Abstract
MicroRNAs (miRNAs) control tissue development, but their mechanism of regulation is not well understood. We used a gene complementation strategy combined with microarray screening to identify miRNAs involved in the formation of erythroid (red blood) cells. Two conserved miRNAs, miR 144 and miR 451, emerged as direct targets of the critical hematopoietic transcription factor GATA-1. In vivo, GATA-1 binds a distal upstream regulatory element to activate RNA polymerase II-mediated transcription of a single common precursor RNA (pri-miRNA) encoding both mature miRNAs. Zebrafish embryos depleted of miR 451 by using antisense morpholinos form erythroid precursors, but their development into mature circulating red blood cells is strongly and specifically impaired. These results reveal a miRNA locus that is required for erythropoiesis and uncover a new regulatory axis through which GATA-1 controls this process.
Keywords: erythrocyte, hematopoiesis, transcription factor, anemia, gene expression
The development of mature tissues from undifferentiated stem cells and progenitors is regulated by nuclear proteins, which coordinate lineage-specific programs of gene expression. GATA-1 is a hematopoietic transcription factor essential for the formation of platelets, eosinophils, mast cells, and erythrocytes (red blood cells) (reviewed in ref. 1 and 2). Mice lacking GATA-1 die of severe anemia during embryonic development (3). In addition, several human disorders are caused by mutations in the X chromosome-linked GATA1 gene (reviewed in ref. 2). For example, germ-line GATA1 mutations cause inherited anemias and thrombocytopenias, and somatic GATA1 mutations contribute to the development of acute megakaryoblastic leukemia (M7 AML) in children with trisomy 21 (Down syndrome). These studies establish roles for GATA-1 in normal and malignant hematopoiesis.
Gene ablation in mice and zebrafish has demonstrated that GATA-1 acts at unique stages in the development of specific lineages (4–8). Without GATA-1, lineage-committed erythroid precursors form but undergo developmental arrest and apoptosis. GATA-1 coordinates erythropoiesis by activating and repressing genes involved in cell division, apoptosis, and terminal maturation (9). Numerous erythroid genes are regulated directly by GATA-1 in combination with other lineage-specific and general transcription factors (reviewed in ref. 1). In turn, these actions initiate indirect genetic cascades that are less well defined.
One recently discovered mechanism through which lineage-specific transcription factors regulate tissue development is via microRNAs (miRNAs), a class of small (≈22 bp) noncoding RNAs that modulate the expression of protein-encoding mRNAs (10–12). MiRNAs bind complementary sequences in the 3′ untranslated region (UTR) of target mRNAs to induce nucleolytic degradation and/or inhibit translation. MiRNAs are conserved in evolution and function in the development of most or all vertebrate tissues, including hematopoiesis (13). During erythropoiesis, numerous miRNAs are induced or repressed, but little is known about their in vivo function or regulation (14–16). We discovered a GATA-1-regulated miRNA locus that is essential for erythropoiesis, thereby identifying a new regulatory hierarchy through which a lineage-specific transcription factor regulates tissue development.
Results and Discussion
Identification of a GATA-1-Regulated miRNA Locus.
To search for GATA-1-regulated erythroid miRNAs, we used the Gata-1− erythroblast line G1E (17). These cells proliferate in culture as immature erythroid precursors and undergo terminal maturation when GATA-1 activity is restored. G1E-ER4 is a subline stably expressing an estrogen-activated form of GATA-1 (GATA-1 fused to the ligand-binding domain of the estrogen receptor) (18). Treatment of G1E-ER4 cells with estradiol induces a GATA-1-regulated program of gene expression with concomitant cellular maturation (9). We used a microarray to evaluate the expression of 292 different miRNAs in G1E-ER4 cells at 0 versus 24 h after GATA-1 activation (19). Eleven miRNAs were altered by at least two-fold during GATA-1-mediated erythroid maturation [supporting information (SI) Table 1]. All of these miRNAs were induced, consistent with findings that miRNA expression is generally higher in differentiated tissues (15). We focused on miR 144 and miR 451, which exhibited the strongest changes in expression (miRBase accession numbers MIMAT0000156 and MIMAT0001632) (20). Northern blots confirmed that these miRNAs were strongly induced by GATA-1 in G1E cells (Fig. 1A). MiR 144 and miR 451 were up-regulated during induction of erythroid maturation of human CD34+ cells and murine erythroleukemia (MEL) cells (SI Fig. 6 and data not shown) (14, 16, 21, 22). Analysis of multiple mouse tissues showed that miR 144 and miR 451 were most highly expressed in blood (Fig. 1B), consistent with previous reports that miR 451 is present in human erythrocytes (23) and in circulating blood of zebrafish (24). In mouse spleen, miR 144 and miR 451 were highly enriched in cells expressing Ter119, an erythroid-specific maturation marker (Fig. 1C and data not shown). Cultured megakaryocytes expressed ≈20-fold less mature miR 451 compared with Ter119+ erythroid cells (data not shown). Our findings, combined with the work of others, indicate that expression of miR 451 is largely restricted to the erythroid lineage.
Fig. 1.
MiRNAs 144 and 451 are abundant in erythroid tissues and GATA-1-regulated. (A) Northern blot showing induction of miR 144 (Top) and miR 451 (Middle) after activation of a GATA-1-estrogen receptor fusion protein in G1E-ER4 murine erythroblasts. Five micrograms of total cellular RNA was analyzed in each lane. (Bottom) Ethidium bromide staining of 5.8S and 5S rRNAs used as a loading control. (B) Multitissue Northern blot of miR 144 and miR 451 expression in mice. MiR 194, which is expressed in nonhematopoietic tissues, was examined as a control. Five micrograms of total cellular RNAs from the following murine tissues were analyzed: Bl, whole blood; Br, brain; Ht, heart; Kn, kidney; Lv, liver; Lg, lung; Mu, muscle; Sp, spleen; Te, testes. (C) Splenocytes were fractionated according to expression of the erythroid-specific surface antigen Ter119 and analyzed for miR 451 expression by Northern blotting. The third lane labeled “oligo” represents 0.1 pmol of a miR 451“sense” DNA oligonucleotide used as a hybridization control. (D) Expression of miR 451 and miR 144 in zebrafish embryos at 26 hpf. Wild-type and gata-1− (vlad tepes) embryos were analyzed by WISH by using miR 144 and miR 451 probes. Expression of both miRNAs was observed in the blood island of the ICM, shown in the boxed regions. Higher-magnification views of the ICM (arrow) are displayed in the Insets. Expression of miR 451 and miR 144 is markedly reduced in the ICM of vlad tepes embryos.
In zebrafish whole-mount in situ hybridization (WISH) studies, miR 451 and miR 144 were detected exclusively in the developing blood island of the intermediate cell mass (ICM) in a pattern identical to that of gata-1 (Fig. 1D). The onset of miR 451 and miR 144 expression was first detected at the 18-somite stage with increasing expression until 26 h postfertilization (hpf), whereas gata-1 expression was initiated slightly earlier, at the 5-somite stage (SI Fig. 7). Expression of miR 144 and miR 451 was greatly reduced in the gata-1-deficient zebrafish mutant vlad tepes (Fig. 1D and SI Fig. 7) (8). Together, data from multiple species demonstrate that the miR 144/451 locus is specifically activated during erythroid maturation in a GATA-1-dependent manner.
MiR 144 and miR 451 are encoded ≈100 bp apart on mouse chromosome 11 and are highly conserved in evolution (SI Fig. 8A). Prior studies suggest that the two miRNAs are transcribed on a single precursor RNA (pri-miRNA) (12, 24–26). We verified this and localized the 5′ end of the common pri-miRNA using RT-PCR and rapid amplification of cDNA ends (RACE) (SI Fig. 8 B and C).
The miR 144/451 Gene Is a Direct Transcriptional Target of GATA-1.
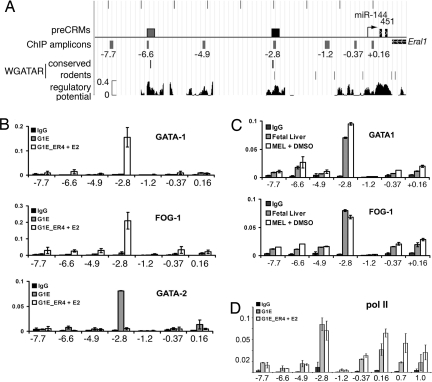
To investigate whether the miR 144/451 gene is induced directly by GATA-1, we searched for active GATA binding motifs. Erythroid cis-regulatory modules (CRMs), such as enhancers, can be predicted in aligned mammalian genomic DNA sequences by the presence of one or more conserved GATA consensus binding motifs within regions whose alignment patterns are similar to those found in a training set of known regulatory regions (27–29). Using thresholds established from previous work (30), we found two predicted erythroid CRMs (preCRMs), located 2.8 and 6.6 kb upstream of the miR 144/451 transcriptional start (Fig. 2A).
Fig. 2.
The miR 144/451 locus is directly activated by GATA-1. (A) Features of the miR 144/451 locus and 5′ flanking DNA. A 10-kb region of mouse chromosome 11 (mm8 assembly) is annotated with the DNA encoding miRNAs (thin black rectangles), the transcription start (bent arrow), and the 3′ end of the adjacent Eral1 gene located telomeric to the miRNA locus. PreCRMs (27) and amplicons used in ChIP assays in B and C are shown as rectangles above and below the line, respectively, with the positions of the amplicons relative to the transcription start given in kilobases (SI Table 3). The −2.8-kb preCRM validated in ChIP (B–D) and enhancer assays (Fig. 3) is shown in black. The positions of GATA consensus binding motifs (WGATAR), either conserved in mammals or present only in rodents, are indicated as vertical lines. The track labeled “regulatory potential” plots sequence similarity to alignment patterns in known regulatory regions (28). (B) Quantitative ChIP analysis of the locus in G1E (no GATA-1) and G1E-ER4 cells treated with estradiol (E2) for 24 h (activated GATA-1). The relative occupancies of GATA-1, FOG-1, and GATA-2 are indicated as vertical bars. As a negative control, ChIP experiments were performed with isotype-matched preimmune IgG. The bar graphs show averages of three independent ChIP experiments. Error bars represent standard deviation. (C) Quantitative ChIP analysis for GATA-1 and FOG-1 in primary fetal liver cells (embryonic day 14) and MEL cells induced to mature with HMBA, performed as in B. (D) Quantitative ChIP analysis of RNA pol II binding to the miR 144/451 locus, performed as in B. Amplicons labeled 0.7 and 1.0, not shown in A, extend into the transcribed region and are designated according to the distance (in kilobases) from the start of transcription.
Chromatin immunoprecipitation (ChIP) showed a strong signal for GATA-1 occupancy at the −2.8-kb preCRM region in estradiol-treated G1E-ER4 cells (Fig. 2B), MEL cells, and embryonic day 14 murine fetal liver, which contains mainly erythroid precursors (Fig. 2C). Consistent with these findings, the −2.8 preCRM contains two highly conserved GATA binding motifs (SI Fig. 9). The GATA-1 cofactor FOG-1 also occupies the −2.8-kb preCRM in estradiol-treated G1E-ER4 cells, MEL cells, and fetal liver (Fig. 2 B and C). In parental G1E cells, which lack GATA-1, the −2.8-kb preCRM is occupied by GATA-2, a related factor that functions in hematopoietic stem cells, multipotential progenitors, and early erythroid precursors (Fig. 2B) (31–33). Thus, GATA-2 binds the miR 144/451 locus but does not activate transcription. Restoration of GATA-1 activity in G1E cells induces an exchange of nuclear factors at the −2.8-kb preCRM whereby GATA-2 is released and GATA-1/FOG-1 become bound coincident with gene activation. This sequence of events likely approximates normal erythroid differentiation where GATA-2 is expressed at relatively high levels in early precursors and is gradually replaced by GATA-1 during later stages of maturation. To test the miR 144/451 preCRMs functionally, we linked them to a minimal erythroid promoter and luciferase reporter and introduced the constructs into MEL and K562 erythroleukemia cells (Fig. 3A). The −2.8-kb preCRM specifically augmented promoter activity, indicating that this region represents an erythroid enhancer (Fig. 3B). The −6.6-kb preCRM was occupied at relatively low levels by GATA-1 and FOG-1, particularly in MEL and fetal liver cells (Fig. 2C), but this region did not show enhancer activity (Fig. 3B). In addition, relatively low-level GATA-1/FOG-1 occupancy was detected at the miR 144/451 promoter, where a rodent-specific GATA binding motif resides (Fig. 2 A and B).
Fig. 3.
Enhancer activity of miR 144/451 preCRMs. (A) Design of the reporter construct. PreCRMs were positioned upstream of a γ-globin gene promoter linked to a firefly luciferase cDNA. The bent arrow indicates the start of the luciferase open reading frame. (B) Luciferase activities, reported as the fold increase over that of the parent plasmid, are shown from three separate transient transfection experiments, represented as separate bars. Error bars indicate standard deviation from quadruplicate luciferase determinations from a single experiment. Reporter assays were performed in human K562 erythroleukemia cells and in MEL cells treated with HMBA to induce erythroid maturation. Firefly luciferase activities are normalized to Renilla luciferase activities expressed from a cotransfected control plasmid. The DNA segment Zfpm1R13 is a positive control for a known enhancer (30).
Many miRNAs are transcribed by RNA polymerase (pol) II (34), and this should be the case for a GATA-1-regulated locus. We used ChIP to examine RNA pol II occupancy at the miR 144/451 gene in G1E cells. In the absence of GATA-1, RNA pol II bound the −2.8-kb enhancer and the proximal promoter region (Fig. 2D). GATA-1-independent recruitment of RNA pol II to these regions is presumably mediated by other transcription factors, including GATA-2. Of note, RNA pol II binds transcriptional enhancers at various other loci, including those expressed in erythroid cells (35). Activation of miR 144/451 transcription by GATA-1 was accompanied by increased RNA pol II occupancy within the transcribed region. Thus, GATA-1 may activate this locus by facilitating RNA pol II transcriptional elongation. Together, the ChIP studies demonstrate that GATA-1 binds the miR 144/451 locus at the promoter and an upstream enhancer at −2.8 kb, displacing GATA-2 and recruiting the cofactor FOG-1.
In summary, several lines of evidence indicate that the miR 144/451 locus is directly activated by GATA-1. First, miR 144/451 requires GATA-1 expression in G1E cells and zebrafish. Second, restoration of GATA-1 rapidly induces miR 144/451 expression in G1E cells. Third, a conserved enhancer in the miRNA locus binds GATA-1 and FOG-1 in erythroid cells.
The miR 144/451 Locus Is Essential for Erythropoiesis in Zebrafish.
Manipulation of miR 451 expression influences erythroid maturation of MEL cells (16). To investigate the functions of miR 144/451 in vivo, we used antisense morpholino oligomers (MOs) to interfere with miRNA function in zebrafish embryos (36). Embryos injected with miR 451-MO or miR 144-MO exhibited marked reductions in expression of the corresponding endogenous miRNAs (SI Fig. 10). At 24 hpf, MO-injected embryos showed no differences in expression of gata-1 or the erythroid markers βe1-globin and band3 within the ICM (Fig. 4A). At this stage of embryonic development, erythroid precursors are immature (37). Hence, miR 451 deficiency does not affect erythroid lineage specification or early maturation. However, 24 h later (48 hpf), miR 451 morphant embryos showed markedly decreased expression of erythroid markers (Fig. 4B and SI Fig. 11A). In normal embryos at this stage, erythroid precursors mature and enter the circulation, as evidenced by the appearance of hemoglobin staining cells in the ducts of Cuvier and cardinal vein (Fig. 4C, first and second panels). In contrast, loss of miR 451 expression caused severe anemia, indicated by reduced hemoglobinized cells throughout the embryo (Fig. 4C, Right). Similar but more severe effects were caused by MO-induced loss of gata-1 (Fig. 4C, third panel). Thus, miR 451 is required for maintenance and/or late-stage maturation of committed erythroid precursors in zebrafish. Studies to determine the effects of miR 144/451 ablation in mice are needed. In murine Gata-1− G1E cells, overexpression of miR 144 and miR 451, either separately or together, failed to induce globin expression or other markers of terminal erythroid maturation (data not shown). These findings support a model in which some important GATA-1 activities are regulated through miR 144/451. However, these miRNAs cannot substitute for loss of GATA-1, which has many additional critical target genes.
Fig. 4.
MiRNA 451 is required for maintenance of erythroid cells in zebrafish. (A) WISH analysis for expression of erythroid markers in wild-type and miR 451 morphant embryos at ≈24 hpf. At this stage, committed erythroid cells are relatively immature and blood circulation is not yet established. The expression of erythroid markers in morphant embryos is unchanged, indicating normal specification of primitive erythropoiesis. The morphant embryos showed strongly reduced expression of miR 451 (SI Fig. 10). (B) WISH analysis for expression of βe1-globin in wild-type and miR 451 morphant embryos at ≈48 hpf. At this stage, mature erythroid cells have entered the circulation. The ducts of Cuvier on the yolk (square bracket, 1) and the cardinal vein (arrow, 2) are indicated, and higher-magnification views are shown in Lower. (C) o-dianisidine staining for hemoglobin in morphant embryos at 48 hpf.
Given the close inter-relationship between vertebrate erythroid and megakaryocytic developmental programs, we evaluated the function of miR 451 on thrombocyte (platelet-equivalent) development in zebrafish. We injected miR 451-MO into transgenic embryos that express green fluorescent protein (GFP) regulated by the megakaryocyte/thrombocyte-specific CD41 promoter [Tg(CD41:GFP)] (38). No differences in GFP+ thrombocytes were observed between control embryos and morphants injected with miR 451-MO, indicating that miR 451 is dispensable for zebrafish thrombopoiesis (SI Fig. 11B). In addition, miR 451 knockdown does not reduce granulocytic (myeloperoxidase-positive) cells in zebrafish embryos (SI Fig. 11C). Thus, the deleterious effects of miR 451-MO are restricted to the erythroid lineage and not caused by generalized toxicity. Aside from anemia and mild pericardial edema (probably attributable to anemia), no other morphological abnormalities were detected in miR 451 morphant embryos (Fig. 4 and data not shown). Injection with miR 144-MO failed to elicit any detectable abnormalities in embryo morphology, erythropoiesis, or thrombopoiesis (data not shown); further studies are required to elucidate the function of this miRNA.
Predicted miR 144/451 Targets Are Overrepresented Among GATA-1-Repressed Transcripts.
GATA-1 controls an extensive program of mRNA repression, most likely via multiple mechanisms (9). Messenger RNAs that are rapidly repressed after restoration of GATA-1 in G1E cells are more likely to represent direct transcriptional targets, as shown for the Gata2 gene (39). In contrast, transcripts down-regulated with delayed kinetics are candidates for repression by GATA-1-induced miRNAs, which likely promote degradation of some targets (40). We identified G1E cell mRNAs that are repressed between 7 and 21 h after GATA-1 function is restored (9). During this time interval, miR 144 and miR 451 become induced and accumulate to maximal levels (see Fig. 1A). We extracted potential miR 144/451 target mRNAs from miRBase (http://microrna.sanger.ac.uk/targets/v4), which identifies sequence complementarity between the 5′ end of the miRNA (“seed sequence”) and the 3′ UTR of cognate mRNAs (20, 41, 42). Among transcripts repressed between 7 and 21 h, predicted targets for both miR 144 and miR 451 were preferentially enriched compared with non-target mRNAs (Fig. 5). Between 0 and 3 h, when miR 144 and miR 451 are not expressed (see Fig. 1A), we found no evidence for overrepresentation of predicted targets among repressed transcripts (data not shown). Therefore, once miR 144 and miR 451 become expressed, their predicted target mRNAs become preferentially repressed. One biological implication is that there are likely to be numerous miR 144/451 targets during erythroid development, in accord with findings that some other tissue-specific miRNAs, such as miR 124 (brain) and miR 1 (muscle), each repress many target mRNAs (40). However, functional annotation of predicted targets for miR 144 and miR 451 showed significant enrichment for genes classified to encode “nuclear proteins” (http://david.abcc.ncifcrf.gov/). Accordingly, one function of miR 144/451 may be to promote terminal maturation by clearing cells of proteins that regulate gene expression, similar to what has been postulated for miRNAs in plants (43). Predicted miR 144 and miR 451 target genes that are down-regulated during GATA-1-induced erythroid maturation of G1E cells are shown in SI Tables 4 and 5, respectively. It is likely that these groups contain numerous genes whose down-regulation is critical for erythropoiesis. For example, overexpressed Myc, a predicted miR 451 target, inhibits erythroid development (44–46). We are investigating whether Myc and other predicted targets are directly repressed by miR 451 and miR 144.
Fig. 5.
Predicted miR 144/451 mRNA targets are overrepresented among repressed genes during later stages of GATA-1-induced erythroid maturation. Messenger RNAs that are repressed during GATA-1-induced erythroid maturation of G1E cells were identified from a previously described dataset (9). We focused on repression between 7 and 21 h after GATA-1 activation, when miR 144/451 becomes induced and reaches maximal levels (see Fig. 1A). The repressed transcripts were examined for expression of predicted miR 144 and 451 targets identified by miRBase (http://microrna.sanger.ac.uk/targets/v4). Fisher's exact test was used to evaluate whether predicted miRNA targets are overrepresented among repressed mRNAs relative to nonrepressed ones. One-sided P values indicating the significance of differences between potential targets and nontargets are shown. An odds ratio of 1.0 indicates that targets are more likely to be repressed within the tested time interval.
MiRNAs play important roles in normal and malignant hematopoiesis (13). Our discoveries that miR 451 is essential for zebrafish erythropoiesis in vivo and regulated directly by GATA-1 are consistent with the emerging concept that “master” transcriptional regulators control tissue development and physiology in part by modulating the expression of miRNAs (47–53). The kinetics of miR 451 induction during mammalian and zebrafish erythropoiesis and the phenotype of morphant embryos demonstrate that this miRNA is dispensable for establishment of the erythroid lineage but required for subsequent survival and/or maturation, similar to what has been observed for GATA-1 (7, 54). These findings predict that the miR 144/451 locus is a major downstream effector of GATA-1 in erythroid cells. Accordingly, it is possible that altered levels of these miRNAs, either through naturally occurring genetic changes or through pharmacologic manipulation, could impact red blood cell production in various diseases.
Materials and Methods
Cell Culture.
MEL cells were cultured in Iscove's modified Dulbecco's medium with 10% FCS. Erythroid maturation was induced by addition of 5 mM N,N′-hexamethylene-bis-acetamide (HMBA). G1E and G1E-ER4 cells were cultured as described (17, 18).
miRNA Northern Blots.
Five micrograms of total RNA was electrophoresed on a 15% urea/polyacrylamide/Tris–borate–EDTA gel and transferred to a BrightStar nylon membrane (Ambion) by semidry electroblotting. Membranes were UV-cross-linked (StrataLinker; Stratagene), rinsed in 5× SSC, and probed with 32P-end-labeled miRCURY locked nucleic acid (LNA) probes (Exiqon) at 42°C (miR 144 and miR 194 probes) or 54°C (miR 451 probe).
MiRNA Microarray Studies.
RNA samples were analyzed by Exiqon A/S by using the miRCURY Hy3/Hy5 labeling kit and the miRCURY LNA array (version 8.0). Detailed methods are provided in SI Materials and Methods. Raw data for the microarray study have been deposited in the Gene Expression Omnibus (GEO) database, accession no. GSE10134.
Enhancer Assays.
These assays were performed as described previously (30) and in SI Materials and Methods.
ChIP.
ChIP assays were performed as described (55, 56). PCR products were quantified by using SYBR green dye on an ABI 7000 real-time PCR machine. Signals were quantified by comparison with a dilution series of the relevant input DNA. Primer pairs used for PCR are described in SI Table 3. Antibodies used for ChIP were obtained from Santa Cruz Biotechnology: GATA-1 (N-6), GATA-2 (H-116), FOG-1 (M-20), and RNA pol II (N-20).
Zebrafish Studies.
Procedures for WISH and morpholino miRNA knockdown studies are described in SI Materials and Methods.
Bioinformatic Analysis.
Methods for analysis of potential miRNA targets and G1E gene expression microarrays are provided as SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Hui-Feng Lin and Robert Handin for the Tg(CD41-GFP) transgenic zebrafish line and Joseph Italiano for access to the Orca IIER CCD camera and Metamorph software program. We also thank Shana Gilbert-Gregory, Alex Valvezan, and Sarah Muse for technical assistance. This work was supported in part by the Cooley's Anemia Foundation (M.J.W.), the Leukemia and Lymphoma Foundation (M.J.W.), the March of Dimes Foundation (B.H.P.), and the National Institutes of Health (R.C.H., B.H.P., and M.J.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE10134).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712312105/DC1.
References
- 1.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migliaccio AR, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1(low) mouse mutant. J Exp Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu C, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1− embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 8.Lyons SE, et al. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc Natl Acad Sci USA. 2002;99:5454–5459. doi: 10.1073/pnas.082695299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch JJ, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 12.Fukao T, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 13.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song CZ. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsang AP, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 19.Castoldi M, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruchova H, Yoon D, Agarwal AM, Mendell J, Prchal JT. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35:1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509–514. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 23.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 24.Kloosterman WP, et al. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006;34:2558–2569. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altuvia Y, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Elnitski L, et al. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova KD. Strong and weak male mutation bias at different sites in the primate genomes: Insights from the human-chimpanzee comparison. Mol Biol Evol. 2006;23:565–573. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- 29.Taylor J, et al. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minegishi N, et al. Expression and domain-specific function of GATA-2 during differentiation of the hematopoietic precursor cells in midgestation mouse embryos. Blood. 2003;102:896–905. doi: 10.1182/blood-2002-12-3809. [DOI] [PubMed] [Google Scholar]
- 32.Tsai F.-Y, et al. An early hematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 33.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 34.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson KD, et al. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 38.Lin HF, et al. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grass JA, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 41.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 44.Coppola JA, Cole MD. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 45.Prochownik EV, Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. Nature. 1986;322:848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- 46.Rylski M, et al. GATA-1-Mediated Proliferation Arrest During Erythroid Maturation. Mol Cell Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 49.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 51.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sylvestre Y, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 53.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon MC, et al. Rescue of erythroid development in gene targeted GATA-1− mouse embryonic stem cells. Nat Genet. 1992;1:92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- 55.Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci USA. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a Tissue-Specific Histone Acetylation Pattern by the Hematopoietic Transcription Factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.