 1 ICD and a shorter form spanning the seven ankyrin repeats, ANK 1–7 (Fig. 1A), displayed robust activity in the in vitro hydroxylation assay, whereas a truncated version containing the RAM domain alone did not (Fig. 1B). Mass spectrometry using a tandem instrument comprising linear and orbital traps (LTQ-Orbitrap) revealed FIH-1-dependent hydroxylation on a tryptic peptide encompassing residues 1937–1952 [supporting information (SI) Fig. 6A]. Data from tandem mass spectrometry (MS/MS) and MALDI-TOF/TOF MS/MS identified the addition of an oxygen atom uniquely in FIH-1-treated samples to hydroxylation on N1945. In addition, a lower level of hydroxylation was detected in a tryptic peptide encompassing residues 1995–2019 at N2012 (SI Fig. 6B).
Fig. 1.
Hydroxylation of Notch ICD at 2 asparagine residues by FIH-1. (A) Schematic diagram showing key features of Notch. (B) Notch 1 ICD, ANK 1–7, and RAM proteins were analyzed in a 14CO2 capture assay. Data are mean relative activity ± range (n = 2) and are representative of >3 independent experiments. (C) Purified wild-type (WT) or Asn mutant ANK 1–7 proteins or (D) mNotch 1–4 ANK 1–7 proteins were compared for their abilities to promote FIH-1-mediated 2-oxoglutarate turnover. Data are presented as mean ± range (n = 2) and are representative of three independent experiments. (E) Kinetic CO2 capture assay with varying HIF-CAD or mNotch1 substrate concentrations, with data fitted to a hyperbolic curve using PRISM software. The HIF-CAD apparent Km is 36 μM, and the apparent Vmax is 4.7 nmol/mg per min, whereas Notch hydroxylation rate is near-maximal (≈0.7 nmol/mg per min), and the apparent Km cannot be determined accurately (<4 μM). Data are mean nmol/mg per min ± range (n = 2), representative of >3 independent experiments.
We next addressed whether hydroxylation occurred in a cellular context. ANK 1–7 was expressed in 293T cells constitutively overexpressing FIH-1 in the presence or absence of the FIH-1 inhibitor dimethyloxalylglycine (DMOG). MALDI-TOF/TOF-MS revealed a similar pattern of modifications as observed in vitro both under hypoxic and normoxic conditions. Exposure to DMOG essentially abolished formation of the peptides corresponding to hydroxylation of N1945 and N2012 (SI Fig. 6 C and D). These results demonstrate that ANK 1–7 is hydroxylated at both positions N1945 and N2012 within mammalian cells.
To corroborate the MS/MS data, wild-type and Asn mutant forms of ANK 1–7 were analyzed for interaction with FIH-1 and for in vitro hydroxylation. Both the wild-type and mutant constructs interacted with FIH-1 (SI Fig. 7A). The wild-type and N2012A constructs showed potent FIH-1-dependent decarboxylation activity, whereas N1945A was considerably reduced. In the case of N1945A/N2012A, activity was abolished, indicating that N1945 and N2012 are likely to be the only two sites of FIH-1-dependent hydroxylation within the complete ankyrin repeat domain and that N1945 is hydroxylated more efficiently by FIH-1 than N2012 (Fig. 1C).
Notably, N1945 and N2012 in Notch 1 are conserved in all Notch 1 orthologs (SI Fig. 8) and in mammalian Notch 2, 3, and 4, with the exception of site 2 (N2012), which is a glycine residue in Notch 4. We tested whether the other Notch paralogs could also be hydroxylated by FIH-1. Trx-H6 fusion proteins corresponding to ANK 1–7 repeats from mNotch 2, 3, and 4 (mN2–4) were expressed, purified, and analyzed by the in vitro hydroxylation assay. mN2 ANK 1–7 and mN3 ANK 1–7 gave similar FIH-1-dependent activity to mN1 ANK 1–7, whereas mN4 ANK 1–7 displayed no activity (Fig. 1D), suggesting that unlike Notch 1, 2, and 3, Notch 4 is not hydroxylated by FIH-1.
A kinetic analysis of mN1 ANK 1–7 hydroxylation by FIH-1 demonstrated that the apparent Vmax is ≈7-fold lower than the apparent Vmax with the HIF-CAD substrate (Fig. 1E). The apparent Km for mN1 ANK 1–7 is obviously at least an order of magnitude lower than the Km for HIF-CAD, although it could not be exactly determined because of limitations with the sensitivity of this assay. In vitro pulldown experiments demonstrated that FIH-1 interacted with purified mN1 ANK 1–7 with much higher affinity than with purified HIF-CAD (SI Fig. 7B), supporting the results from the kinetic assay. In summary, the data reveal that Notch ICD is a substrate for FIH-1 hydroxylation and that the two hydroxylated Asn residues are located at positions similar to those found in other recently discovered ankyrin repeat-containing proteins hydroxylated by FIH-1 (10).
N1945 and N2012 Residues Are Important for Notch-Mediated Transcriptional Activation.
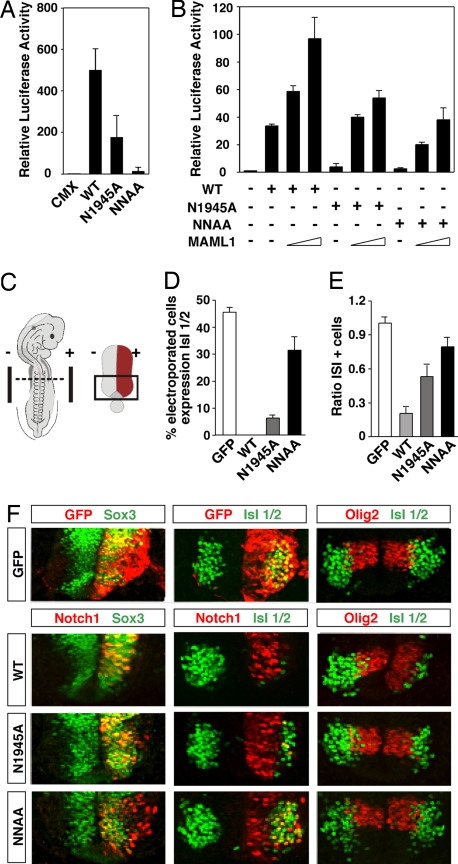
To investigate the role of residues N1945 and N2012 in Notch ICD function, we performed CSL-driven reporter gene assays in P19 cells. Alanine mutation of N1945 had a major impact on gene activation, and mutation of both N1945 and N2012 almost abolished the activity (Fig. 2A). Western blot analysis showed comparable expression levels for the wild type and Notch mutants (data not shown). Although the mutants showed significantly reduced activity, their functionality could partially be rescued by coexpression of the coactivator MAML1 (Fig. 2B), indicating that the interaction with this coactivator is preserved in the mutated forms. In conclusion, mutation of the two Asn residues identified as targets of hydroxylation by FIH-1 substantially reduces the transactivation capacity of Notch ICD.
Fig. 2.
Mutation of Notch 1 ICD N1945 and N2012 reduces the transactivation potency and impairs Notch-mediated repression of neuronal differentiation. (A) Notch N1945/N2012 is a very weak transcriptional activator. P19 cells were transfected with 0.5 μg of a CSL-driven luciferase plasmid and 100 ng of CMX or plasmids encoding wild-type (WT), N1945A, or N1945A/N2012A (NNAA) mutant ICD. (B) Activity of Notch Asn mutants is rescued by expression of MAML1. HeLa cells were transfected with 0.5 μg of 12×CSL-Luc plasmid together with 100 ng of plasmids encoding WT or mutant forms of Notch 1 ICD in the presence of increasing concentrations of an expression vector (50 ng and 200 ng) encoding MAML1. Data are presented as luciferase activity relative to cells transfected with 12×CSL-Luc. Values represent the mean ± SE of three independent experiments performed in duplicate. (C) Schematic representation of a harvested chicken embryo after electroporation. Transverse sections at the forelimb level (dashed line) were analyzed. In the electroporated spinal cord, only one of the sides was electroporated (red region). The analysis was focused on the most ventral region of the spinal cord (boxed region). Quantification of the effect on motor neuron generation after expression of GFP or WT; N1945A, or NNAA Notch1 ICD is shown. The percentage of cells electroporated with the indicated constructs and expressing the postmitotic motor neuron marker Isl1/2 (D), and the ratio of the total number of motor neurons between the electroporated vs. control (el/co) side (E) were determined. (F) Effect of wild-type and mutant forms of Notch on neurogenesis in the ventral spinal cord. Embryos were electroporated with GFP or with wild-type or mutant Notch1 ICD as indicated. The ability of the different constructs to maintain cells in a progenitor stage or to repress motor neuron generation was analyzed by immunohistochemistry using antibodies against Sox3 or Isl1/2, respectively. Expression of the different constructs did not affect the motor neuron progenitor pool expressing Olig2 (F), arguing that the observed role of Notch does not result from progenitor pool depletion. +, electroporated side; −, control side.
Mutation of Notch ICD N1945 and N2012 Residues Compromises Notch Function in Neurogenesis and Myogenesis.
In vertebrates, Notch signaling inhibits neuronal differentiation by promoting the maintenance of the progenitor state or glial differentiation (11). To explore the role of residues N1945 and N2012 in the control of neuronal differentiation, we introduced wild-type or mutated Notch ICD forms into the developing chick CNS by in ovo electroporation (Fig. 2C), and we examined their effects on motor neuron generation as a readout of neurogenesis (SI Fig. 9). Wild-type Notch 1 ICD significantly blocked motor neuron differentiation, i.e., the number of cells expressing the motor neuron-specific transcription factor Isl1/2 was reduced by 80% (Fig. 2E). Very few Notch 1 ICD-expressing cells coexpressed Isl1/2, whereas, in contrast, Notch 1 ICD colocalized extensively with the progenitor marker Sox3 (Fig. 2 D and F). In the case of the N1945A and N1945A/N2012A mutants, we noted that the reduction in the number of cells expressing Isl1/2 was progressively smaller (47% and 21%, respectively) compared with wild-type Notch 1 ICD. Furthermore, fewer N1945A- and N1945A/N2012A-expressing cells colabeled with Sox3 compared with wild-type Notch 1 ICD and began to coexpress Isl1/2 (Fig. 2 D and F).
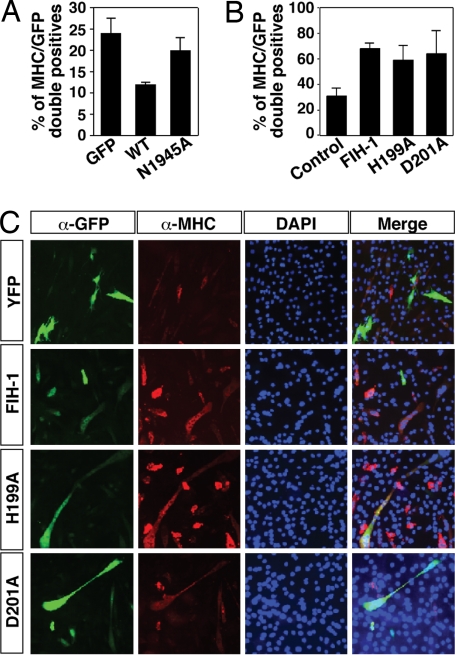
We next tested the effect of mutating N1945 in the control of myogenic differentiation. To this end, we used the myogenic cell line C2C12, which can be induced to differentiate from a myoblast-like stage to form multinucleated myotubes after serum differentiation, and Notch signaling can abrogate this differentiation process (12, 13). Transfection of Notch 1 ICD-GFP led to a reduced number of C2C12 cells expressing myosin heavy chain (MHC) compared with GFP expression alone. Expression of Notch 1 ICD N1945A-GFP led to a much more modest reduction in the number of differentiated, MHC-positive cells compared with wild-type Notch 1 ICD-GFP (Fig. 3A). In conclusion, our results indicate that the function of Notch in repressing neuronal and myogenic differentiation is compromised when the critical Asn residues are mutated, consistent with the reduced transactivation function of the mutants.
Fig. 3.
FIH-1 increases myogenic differentiation. (A) Quantification of the percentage of GFP-positive cells (expressing either GFP, Notch 1 ICD-GFP, or Notch 1 ICD N1945A-GFP) that are also MHC-positive. (B) Quantification of the percentage of YFP-positive cells that are also MHC-positive (see below). (C) Differentiated (MHC-positive, red) C2C12 cells costained for expression of YFP, FIH-1-YFP, FIH-1H199A-YFP, or FIH-1D201A-YFP (green).
FIH-1 Negatively Regulates Notch ICD and Accelerates Myogenic Differentiation.
We have observed that wild-type FIH-1 negatively regulates transactivation by Notch ICD (6). To explore the functional significance of FIH-1 for Notch signaling in myogenic differentiation, yellow fluorescent protein (YFP)-tagged FIH-1 was transiently expressed in C2C12 cells cultured under differentiation-promoting conditions. Four days after transfection of FIH-1-YFP we observed an increased proportion of YFP-expressing cells coexpressing MHC compared with transfection of YFP alone. Interestingly, a similar increase in the proportion of differentiated C2C12 cells was observed also after transfection of the catalytically inactive H199A or D201A forms of FIH-1 (Fig. 3 B and C). In conclusion, our data suggest that both wild-type and mutant FIH-1 can accelerate myogenic differentiation, consistent with negative regulation of Notch ICD function independent of hydroxylation and consistent with the very high affinity observed between Notch ICD and FIH-1.
To study this mode of regulation further, we transfected wild-type, H199A, and D201A FIH-1 together with Asn mutant Notch 1 ICD in 293T cells. Wild-type FIH-1, and FIH-1 H199A and D201A reduced the activity of CSL-dependent reporter activity driven by N1945A and N1945A/N2012A Notch 1 ICD (SI Fig. 7 C and D), suggesting that the enzymatic activity is dispensable for negative regulation of Notch. However, in vitro pulldown assays demonstrated that FIH-1 interacts with nonhydroxylated Notch 1 ANK 1–7 with a much higher affinity than when it is hydroxylated (SI Fig. 7E). Hence, hydroxylation may indirectly modulate negative regulation of the ICD by reducing affinity for FIH-1.
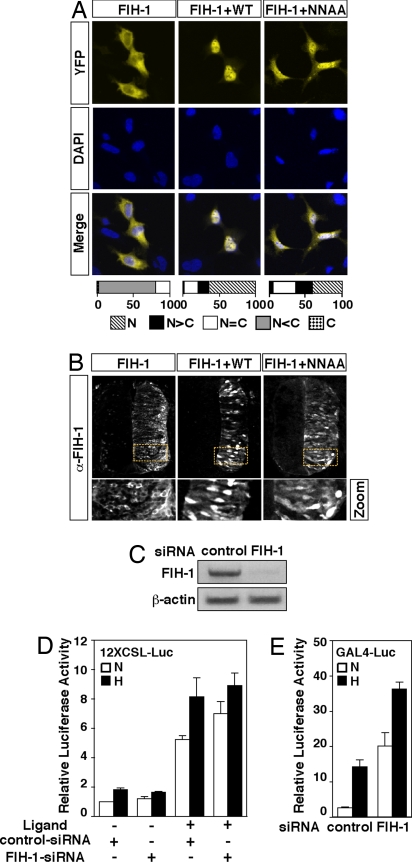
To assess the interaction between Notch ICD and FIH-1 in vivo, we studied whether Notch ICD could influence the intracellular localization of FIH-1. Expression of YFP-tagged FIH-1 revealed a predominantly cytoplasmic distribution. In contrast, coexpression of Notch 1 ICD with YFP-tagged FIH-1 changed the localization of FIH-1 to the nucleus in 64% of the fluorescent cells (Fig. 4A). Expression of Notch ICD N1945A/N2012A also recruited FIH-1 to the nucleus, although to a lesser extent. A similar Notch ICD-dependent shift in intracellular localization of FIH-1 was observed in the chick neural tube after coelectroporation of FIH-1 with Notch 1 ICD. Wild-type Notch ICD resulted in nuclear localization of FIH-1, whereas Notch ICD N1945A/N2012A produced an intermediary distribution pattern (Fig. 4B). These data support the notion that Notch ICD and FIH-1 interact in vivo but may also suggest that Asn mutant forms of Notch bind FIH-1 less effectively in vivo, given the more partial nuclear translocation of FIH-1 by Notch Asn mutants.
Fig. 4.
Notch ICD induces nuclear accumulation of FIH-1. (A) 293A cells were transfected with 0.4 μg of YFP-fused FIH-1 alone or together with 0.6 μg of plasmids encoding wild-type Notch ICD (WT) or ICD N1945A/N2012A (NNAA). Cells were analyzed by confocal microscopy, and intracellular distribution was categorized as described in ref. 17. Nuclear compartment was identified by DAPI staining. (B) Notch affects cellular localization of FIH-1 in vivo. Chick neural tubes were electroporated with FIH-1 alone or together with wild-type or NNAA mutant Notch1 ICD. Cellular localization of FIH-1 was determined by immunohistochemistry (α-FIH-1). The zoom panels on each column correspond to magnification of the dashed yellow boxes. (C) Levels of FIH-1 mRNA in cells transfected with control siRNA and FIH-1-specific siRNA and analyzed by quantitative RT-PCR. (D) siRNA targeting FIH-1 produces a modest effect on Notch-dependent luciferase activity. 293T cells stably expressing full-length Notch and plated on IgG (ligand −) or Jagged-1-coated plates (ligand +) were first transfected with 100 nM negative control or a FIH-1-specific siRNA and 24 h later were transfected again with 0.5 μg of 12×CSL-Luc plasmid. (E) Inhibition of FIH-1 expression by siRNA mediates derepression of HIF-1α CAD function. Twenty-four hours after transfection with siRNAs, a GAL4-driven luciferase plasmid (0.5 μg) and an expression plasmid encoding GAL4-fused HIF-1α C-TAD (50 ng) were transfected. Data are presented as luciferase activity relative to cells transfected with reporter gene alone. Values represent the mean ± SE of three independent experiments performed in duplicate.
To examine the effect on Notch signaling by reducing endogenous FIH-1 levels, we transiently transfected a cell line stably expressing full-length Notch 1 with siRNAs targeting FIH-1 (Fig. 4C). Notch function was activated by using immobilized Notch ligand (Jagged). A modest but significant (30%) increase in CSL-reporter gene activity was observed in cells transfected with FIH-1-specific siRNA under normoxic conditions (Fig. 4D). Under hypoxic conditions, this difference was not evident. As a control for the efficacy of FIH-1 siRNA, inhibition of FIH-1 expression led to a dramatic increase of HIF-1-dependent activity at normoxia (Fig. 4E).
Activation of the Notch Signaling Pathway Increases Recruitment of HIF-1α to HREs of Canonical HIF Target Genes.
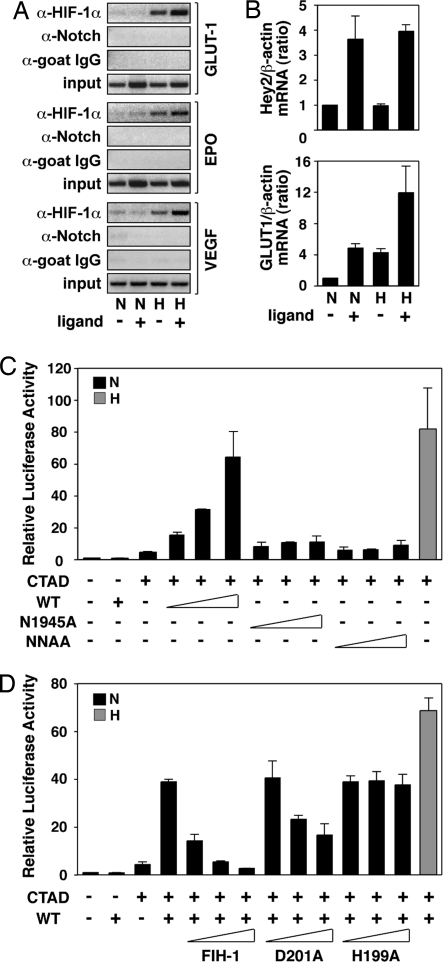
In the canonical hypoxic response, target genes, such as GLUT1, EPO, and VEGF, are directly controlled by binding of HIF-1α to HREs within their promoters (1). To test whether active Notch signaling influences recruitment of HIF-1α to these promoters, we analyzed HIF-1α and Notch ICD promoter occupancy by ChIP assays under various conditions of hypoxia and Notch activation. C2C12 cells stably expressing full-length Notch 1 were cocultured with cells stably expressing the Notch ligand Jagged-1. Ligand-dependent activation of Notch signaling under hypoxic conditions led to an increased recruitment of HIF-1α to the HRE-containing regions of the GLUT1, EPO, and VEGF promoters, whereas recruitment of Notch 1 ICD to the same promoter regions could not be detected (Fig. 5A). These results extend our previous observations on the HIF-1α target gene PGK1 (6) and indicate that Notch-mediated potentiation of HIF-1α recruitment is a mechanism common to many, if not all, HIF-1α target genes.
Fig. 5.
Activation of Notch increases binding of HIF-1α to target genes and causes derepression of HIF-1α function. (A) Binding of HIF-1α to target gene HREs by ChIP analysis in the absence or presence of activated Notch. C2C12 cells stably expressing full-length Notch were cultured in the presence of control 293T cells (ligand −) or 293T cells stably expressing Notch ligand Serrate-1 (ligand +) and kept at normoxia (N) or exposed to hypoxia (H). ChIP assays were performed using the indicated antibodies, and RT-PCR analysis of target gene HREs was carried out. (B) Activation of the Notch pathway increases HIF-1 target gene expression. 293T cells stably expressing full-length Notch were cultured on plates coated with IgG (ligand −) or Jagged-1 (ligand +) and kept at normoxia (N) or hypoxia (H). Total RNA was prepared, and quantitative RT-PCR was performed. (C) Notch ICD derepresses HIF-1α CAD function. P19 cells were transfected with 0.5 μg of a GAL4-driven luciferase plasmid and 50 ng of GAL-fused HIF-1α C-TAD expression plasmid in the presence of increasing amounts (50, 100, and 200 ng) of plasmids encoding wild-type or Asn mutant Notch1 ICD. (D) Expression of wild-type FIH-1 restores repression of CAD function at normoxia. P19 cells were transfected with 0.5 μg of a GAL4-driven luciferase plasmid together with a GAL-fused HIF-1α CAD expression plasmid (50 ng) and a plasmid encoding wild-type Notch 1 ICD (200 ng) in the presence of increasing amounts (50, 100, 200 ng) of plasmids encoding wild-type or mutant (D201A and D199A) FIH-1. Data are presented as luciferase activity relative to cells transfected with GAL4-Luc. Values represent the mean ± S.E. of three independent experiments performed in duplicate (C and D).
We next investigated whether increased HRE binding of HIF-1α had any effect on HIF-1α target gene expression. To address this question, we analyzed by quantitative RT-PCR RNA from 293T cells stably expressing full-length Notch 1 in the presence or absence of Notch-specific ligand under normoxic or hypoxic conditions. As a reference, mRNA expression of the immediate Notch target gene Hey2 was up-regulated in cells treated with the Notch ligand Jagged, paralleled by activation of the HIF-1α target gene GLUT1, where mRNA expression increased to levels comparable with the hypoxic induction response (Fig. 5B). Under hypoxic conditions, exposure to Notch ligand further enhanced the induction response, correlating with increased recruitment of HIF-1α to the GLUT1 promoter. These observations argue that activation of Notch pathway results in enhanced HIF-1α target gene mRNA expression.
Notch ICD-Mediated Derepression of HIF-1α Function Is Abrogated by Elevated Levels of FIH-1.
The fact that Notch-mediated increased recruitment of HIF-1α to the HRE of canonical hypoxia-activated genes occurs without apparent recruitment of Notch ICD (Fig. 5A) suggests that Notch exerts its effect to derepress HIF-1α in a more indirect manner, with no apparent association with the active transcriptional complex. Given the high affinity of FIH-1 for Notch 1 ICD compared with HIF-1α, it may be hypothesized that Notch ICD could act as a molecular “sink” and recruit FIH-1 away from HIF-1α, resulting in transcriptional derepression of HIF1α-dependent promoters. To test this hypothesis and to monitor the activity of only the portion of HIF-1α that is the target of FIH-1 activity, i.e., the CAD (5), we transiently transfected P19 cells with an expression vector encoding GAL4-fused HIF-1α CAD in the presence of increasing amounts of wild-type and Asn mutant Notch ICD expression vectors. Expression of wild-type Notch ICD resulted in a dose-dependent increase in CAD-mediated activity at normoxia, as determined by a GAL4-driven reporter gene, whereas the Asn single and double mutants expressed at similar levels (data not shown) had very modest effects (Fig. 5C). At hypoxia, no significant effect was observed in the presence of either wild-type or mutant Notch proteins (data not shown). We next observed that overexpression of FIH-1 abrogated Notch ICD-dependent activation of HIF-1α CAD function at normoxia (Fig. 5D). The catalytically inactive FIH-1 mutants D201A and H199A were partially or completely unable to restore inhibition of HIF-1α CAD activity in the presence of overexpressed Notch 1 ICD, consistent with the critical role of hydroxylation of N803 for inhibition of CAD function (5). Our results indicate that Notch may enhance HIF-1α function by competing away FIH-1 from HIF-1α CAD and that repression of HIF-1α CAD at normoxia requires the full catalytic function of FIH-1. Why the Asn mutant Notch ICDs were less effective cannot be fully explained but may reflect the reduced interaction with FIH-1 under in vivo conditions where they are less effective than the wild-type protein at inducing nuclear localization of FIH-1 (Fig. 4A).
Conclusions
Notch signaling and the cellular hypoxic response have recently been shown to be functionally integrated (6). In this work, we reveal an additional layer of complexity in the cross-coupling of the two signaling pathways, involving the FIH-1 protein. In the control of the Notch downstream response, FIH-1 hydroxylates N1945 and N2012 of Notch 1 ICD.‖ Hydroxylation is a type of posttranslational modification of Notch ICD, and mutation of the two Asn residues attenuates Notch function in transcriptional assays and in control of neuro- and myogenesis. FIH-1 acts as a negative regulator of Notch signaling in transcriptional assays and also accelerates myogenic differentiation in C2C12 cells. These results are in excellent agreement with negative control of Notch signaling, and other negative regulators of Notch, such as Numb and Numblike, similarly promote myogenic differentiation (14).
The two sites of hydroxylation, N1945 and N2012, are located at similar positions before the β-hairpin turns separating ICD ANK 2 and 3 repeats, and 4 and 5 repeats, respectively (SI Fig. 10). Within these domains lie the primary contacts mediating formation of dimeric Notch complexes that are cooperatively formed on promoters with paired CSL-binding sites (15). Importantly, it was observed that mutations that disrupt the predicted dimerization still allow monomeric ICD to form ternary complexes with MAML1 (15). In our experiments, the transcriptional activity of Notch 1 ICD is severely impaired by mutations of one or both of the Asn residues but is restored in the presence of MAML1. It will therefore be important to investigate the role of the Asn residues in formation of higher-order ICD complexes. The two Asn residues are highly conserved among Notch paralogs and in different species, with the exception of Notch 4, which escapes Asn hydroxylation and FIH-1 interaction. This finding is, to our knowledge, a unique differential enzymatic modification in the Notch receptor family, and it will be interesting to learn whether it contributes to functional differences between different Notch receptors.
The present data show that FIH-1 also plays a role in how Notch potentiates the cellular hypoxic response. We observed that Notch 1 ICD-mediated derepression of HIF-1α was abrogated by increasing the level of FIH-1. Given that FIH-1 has higher affinity for Notch 1 ICD than for HIF-1α, it is an interesting idea that Notch ICD can sequester FIH-1 away from HIF-1α, resulting in derepression of HIF-1α function (SI Fig. 11). This mode of regulation occurred only if FIH-1 was present in limiting amounts, but not under conditions of excess FIH-1. To rerepress HIF-1α, normoxic conditions were necessary, consistent with the requirement of the oxygen-dependent catalytic function of FIH-1. This mechanism may therefore be particularly relevant in situations where HIF-1α is stabilized under normoxic conditions, for example, in the von Hippel–Lindau syndrome (16). In summary, the data on the role of FIH-1 in regulating Notch signaling reveals a more dynamic and complex relationship between Notch and hypoxia, suggesting that a fine-tuned control of signaling outputs from the two mechanisms is required for modulating the cellular response to low oxygen.
Materials and Methods
Details for the generation of plasmid constructs; the expression, purification, and MS analyses of Notch ICD functional domains; cell culture and transient transfection conditions; chick in ovo electroporation and immunohistochemistry; immunoblotting; intracellular localization; RNA interference; ChIP; and quantitative RT-PCR assays are given in SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Anne Chapman-Smith for assistance with the preparation of the supporting information. This work was supported by the National Health and Medical Research Council of Australia, the National Heart Foundation of Australia, the National Natural Science Foundation of China, the Swedish Cancer Society, the Swedish Foundation for Strategic Research, the Göran Gustafsson Foundation, the Swedish Research Council, and the European Union.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711591105/DC1.
During the preparation of this paper, a study describing similar data for the modification of Notch by FIH-1 was published (18). In this study, hydroxylation of N1945 and N2012 was identified, and a cocrystal structure of Notch ICD and FIH-1 was presented.
References
- 1.Semenza GL. Life with oxygen. Science. 2007;318:62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- 2.Mahon PC, Hirota K, Semenza GL. FIH-1: A novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewitson KS, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 5.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: A hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 6.Gustafsson MV, et al. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Pear WS, Simon MC. Lasting longer without oxygen: The influence of hypoxia on Notch signaling. Cancer Cell. 2005;8:435–437. doi: 10.1016/j.ccr.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Bray SJ. Notch signaling: A simple pathway becomes complex. Nature Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 9.Krejcí A, Bray SJ. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockman ME, et al. Posttranslational hydroxylation of ankyrin repeats in IκB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor-inhibiting HIF (FIH). Proc Natl Acad Sci USA. 2006;103:14767–14772. doi: 10.1073/pnas.0606877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 12.Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: A constitutively activated repressor of myogenesis directed at the basic helix–loop–helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 13.Dahlqvist C, et al. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 2003;130:6089–6099. doi: 10.1242/dev.00834. [DOI] [PubMed] [Google Scholar]
- 14.Chapman G, Liu L, Sahlgren C, Dahlqvist C, Lendahl U. High levels of Notch signaling down-regulate Numb and Numblike. J Cell Biol. 2006;175:535–540. doi: 10.1083/jcb.200602009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci USA. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaelin WG., Jr von Hippel–Lindau disease. Annu Rev Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 17.Zheng X, et al. Cell type-specific regulation of degradation of hypoxia-inducible factor 1α: Role of subcellular compartmentalization. Mol Cell Biol. 2006;26:4628–4641. doi: 10.1128/MCB.02236-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman ML, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.