Abstract
A generic method is described for the fluorescence “readout” of the activity of single redox enzyme molecules based on Förster resonance energy transfer from a fluorescent label to the enzyme cofactor. The method is applied to the study of copper-containing nitrite reductase from Alcaligenes faecalis S-6 immobilized on a glass surface. The parameters extracted from the single-molecule fluorescence time traces can be connected to and agree with the macroscopic ensemble averaged kinetic constants. The rates of the electron transfer from the type 1 to the type 2 center and back during turnover exhibit a distribution related to disorder in the catalytic site. The described approach opens the door to single-molecule mechanistic studies of a wide range of redox enzymes and the precise investigation of their internal workings.
Keywords: electron transfer, redox enzyme, Förster transfer, nitric oxide, fluorescent label
In the past few years, single-enzyme studies have revealed numerous hidden aspects of enzyme behavior (1). The huge potential of these studies to unravel the intricate kinetics and precise workings of enzymes, which are often hidden within the ensemble properties, is somewhat restricted by current approaches. The majority of existing single-molecule enzymatic assays are based on fluorescence and have been limited to the flavoenzymes, which contain a fluorescent cofactor (2–5), or to enzymes for which a suitable fluorogenic substrate could be designed (6–9). Recently, it was shown how redox enzyme activity in the bulk can be studied by Förster resonance energy transfer (FRET) from an attached fluorescent label to the enzyme cofactor (10, 11). Here, we report, first, how this technique can be successfully applied to study the enzymatic turnover of single surface-confined copper-containing nitrite reductase (NiR) molecules labeled with ATTO 655 using scanning confocal fluorescence microscopy. Second, it is shown how the kinetics of the fluorescence time traces can be connected with the kinetic parameters that describe the ensemble behavior of the enzyme. Finally, the rate of the electron transfer between the types 1 and 2 Cu centers during turnover could be established. The observed distribution of rates is connected to the partial structural disorder in the catalytic site that has been observed in crystallographic studies.
Results
Enzyme Mechanism.
Dissimilatory copper-containing nitrite reductase (NiR) from Alcaligenes faecalis S-6 converts nitrite into nitric oxide. The enzyme is a homotrimer, each monomer containing one type 1 and one type 2 copper site (Fig. 1). The type 1 copper accepts an electron from the physiological donor and transfers it to the type 2 copper, where nitrite is reduced to nitric oxide (NO). The midpoint potentials of the types 1 and 2 Cu centers are close, resulting in a redox equilibrium constant for the two sites that is close to 1 (12, 13). Regarding the enzyme mechanism, it has been stated (14, 15) that, after reduction of the type 1 Cu site, the electron is passed on to the type 2 Cu site, after which nitrite binds to this site and is converted (“reduction first”). Others have found that nitrite first binds to the oxidized type 2 site, after which an electron is transferred from the type 1 to the type 2 site and nitrite conversion takes place (“binding first”) (12). More recent investigations show that the enzyme operates according to a “random sequential mechanism” in which both pathways run in parallel, although either one of them may prevail depending on the experimental conditions (16) [see supporting information (SI) Appendix, SI Scheme 2]. The choice of pH and nitrite concentration in the present study favor the “reduction first” pathway (16).
Fig. 1.
Sequence of events during the turnover of NiR labeled on the N terminus with ATTO 655. (Left) In the resting enzyme, both types 1 (blue) and Cu (red) are oxidized, and the ATTO 655 fluorescence is low because of FRET to the oxidized type 1 Cu. (Center) Upon reduction of type 1 Cu by an external electron donor, the dye fluorescence goes up, because FRET from ATTO 655 to the reduced type 1 Cu (gray) is not possible. (Right) An electron is transferred from types 1 to 2 Cu, which becomes reduced (gray). The label fluorescence is quenched by the oxidized type 1 Cu. Nitrite binds to the reduced type 2 Cu, is converted into nitric oxide, and dissociates from the enzyme.
Fluorescent Labeling.
When oxidized, the type 1 Cu site has two broad absorption bands at 450 and 590 nm (SI Appendix, SI Fig. 5) that disappear upon reduction. [The contribution of the type 2 copper to the NiR absorption is negligible (17)]. This property was used to monitor the enzyme turnover by attaching a fluorescent label to the enzyme. If spectral overlap and distance between label and cofactor allow Förster resonance energy transfer (FRET), the label fluorescence will reflect the progress along the catalytic cycle.
The ATTO 655 chromophore was chosen as a label. The absorption of oxidized type 1 Cu has a significant spectral overlap with the ATTO 655 emission (SI Appendix, SI Fig. 5), corresponding with a Förster radius of 3.5 nm (see SI Appendix, SI Text). The donor-acceptor distance from ATTO 655 attached to the N terminus of NiR to the type 1 Cu of the same monomer is 3.9 ± 0.5 nm, as estimated from the NiR crystal structure (18). This leads to an estimated efficiency of FRET from the fluorophore to the oxidized type 1 copper of 30–45%. Because reduced type 1 Cu has no absorption in the visible region, the fluorescence of ATTO 655-labeled NiR will be significantly higher for the protein with a reduced than with an oxidized type 1 Cu center. During enzymatic turnover of the labeled NiR, the fluorescence intensity of ATTO 655 is expected to switch, therefore, between two distinct levels: high (corresponding to reduced type 1 Cu) and low (oxidized type 1 Cu) (Fig. 1).
To observe the activity of only one monomer in the NiR trimer, the labeling conditions were chosen to yield on average no more than one dye molecule per trimer. Reaction conditions favored labeling of the N terminus over lysine labeling (see Methods). As a control, an H145A mutant of NiR was used, in which one of the histidines (His-145) coordinating the type 1 copper is replaced by alanine. This mutation leads to a large increase in the midpoint potential of the type 1 site, resulting in almost complete loss of catalytic activity (17). An electron donor excess sodium ascorbate with phenazine ethosulfate (PES) as a redox mediator was used to initiate the NiR turnover (19). The ascorbate/PES also has the advantage of keeping the system anaerobic, preventing NiR inactivation (20).
Single-Molecule Experiments.
For the single-molecule confocal fluorescence microscopy experiments, the enzyme was immobilized on silanized glass through a homobifunctional thiol-reactive polyethyleneglycol linker (Fig. 2). A typical confocal fluorescence microscope image of ATTO 655-labeled immobilized L93C NiR is shown in Fig. 3A. To study an individual enzyme, a single labeled NiR molecule was positioned in the focus of the microscope (at the laser excitation spot), and its emission intensity time trace was measured (20 to >300 sec). Molecules that were turning over were always measured for at least 100 sec.
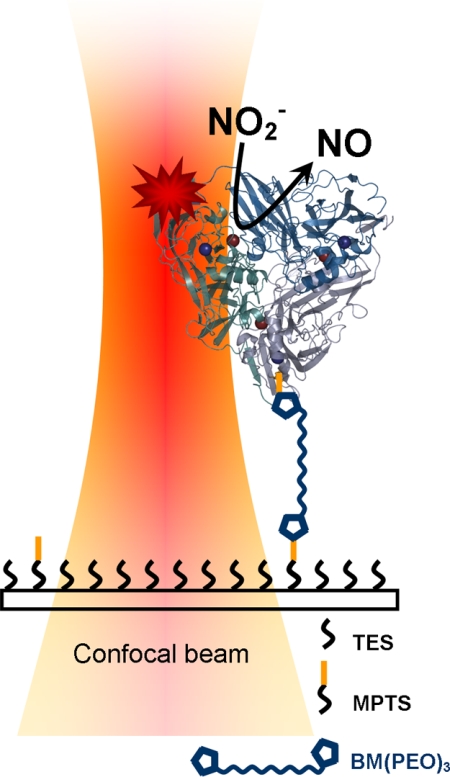
Fig. 2.
Enzyme immobilization scheme. A molecule of L93C NiR is bound through the BM(PEO)3 linker to the glass coverslip modified with a 100:1 mixture of triethoxysilane (TES):mercaptopropyl trimethoxysilane (MPTS). This gave reproducible results and prevented protein aggregation at the surface. The BM(PEO)3 linker is attached to the protein via the exposed Cys 93. The glass coverslip with immobilized protein was covered with buffer solution and placed on top of the objective of a scanning confocal fluorescence microscope with the exciting laser beam (λex = 620 nm) coming from below. The substrate and then the reductant were added directly into the buffer, and several images were taken to ensure that the protein was immobilized.
Fig. 3.
Single-molecule fluorescence experiment on immobilized NiR molecules. (A) Scanning confocal fluorescence picture of L93C NiR immobilized on silanized glass (10 × 10 μm). (B) Fluorescence time trace and fluorescence intensity distribution histogram for ATTO 655-labeled L93C/H45A NiR in buffer. (Left) Background fluorescence (“bg”). (Center) Fluorescence of a single NiR molecule (“dye blinking”). (Right) Intensity histogram (circles) together with the best fit (blue line) by two Poissonian distributions (red and green). Black line, residuals; gray line, fit for background fluorescence. (C) Same for L93C NiR in the presence of ascorbate/PES and 5 mM NaNO2 (labeled “bg” and “turnover”). The length of the observed time traces (of which only sections are shown) amounted to 40, 140, 30, and 140 sec (B, “bg” and “dye blinking;” C, “bg” and “turnover,” respectively).
Two types of fluorescence intensity time traces were observed. An example of the first type is shown in Fig. 3B. It was observed for H145A/L93C NiR in the absence of ascorbate/PES and nitrite. Similar traces were observed with the active L93C NiR (no ascorbate/PES, no nitrite) and with glass slides spin-coated with an ATTO 665 solution in 1% polyvinyl alcohol. Periods of high fluorescence alternate on a time scale of seconds with dark periods in which only the background signal is observed; we ascribe this to dye blinking (21–24). The steady nature of the observed fluorescence during the “on” periods is consistent with crystallographic and NMR evidence, indicating absence of large-scale conformational fluctuations (18, 25).¶ In passing, we note that the one-step bleaching (data not shown) and blinking observed under nonturnover conditions confirmed we were monitoring single molecules.
Addition of ascorbate/PES and nitrite had no effect on the signal of H145A/L93C NiR except for an increased prevalence of the “off” periods (SI Appendix, SI Fig. 6). A completely different type of fluorescence trace was observed when nitrite and ascorbate/PES were added to immobilized active L93C NiR (Fig. 3C). The corresponding fluorescence intensity histogram can be fit with two Poissonian distributions, showing the molecules are switching between two fluorescence levels, which are clearly above the background. These traces we attribute to catalytically active ATTO 655-labeled L93C NiR, the state with the lower fluorescence corresponding to intermediates in the NiR catalytic cycle where the type 1 copper is oxidized and the state with the higher fluorescence to intermediates where the type 1 copper is reduced.‖ The virtual disappearance of blinking under these conditions was not further investigated and is the subject of further study.
Effect of Substrate Concentration.
The occurrences of the high- and low-fluorescent states varied with the substrate concentration. In Fig. 4A, the relative occurrence, P, of the high fluorescent state is presented as a function of the nitrite concentration. Here, P = Ihigh/(Ihigh + Ilow), where Ihigh (Ilow) is the integral under the Poissonian histogram corresponding to the high (low) fluorescent state. At high [NaNO2], the probability of finding the type 1 Cu center reduced levels off at ≈30%. This is consistent with the expectation that the type 1 Cu on average will stay longer in the reduced form at low substrate concentrations, because the electron flow from it will be limited by the availability of substrate. The same value is found in bulk fluorescence experiments at 5 mM sodium nitrite. At the same time, the ratio of the high- and low-fluorescence intensities was similar in all experiments, with (Fhigh−Flow)/Fhigh in the range of 0.3–0.6 with an average of 0.45 ± 0.08. This value fits well with the (Fred−Fox)/Fred = 0.49 we observe in bulk experiments (data not shown) and with the 0.3–0.5 range predicted from the spectral overlap (see above).
Fig. 4.
Autocorrelation analysis of fluorescence time traces. (A) Relative occurrence, P, of the states with reduced type 1 Cu during the enzyme turnover at different substrate concentrations. Each point is an average over at least eight individual molecules. Error bars present standard deviations. (B) Fluorescence intensity autocorrelation functions calculated from individual traces of ATTO 655-labeled NiR molecules at 5 μM (black), 50 μM (green), 500 μM (blue), and 5 mM (red) NaNO2. Normalized experimental data (circles) were fit to stretched exponentials (continuous lines), G(t) = exp(−(t/τ0)β). The fitting parameters were: for 5 μM NaNO2 τ0 = 70 ms, β = 0.81; for 50 μM NaNO2 τ0 = 34 ms, β = 0.72; for 500 μM NaNO2 τ0 = 22 ms, β = 0.60; for 5 mM NaNO2 τ0 = 17 ms, β = 0.61. (Inset) Same curves in log(−log(G(t))) vs. log(t) representation. In this representation, a stretched exponential is linear with slope β. A line with a slope of 1 is plotted for comparison. (C) Inverse of average decay times, 1/<τ>, of ATTO 655-labeled NiR molecules (blue squares) for different nitrite concentrations, Eq. 5. Each point is an average over seven (5, 50, and 500 μM NaNO2) or five individual molecules (5 mM NaNO2). Error bars present standard deviations. The continuous lines in A and C correspond to the best global fit to Eqs. 6 and 5 (see text).
Autocorrelation Analysis.
Because the Poissonian distributions corresponding to the high- and low-fluorescent states overlap, the distinction between the two states necessary to calculate the probability density functions of “on” and “off” times becomes blurred. Instead, to obtain the kinetics of the enzyme turnover, we calculated (see Methods for details) the fluorescence intensity autocorrelation functions, G(t), for the emission traces at different substrate concentrations and fitted the normalized functions to single stretched exponentials, G(t) = exp(−(t/τ0)β). The stretched exponential function can be considered as a sum of single exponentials with decay times τ after a distribution ρ(τ): exp(−(t/τ0)β) = ∫ρ(τ)exp(−t/τ)dτ (26, 27). The analytical expression for ρ(τ) depends only on τ0 and β (27). Single-exponential behavior is observed for β = 1, whereas for β<1, the distribution of τ values becomes broader the more β deviates from 1. The average value of τ, which will be used in the subsequent analysis, is given by <τ> = ∫ρ(τ) τ dτ = (τ0/β) Γ(1/β) that can be evaluated numerically for given τ0 and β (26, 27).
Fig. 4B shows four characteristic autocorrelation curves corresponding to different concentrations of NaNO2. (Autocorrelation graphs of more than one individual molecule have been plotted in SI Appendix, SI Figs. 7 and 8 to give an idea of the spread of the observed traces.) It can be clearly seen that the characteristic time τ0 (see Fig. 4B legend) is longer at low substrate concentrations and shows saturation behavior toward high concentration. The dependence of 1/<τ> on substrate concentration is shown in Fig. 4C (squares).
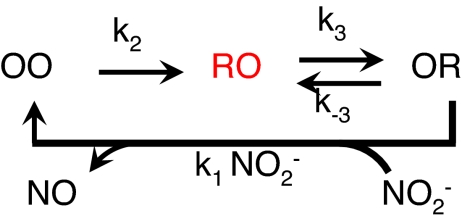
Scheme 1 (for a discussion of alternative models, see SI Appendix, SI Text) was used to analyze the data, in which OR denotes the state of the enzyme with the type 1 site oxidized (O) and the type 2 site reduced (R); similar definitions apply to OO and RO. The RO state has high, the others low fluorescence intensity.
Scheme 1.
Enzyme cycle of NiR.
Qian and Elson (28) have shown that for a cyclic reaction scheme as in Scheme 1, the occupation probabilities for the three states under nonsteady-state conditions exhibit biexponential kinetics, i.e., the change in occupation probability is the sum of two exponentials with different amplitudes for each state. The same is true for the first-order autocorrelation function relating to the occupational probability of each state under steady-state conditions. In our case, the autocorrelation function of the fluorescence corresponds with the occupation probability of the state “RO” in Scheme 1. Thus, the fluorescence correlation function becomes (28),
The expression for λ1,2 in terms of the rate constants in Scheme 1 can be found directly from the expressions derived by Qian and Elson and is given by
with
and with S the substrate concentration.
The decay time 1/ λ1 is too short for the exp(λ1t) component to give a measurable contribution to G(t) within the 1-ms binning time used for the present analysis of the fluorescence traces. Consequently, G(t) simplifies to
with
Although Eq. 4 indicates that the fluorescence autocorrelation function consists of a single exponential with a single rate, the experiment shows that we are dealing with a sum of such exponentials (i.e., a stretched exponential) with a distribution of rates. The distribution is characterized by an average decay time <τ>, which has been experimentally determined (Fig. 4C). We will use Eq. 5 for the analysis of the 1/<τ> data as a function of the nitrite concentration (Fig. 4C) by replacing |λ2| by 1/<τ>. The corresponding values of k1, k2, etc., then reflect distribution averaged parameters.
Finally, the steady-state probability of the on state, PSS(RO), can be found directly from the expressions derived by Qian and Elson and is given by
A global fit of the experimental data in Fig. 4 A and C to Eqs. 6 and 5, respectively, produced the continuous lines in Fig. 4 A and C. The corresponding values of the rate constants are k1 = (4 ± 2) · 105 M−1·s−1, k2 = 10 ± 3 s−1, k3 = 21 ± 6 s−1, k−3 = 14 ± 4 s−1.
It should be noted that |λ2| does not represent the turnover rate of the enzyme, and that the dependence of λ2 on the substrate concentration does not exhibit Michaelis–Menten characteristics. For the connection of the single-molecule kinetic parameters of Scheme 1 with Michaelis–Menten kinetics, see SI Appendix, SI Text.
Discussion
We first investigate whether and how the data obtained from the present single-molecule experiments can be related to the enzymological parameters that have been reported for NiR in the literature. Subsequently, we comment on possible causes for the distribution in rate parameters.
The rate constants of forward and backward electron transfer during catalysis between the types 1 and 2 Cu centers in L93C NiR from A. faecalis amount to k3 = 21 ± 6 s−1 and k−3 = 14 ± 4 s−1. There are no data in the literature by which these values can be compared directly. The data that come closest are provided by pulse radiolysis experiments on a range of NiRs. These have provided values for kET = (k3 + k−3) that range from 450 to 2,100 s−1. However, they refer to experiments in the absence of nitrite. Suzuki et al. (29) have reported that these rates drop, in some cases by >1 order of magnitude, in the presence of nitrite (but still in the absence of turnover). It is generally accepted now that binding and turnover of nitrite may cause subtle structural changes in the protein framework around the catalytic site that may considerably affect the rate of intramolecular electron transfer.
A better set of data by which to compare the present results is provided by the Michaelis–Menten parameters of the enzyme, provided the microscopic parameters of Scheme 1 can be connected to the macroscopic parameters observed in the bulk. It can be shown (see SI Appendix, SI Text) that this connection is given by KM = k2(k3 + k−3)/(k1(k2 + k3)) and Vmax = k2 k3/(k2 + k3), which, in the present case, leads to KM = 31 ± 17 μM and Vmax = 6.5 ± 2 s−1. Here, KM is the Michaelis–Menten constant, and Vmax is the maximal turnover rate. With pseudoazurin, the turnover rate in the bulk at pH 7 and 298 K has been reported as 131 s−1 per monomer with KM = 36 μM (30) in agreement with data from Farver et al. on Alcaligenes xylosoxidans NiR (13). Ulstrup and coworkers (31), in electrochemical experiments on NiR immobilized on a methyl-bezenethiol SAM on Au, found a KM of 44 μM and a turnover rate of 30 s−1, although they could not exclude that electron transfer from electrode to enzyme might partially limit turnover.
Attempts by us to use pseudoazurin as a reductant in a single-molecule experiment have failed so far, possibly because of preferential absorption of the pseudoazurin to the hydrophobic glass surface. Instead, we checked the catalytic rate of the ATTO 655-labeled L93C NiR in bulk by determining the rate of NO formation under the same conditions (see Methods), as used in the single-molecule experiments (data not shown). The KM measured in these experiments was 50 ± 20 μM, and the Vmax was 8 ± 1 s−1. Similar activities were found for unlabeled NiR. Although the data appear in good agreement with the values extracted from the single-molecule experiments, further attempts are underway to study the effect of reductant and pH on the rates of internal ET. This is of interest, because the turnover rate is known to depend on pH.
In the present experiments, we find that β decreases with increasing substrate concentration (Fig. 4B) to reach a value of β = 0.6 at saturating nitrite concentrations. Using the approach of Lindsey and Patterson (27), this value of β corresponds to a distribution by which 80% of the decay times lie between 0.5·τ0 and 5·τ0, i.e., a spread by 1 order of magnitude. According to Eq. 2, at saturating substrate concentrations, the characteristic decay time of the autocorrelation function only depends on k2 and k3, i.e., on the rates of reduction of the types 1 and 2 sites. In the following, we discuss possible causes for the spread in these rates.
The overall structure of NiR appears remarkably stable, with little indication for conformational flexibility. X-ray diffraction data show no regions in the structure with conspicuously high-temperature factors (12, 32).** The structures of reduced, oxidized, and substrate- and product-bound NiR are virtually superimposable. The structures of NiRs from various sources have been reported, and the Cα rmsds between the various structures are small (33). There is no reason, therefore, to suspect there are variations in the gross structural features of the enzyme that might be connected with the observed rate distribution. Instead, we focus on the types 1 and 2 Cu sites.
Two histidines, a cysteine, and a methionine provide the Cu ligands in the type 1 site. In the structure of Rhodobacter sphaeroides 2.4.3 NiR, this methionine is partially disordered (33). In all NiRs studied to date, the type 2 Cu is immobilized in the protein framework by three histidines. When this site is oxidized, its fourth coordination position is occupied by a water molecule or a hydroxide ion at different positions, depending on pH (34). When the site is reduced, this fourth ligand disorders (18, 33). The reduction of the nitrite requires the uptake of two protons, in the delivery of which two residues close to the type 2 site play a crucial role, Asp-98 and His-255 (A. faecalis numbering) (35). They are part of a hydrogen-bonding network involving a number of water molecules, which are likely to play a role in the transport of the protons. [A second network of water molecules in the vicinity of the active site has been identified, but its function is uncertain (36).] The water molecules exhibit partial disorder, as does His-255.†† The relatively large B factor of the Asp-98 side chain points to an increased mobility of this residue (18, 35). The conclusion must be that, at the catalytic heart of the enzyme (i.e., the types 1 and 2 Cu sites), there is partial disorder in the first (types 1 and 2) and the second (type 2) coordination shells of the metals.
Structural and conformational variations in the first and second coordination sphere of a metal are known to cause variations in midpoint potential (34). They can easily amount to plus or minus 50 mV, in line with recent observations on the range of midpoint potentials observed by FRET detected cyclic voltammetry on small clusters of azurin (34, 37). With a typical value for the reorganization energy, λ, of 700 mV and a driving force ΔG ≪ λ, a 100-mV variation in midpoint potential will affect the rate of electron transfer from the type 1 to the type 2 site by a factor of ≈2, as calculated from Marcus' theory. An effect of similar magnitude will arise from a variation of 100 mV in λ. Thus, variations in midpoint potential and reorganization energy caused by structural variations in the types 1 and 2 sites may, at least in part, account for the observed distribution in characteristic times.
The technique described in the present work is based not on the intrinsic fluorescence of the protein or a fluorogenic substrate but on the fluorescence of a label, the emission of which depends on the absorption properties of the enzyme cofactors. The method can dramatically increase the range of redox enzymes for which the turnover can be studied by single-molecule fluorescence methods. It can be potentially used for any enzyme for which the absorption significantly changes during its catalytic cycle.
However, the method may have some drawbacks as well. First, with methods that rely on the fluorescence of a fluorogenic substrate, bleaching of the fluorophore will in general be much less of a problem than when a label is used. We have been fortunate that this complication disappeared to a large extent by working under turnover conditions, but bleaching remains a technical challenge. Second, the limited contrast between on and off states has precluded the thresholding of these states, leading to loss of valuable information. The contrast may be improved by increasing the intensity of the excitation source, although this increases the risk of bleaching. Alternatively, the contrast may be improved first by a judicious choice of the label, i.e., one that optimizes the redox state-dependent difference in spectral overlap between label and cofactor and second, by optimizing the position of the label with respect to the cofactor, taking into account the Förster radii of the on and off states. This remains to be tested by future experiments.
Methods
Protein Labeling and Immobilization.
The L93C and L93C/H145A variants of nitrite reductase from A. faecalis S-6 were expressed and purified as described (17, 38). Both L93C and L93C/H145A NiR mutants were labeled with ATTO 655 succinimidyl ester (ATTO-TEC Biolabeling and Ultraanalytics) on the N terminus using a 5× molar excess of dye over the protein and purified by using Centrispin-10 size-exclusion chromatography spin columns with a 5-kDa cutoff (Princeton Separations) (10, 11). The degree of labeling has been quantified taking ε665 = 125 mM−1·cm−1 for ATTO 655 and ε280 = 46 mM−1·cm−1 per monomer for both nitrite reductase mutants (17). The pH was chosen to favor labeling of the N terminus over lysine labeling (10). The conditions were checked for the case of azurin, for which it was confirmed by electrospray MS that the label was exclusively present on the N terminus (10).
Before modification of the N-terminally labeled L93C NiR with 1–11-bis-male-imidotetraethyleneglycol [BM(PEO)3, Pierce], all of the possible disulfide bridges between the introduced cysteines were reduced by incubating the sample solution for 1 hour with a 10× molar excess of dithiothreitol, which was then removed by using a Centrispin-10 size-exclusion column. The BM(PEO)3 linker was added in a 25-fold excess over the protein, followed by a reaction time of 2 h before a second size-exclusion step. After, the purification the modified enzyme was put immediately on the surface of the glass coverslip to avoid possible oxidation of the free maleimide groups.
Glass cover slips (Menzel-Glaeser) with 24-mm diameter were sonicated in acetone, 10% NaOH/H2O (2×), water (2×), and stored in methanol. The cover slips were then ozone-cleaned (UVP PR-100 UV-ozone photoreactor) for ≈1 h immediately before silanization. The cleaned cover slips were sonicated for 30 min in 0.1% 3-mercaptopropyl-trimethoxysilane (MPTS), 10% triethoxysilane (TES) (Fluka) solution in toluene, rinsed with toluene, and dried in a nitrogen flow. A 10-nM solution of the labeled and BM(PEO)3-modified enzyme in 20 mM Hepes, pH 7.0, was deposited onto the glass cover slips and incubated overnight at 4°C. Before measurements, the protein solution was rinsed off with water, and a fresh buffer solution was put on. Reduction agents and substrate were added from stock solutions to final concentrations of 10 mM sodium ascorbate/1 nM PES and 5 μM, 50 μM, 500 μM, or 5 mM sodium nitrite. The number of enzyme molecules exhibiting switching varied with sample preparation; the data reported here were taken from samples where an estimated 60–80% of the fluorescing molecules exhibited switching.
Fluorescence Measurements in Bulk.
Fluorescence time courses were measured on a Cary Eclipse fluorimeter with λex = 665 nm and λem = 685 nm. The enzyme concentration was 100 nM. All of the other experimental conditions were the same as in the single-molecule experiments. First 5 mM NaNO2 and then 10 mM sodium ascorbate/1 nM PES were added to the protein sample, and the steady-state fluorescence intensity was recorded. The fluorescence intensities corresponding to fully reduced and fully oxidized NiR were determined by reducing the same concentration of labeled NiR by excess ascorbate/PES and oxidizing it by ferrocyanide, respectively.
Enzyme Activity Assay in Bulk.
The rates of NO formation by labeled enzyme were measured in bulk in an anaerobic 20 mM Hepes solution at pH 7.0 by using a 10 mM ascorbate/1 nM PES mixture and 30 nM of enzyme. The concentration of sodium nitrite was varied from 5.4 μM to 4.4 mM. The production of NO was monitored by using a Clark-type electrode (39). It was checked that the labeling did not significantly affect the enzymatic activity by assaying the activities of labeled and unlabeled enzyme in separate experiments. This was confirmed in a second assay by which methylviologen (MV) was used as an electron donor, and the consumption of the MV was followed optically.
Single-Molecule Fluorescence Experiments.
Laser light [Coherent CR599 Dye laser (Rhodamine 6G), 160 mW, 620 nm, pumped by a 5-W Spectra-Physics Millennia Pro] was coupled into a single-mode optical fiber, reflected by a dichroic beam splitter (FT 645), and focused on the sample by an oil-immersion ×100 objective (Zeiss, N.A. = 1.30) mounted on a Zeiss Axiovert 200 inverted microscope. The power density at the sample was 0.5–1 kW·cm−2. Fluorescence emission emerging from the focal volume was collected through the same objective, passed through the beam splitter, filtered through a red cutoff filter, directed through a 50-μm pinhole, and finally focused on an avalanche photodiode (Perkin–Elmer SPCM-AQR-14) connected to a National Instruments PCI-6036E data acquisition card operating at 20 MHz. Samples were mounted onto a Physik Instrumente P-517.2 CL nanopositioner (9). Sample movement and data collection and analysis were controlled by a LabView program.
Data Processing.
The binning, histogram calculations, histogram fitting with Poissonian distributions, and autocorrelation calculations were done by using a home-developed LabView program. For the calculation of the autocorrelation functions, various bin sizes were tried, including a calculation based on the photon arrival times (40, 41). No strong dependence of the results on the bin size or the use of the photon arrival times was observed (SI Appendix, SI Fig. 9). The bin size used eventually for the autocorrelations was 1 and 10 ms for other calculations. The autocorrelation functions were fitted with stretched exponential functions by using Origin 7.5 (OriginLab).
Supplementary Material
ACKNOWLEDGMENTS.
We are grateful to Dr. H. Wijma for useful discussions about the enzymology of NiR; to M. Stampraad and Prof. S. de Vries (Delft Technical University, Delft, The Netherlands) for their help with the NiR assay; and to Dr. A. W. J. W. Tepper for discussions. This work is supported by the Stichting voor Fundamenteel Onderzoek der Materie (FOM) and the Stichting Aard- en Levenswetenschappen (Project ALW 805.47.105) and by the European Community through the EdRox network under contract no. MRTN-CT-2006-035649.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707736105/DC1.
Attempts to observe the fluorescence of single molecules in the absence of nitrite and the presence of reductant so far have failed, due to instability of the response, possibly caused by reductive photochemistry.
Photoinduced electron transfer from tryptophans in the protein to the label can be excluded as a possible cause for fluorescence intensity fluctuations, because such fluctuations are observed neither for active NiR in the absence of turnover (no reductant, only nitrite present in solution) nor for inactive H145A NiR.
The crystallographic temperature factors or B factors are a measure for the accuracy by which the atomic coordinates in a crystal structure have been determined. There are two main contributions to the B factor: increased mobility and static disorder (see also ref. 34).
Partial disorder of water molecules means that in the electron density map, weak electron density is observed at some points that may correspond to water molecules. The water molecules occupy these positions only partially, which is why the density is weak. See also ref. 34.
References
- 1.Nie SM, Zare RN. Optical detection of single molecules. Annu Rev Biophys Biom. 1997;26:567–596. doi: 10.1146/annurev.biophys.26.1.567. [DOI] [PubMed] [Google Scholar]
- 2.Edman L, Foldes-Papp Z, Wennmalm S, Rigler R. The fluctuating enzyme: a single molecule approach. Chem Phys. 1999;247:11–22. [Google Scholar]
- 3.Shi J, Dertouzos J, Gafni A, Steel D, Palfey BA. Single-molecule kinetics reveals signatures of half-sites reactivity in dihydroorotate dehydrogenase A catalysis. Proc Natl Acad Sci USA. 2006;103:5775–5780. doi: 10.1073/pnas.0510482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, et al. Protein conformational dynamics probed by single-molecule electron transfer. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 5.Xie SN. Single-molecule approach to enzymology. Single Mol. 2001;2:229–236. [Google Scholar]
- 6.English BP, et al. Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 7.Flomenbom O, et al. Stretched exponential decay and correlations in the catalytic activity of fluctuating single lipase molecules. Proc Natl Acad Sci USA. 2005;102:2368–2372. doi: 10.1073/pnas.0409039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velonia K, et al. Single-enzyme kinetics of CALB-catalyzed hydrolysis. Angew Chem Int Edit. 2005;44:560–564. doi: 10.1002/anie.200460625. [DOI] [PubMed] [Google Scholar]
- 9.Engelkamp H, et al. Do enzymes sleep and work? Chem Commun. 2006:935–940. doi: 10.1039/b516013h. [DOI] [PubMed] [Google Scholar]
- 10.Kuznetsova S, et al. A Forster-resonance-energy transfer-based method for fluorescence detection of the protein redox state. Anal Biochem. 2006;350:52–60. doi: 10.1016/j.ab.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 11.Schmauder R, et al. Sensitive detection of the redox state of copper proteins using fluorescence. J Biol Inorg Chem. 2005;10:683–687. doi: 10.1007/s00775-005-0020-6. [DOI] [PubMed] [Google Scholar]
- 12.Adman ET, Murphy MEP. In: Handbook of Metalloproteins. Messerschmidt A, Huber R, Poulos T, Wieghardt K, editors. Chichester, UK: Wiley; 2001. pp. 1381–1390. [Google Scholar]
- 13.Farver O, Eady RR, Abraham ZHL, Pecht I. The intramolecular electron transfer between copper sites of nitrite reductase: a comparison with ascorbate oxidase. Febs Lett. 1998;436:239–242. doi: 10.1016/s0014-5793(98)01120-x. [DOI] [PubMed] [Google Scholar]
- 14.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol R. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham ZHL, Smith BE, Howes BD, Lowe DJ, Eady RR. pH-dependence for binding a single nitrite ion to each type 2 copper centre in the copper-containing nitrite reductase of Alcaligenes xylosoxidans. Biochem J. 1997;324:511–516. doi: 10.1042/bj3240511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wijma HJ, Jeuken LJC, Verbeet MP, Armstrong FA, Canters GW. A random-sequential mechanism for nitrite binding and active site reduction in copper-containing nitrite reductase. J Biol Chem. 2006;281:16340–16346. doi: 10.1074/jbc.M601610200. [DOI] [PubMed] [Google Scholar]
- 17.Wijma HJ, et al. Reconstitution of the type 1 active site of the H145G/A variants of nitrite reductase by ligand insertion. Biochemistry US. 2003;42:4075–4083. doi: 10.1021/bi027270+. [DOI] [PubMed] [Google Scholar]
- 18.Murphy MEP, Turley S, Adman ET. Structure of nitrite bound to copper-containing nitrite reductase from Alcaligenes faecalis - Mechanistic implications. J Biol Chem. 1997;272:28455–28460. doi: 10.1074/jbc.272.45.28455. [DOI] [PubMed] [Google Scholar]
- 19.Kashem MA, Dunford HB, Liu MY, Payne WJ, Legall J. Kinetic-Studies of the Copper Nitrite Reductase from Achromobacter-Cycloclastes and Its Interaction with A Blue Copper Protein. Biochem Biophys Res Commun. 1987;145:563–568. doi: 10.1016/0006-291x(87)91357-x. [DOI] [PubMed] [Google Scholar]
- 20.Kakutani T, Watanabe H, Arima K, Beppu T. A Blue Protein As An Inactivating Factor for Nitrite Reductase from Alcaligenes-Faecalis Strain S-6. J Biochem. 1981;89:463–472. doi: 10.1093/oxfordjournals.jbchem.a133221. [DOI] [PubMed] [Google Scholar]
- 21.Buschmann V, Weston KD, Sauer M. Spectroscopic study and evaluation of red-absorbing fluorescent dyes. Bioconjugate Chem. 2003;14:195–204. doi: 10.1021/bc025600x. [DOI] [PubMed] [Google Scholar]
- 22.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 23.Tinnefeld P, Buschmann V, Weston KD, Sauer M. Direct observation of collective blinking and energy transfer in a bichromophoric system. J Phys Chem A. 2003;107:323–327. [Google Scholar]
- 24.Yeow EKL, Melnikov SM, Bell TDM, De Schryver FC, Hofkens J. Characterizing the fluorescence intermittency and photobleaching kinetics of dye molecules immobilized on a glass surface. J Phys Chem A. 2006;110:1726–1734. doi: 10.1021/jp055496r. [DOI] [PubMed] [Google Scholar]
- 25.Impagliazzo A, Ubbink M. H-1, C-13 and N-15 resonance assignment of Cu(I)-pseudoazurin from Alcaligenes faecalis S-6. J Biomol NMR. 2004;29:541–542. doi: 10.1023/B:JNMR.0000034341.96429.c1. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez F, Alegria A, Colmenero J. Relationship Between the Time-Domain Kohlrausch-Williams-Watts and Frequency-Domain Havriliak-Negami Relaxation Functions. Phys Rev B. 1991;44:7306–7312. doi: 10.1103/physrevb.44.7306. [DOI] [PubMed] [Google Scholar]
- 27.Lindsey CP, Patterson GD. Detailed Comparison of the Williams-Watts and Cole-Davidson Functions. J Chem Phys. 1980;73:3348–3357. [Google Scholar]
- 28.Qian H, Elson EL. Single-molecule enzymology: stochastic Michaelis-Menten kinetics. Biophys Chem. 2002;101:565–576. doi: 10.1016/s0301-4622(02)00145-x. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki S, et al. Spectroscopic characterization and intramolecular electron transfer processes of native and type 2 Cu-depleted nitrite reductases. J Biol Inorg Chem. 1997;2:265–274. [Google Scholar]
- 30.Wijma HJ, Canters GW, de Vries S, Verbeet MP. Bidirectional catalysis by copper-containing nitrite reductase. Biochemistry US. 2004;43:10467–10474. doi: 10.1021/bi0496687. [DOI] [PubMed] [Google Scholar]
- 31.Chi QJ, Zhang JD, Jensen PS, Christensen HEM, Ulstrup J. Long-range interfacial electron transfer of metalloproteins based on molecular wiring assemblies. Faraday Discuss. 2006;131:181–195. doi: 10.1039/b506136a. [DOI] [PubMed] [Google Scholar]
- 32.Drenth J. Principles of Protein X-Ray Crystallography. 2nd Ed. Heidelberg: Springer; 2003. [Google Scholar]
- 33.Jacobson F, et al. Structures of the oxidized and reduced forms of nitrite reductase from Rhodobacter sphaeroides 2.4.3 at high pH: changes in the interactions of the type 2 copper. Acta Crystallogr D. 2005;61:1190–1198. doi: 10.1107/S0907444905017488. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson F, et al. pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase. J Biol Chem. 2007;282:6347–6355. doi: 10.1074/jbc.M605746200. [DOI] [PubMed] [Google Scholar]
- 35.Adman ET, Godden JW, Turley S. The structure of copper-nitrite reductase from achromobacter cycloclastes at 5 Ph values, with NO2-bound and with type-II copper depleted. J Biol Chem. 1995;270:27458–27474. doi: 10.1074/jbc.270.46.27458. [DOI] [PubMed] [Google Scholar]
- 36.Ellis MJ, et al. X-ray structure of a blue copper nitrite reductase at high pH and in copper-free form at 1.9 angstrom resolution. Acta Crystallogr D. 2001;57:1110–1118. doi: 10.1107/s0907444901008654. [DOI] [PubMed] [Google Scholar]
- 37.Davis JJ, et al. Monitoring interfacial bioelectrochemistry using a FRET switch. J Phys Chem B. 2006;110:20649–20654. doi: 10.1021/jp0630525. [DOI] [PubMed] [Google Scholar]
- 38.Astier Y, et al. A systematic study of the influence of peptide modification of a gold electrode on the cyclic voltammetry of pseudoazurin from Alcaligenes faecalis strain S-6. Electroanalysis. 2004;16:1155–1165. [Google Scholar]
- 39.Girsch P, de Vries S. Purification and initial kinetic and spectroscopic characterization of NO reductase from Paracoccus denitrificans. BBA Bioenergetics. 1997;1318:202–216. doi: 10.1016/s0005-2728(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 40.Wahl M, Gregor I, Patting M, Enderlein J. Fast calculation of fluorescence correlation data with asynchronous time-correlated single-photon counting. Opt Express. 2003;11:3583–3591. doi: 10.1364/oe.11.003583. [DOI] [PubMed] [Google Scholar]
- 41.Laurence TA, Fore S, Huser T. Fast, flexible algorithm for calculating photon correlations. Opt Lett. 2006;31:829–831. doi: 10.1364/ol.31.000829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.