Abstract
Controversy regarding genetically modified (GM) plants and their potential impact on human health contrasts with the tacit acceptance of other plants that were also modified, but not considered as GM products (e.g., varieties raised through conventional breeding such as mutagenesis). What is beyond the phenotype of these improved plants? Should mutagenized plants be treated differently from transgenics? We have evaluated the extent of transcriptome modification occurring during rice improvement through transgenesis versus mutation breeding. We used oligonucleotide microarrays to analyze gene expression in four different pools of four types of rice plants and respective controls: (i) a γ-irradiated stable mutant, (ii) the M1 generation of a 100-Gy γ-irradiated plant, (iii) a stable transgenic plant obtained for production of an anticancer antibody, and (iv) the T1 generation of a transgenic plant produced aiming for abiotic stress improvement, and all of the unmodified original genotypes as controls. We found that the improvement of a plant variety through the acquisition of a new desired trait, using either mutagenesis or transgenesis, may cause stress and thus lead to an altered expression of untargeted genes. In all of the cases studied, the observed alteration was more extensive in mutagenized than in transgenic plants. We propose that the safety assessment of improved plant varieties should be carried out on a case-by-case basis and not simply restricted to foods obtained through genetic engineering.
Keywords: food safety evaluation, rice, genetically modified organisms, genetic engineering, γ-irradiation
Plant breeding started thousands of years ago, through the unconscious selection of seeds from plants with higher quality and productivity. After sexual plant reproduction was discovered, in the 17th century, people started to use deliberate interbreeding (crossing) of closely or distantly related species to produce new crops with desirable properties (1). With the discovery, in the beginning of the 20th century, that x-rays induced mutations in the fruit fly Drosophila melanogaster and barley, plant breeders and geneticists started to use mutagenesis to rapidly create and increase variability in crop species and ultimately change plant traits. The high efficiency of classical mutagenesis has been widely documented (2), and its global impact for crop improvement has also been evaluated (3). Since the establishment of the joint Food and Agriculture Organization/International Atomic Energy Agency, Division of the Nuclear Techniques in Agriculture (www-infocris.iaea.org/MVD), 1,916 crop and legume varieties were released worldwide (40% γ-irradiated).
Since the 1970s, advances in molecular biology have provided the basis for the development of genetic engineering, leading to the next level of genetic gain in crop cultivars. This technology permits the identification, isolation, and transfer of a gene of interest, originated from any type of organism, to plant cells. Transformed plants are then regenerated from these cells through tissue culture (4).
Contrasting with the readily acceptance of food products obtained through conventional plant breeding, the potential benefits of this new technology have been held largely at bay because of the enormous controversy regarding the food safety of the resulting products (5).
Despite the lack of universal methods for evaluating the potentially hazardous effects of genetic modification, Food and Agriculture Organization and the European Food Safety Authority recommendations call for targeted approaches to evaluate macro-, micro-, and anti-nutrients, toxins, allergens, and secondary metabolites. To increase the chances of detecting unintended effects, some molecular profiling methods have also been proposed (6). One of the mentioned profiling techniques is microarrays. This technology allows for monitoring the expression of thousands of genes simultaneously.
In this study, we used expression microarray analyses to monitor the extension of unexpected transcriptome modifications obtained in rice by conventional plant breeding by γ-irradiation as compared with the ones obtained through genetic engineering. We have analyzed four rice lines (two mutagenized and two transgenic ones) and further compared the stable lines against the recently modified ones.
Results and Discussion
Differentially Expressed Genes Increase with Genetic Instability and from Transgenic to Mutant Lines.
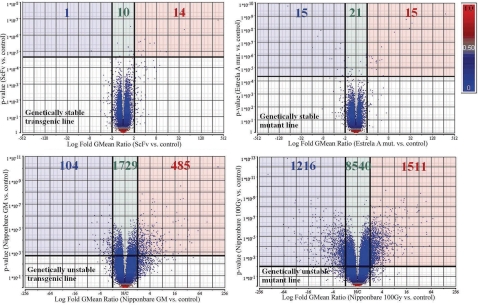
Hierarchical clustering (Fig. 1) of the microarray data of transgenic, mutagenized, and control plants showed that duplicate samples always grouped together and modified genotypes always grouped with the respective unmodified controls [see supporting information (SI) Fig. 3 for Pearson's correlation between samples]. Despite the different type of breeding strategy used, genetically stable samples [transgenic single-chain variable fragment (ScFv) and mutant Estrela A] are more closely grouped with their corresponding controls than nonstable ones. Additionally, in nonstable lines, transgenic Nipponbare [Nip. genetically modified (GM)] is more closely related to its control than the line obtained through 100-Gy γ-irradiation. As visible in volcano plots (Fig. 2), 11,267 genes showed differential expression in the nonstable mutagenized rice line, whereas only 2,318 genes were detected in the nonstable transgenic line (despite the inserted gene being a transcription factor). The number of affected genes was strongly reduced in stable lines (to 51 in the mutant and 25 in the transgenic).
Fig. 1.
Plant material used and hierarchical clustering dendrogram of the different samples.
Fig. 2.
Volcano plots for differentially expressed genes. Differentially expressed genes appear above the thick horizontal lines. Genes induced >2-fold are on the right of the right vertical lines, and the ones repressed >2-fold are on the left of the left vertical line. The numbers corresponding to the differentially expressed genes induced >2-fold for each experiment (red-shadowed area) are red, and those corresponding to the genes repressed >2-fold (blue-shadowed area) are blue. The green-shadowed area corresponds to differentially expressed genes that were up- or down-regulated <2-fold (green-colored numbers). Blue-colored genes are those with P between 0 and 0.5, and red-colored genes are those with P between 0.5 and 1.
The Analyzed Breeding Strategies Cause Stress, and Plants Respond to It by Modifying Transcription for Several Generations.
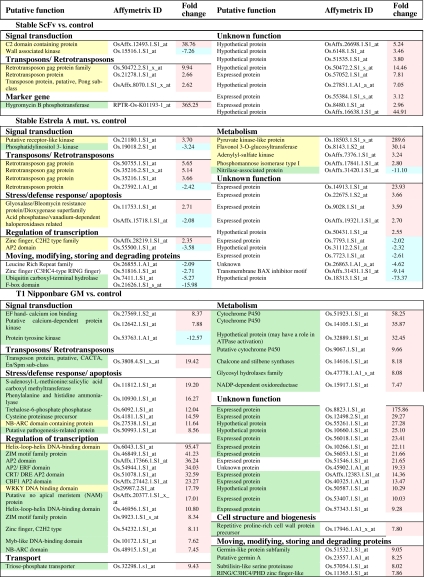
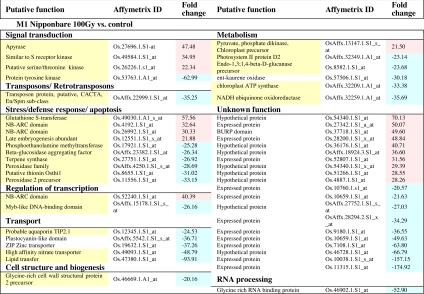
The list of the differentially expressed genes with a cut-off P < 0.05 and with >2-fold change (>2 or <−2) after Log2 transformation (identified as high fold change) is shown in Table 1. For nonstable lines, only the top 50 differentially expressed genes are presented (Table 1). For those two experiments we also present a pie chart with all of the differentially expressed genes with a cut-off P < 0.05 and high fold change (we only considered genes whose function could be retrieved) separated by functional categories (SI Fig. 4). The genes listed in Table 1 were identified and analyzed for their functions by using Affymetrix, TIGR rice genome annotation, National Center for Biotechnology Information, UniProt, and Pfam internet resources. We found that in all of the experiments, the acquisition of the desired traits is accompanied by modifications in transcript levels of untargeted stress-related genes (genes whose altered transcription cannot be directly related with the introduced transgenes or desired traits are yellow-shadowed in Table 1).
Table 1.
Significantly induced and repressed genes (fold change > 2 or < −2) for each experiment
For T1 Nipponbare GM vs. control and M1 Nipponbare 100 Gy vs. control, only the top 50 differentially expressed genes are presented. Yellow shading indicates the genes are directly or indirectly related with stress response. Green shading indicates the genes' altered expression can be associated with the introduced genes or desired traits. Red shading indicates up-regulated genes, and blue shading indicates down-regulated genes.
We have also verified that the stressing event is memorized along several generations, although with a decreasing impact in the number of altered transcripts in each new generation (Table 1). This phenomenon of transgeneration memory of stress could be possibly attributed to epigenetic mechanisms and has been reported by others (7).
Although a complete understanding of plant stress response is far from being reached, various papers reporting molecular and biochemical studies suggest the involvement of at least six classes of genes (a–f): class a, genes implicated in stress/defense signaling-signal perception (several types of receptor-like protein kinases, two-component histidine kinases, G protein-coupled receptors, Ca2+-releasing modules), and signal transduction (protein kinases, protein phosphatases, MAP kinases) (8–10); class b, second messengers, such as reactive oxygen species (ROS), salicylic acid (SA), jasmonic acid (JA), and ethylene, which are involved in the regulatory pathways (11); class c, genes implicated in stress response–ROS network (GST, peroxidases) (12) and the systemic acquired resistance (SAR) response (pathogenesis-related genes) (13); class d, genes implicated in protein modification (methylation, isoprenylation, lipidation, ubiquitination) and scaffolds or adapters [these molecules regulate the activity of stress signaling components (8)]; class e, genes encoding transcription factors that are involved in the temporal and spatial regulation of specific stress genes (14); and class f, genes encoding retrotransposons that represent sensitive markers of plant stress (15).
Genes Whose Altered Transcription Could Be Directly Related to the Introduced Genes or Desired Traits.
Some of the differentially expressed genes found in Table 1 can be directly associated with the transgenes introduced or with the desired new characteristics of the modified plant (green shadowed). One example of these differentially expressed genes is the hygromycin B phosphotransferase gene, used as a marker gene in the ScFv stable transgenic line (Table 1).
Some of the differentially expressed genes found in the stable Estrela A line (Table 1) can be eventually related to a reduced indole-3-acetic acid (IAA) content, because the down-regulation of a nitrilase-associated protein was observed in the mutant/dwarfed line (SI Fig. 5A). Nitrilases are key enzymes in the biosynthesis of the plant hormone IAA (16), which belongs to the auxin class of plant growth regulators. The enzyme phosphatidylinositol 3-kinase, found in the signal transduction functional group, can also be related to this putative reduced IAA content because the phosphatidylinositol signaling pathway is also involved in plant responses to hormones, like auxins (17). We also found, in this experiment, a group of genes implicated in protein modifications whose altered transcription can be related to the hypothetical reduced IAA content. This group consisted of two proteins involved in ubiquitination: one F-box domain-containing protein and the ubiquitin carboxyl-terminal hidrolase. F-box proteins act as adaptor components of the modular E3 ubiquitin ligase SKP1-CUL1-F-box protein (SCF) complex that functions in phosphorylation-mediated ubiquitination. Protein ubiquitination is a precise strategy for regulating gene function, driving tagged proteins for degradation via the proteasome, and it is suggested as an important control system in desiccation tolerance (18). The down-regulation of these two proteins could be explained by the decreased auxin content because auxin regulates transcription by promoting the degradation of a family of transcriptional repressors known as Aux/IAA proteins, this degradation depending on a ubiquitin protein ligase named SCF(TIR1). In the presence of auxin, the F-box protein TIR1 binds to the Aux/IAA proteins, resulting in their ubiquitination and consequent degradation (19).
The unstable transgenic line Nipponbare GM contains one copy of the barley CBF1 gene (BCBF1). C-repeat binding factors (CBFs) specifically interact with the cis-acting dehydration-responsive element-DRE (core motif:G/ACCGAC) and control the expression of many stress-inducible genes (20). Although BCBF1 gene is under the control of a stress-inducible promotor (AtRD29A), preliminary experimental results obtained within our team (unpublished data) reveal a leaky expression of the BCBF1 gene in rice, even in the absence of stress conditions. For this reason, in this particular case, the differential expression of the stress-related genes found in our experiments may be either caused by the stress imposed by the Agrobacterium-mediated genetic modification or, at least in part, by the introduced BCBF1 transcription factor. To clarify this point we decided to analyze the promoter (2 kb upstream of the ATG start codon) of the top 50 differentially expressed genes to search for DRE core motifs. From this study we found that almost all of the top 50 genes (90%) contain several DRE core motifs in their promoter regions (green shadowed in Table 1). Therefore, it seems that the differential expression of these genes may be related mainly to the specific transgene integrated. This result highlights the importance of carefully studying transformants carrying inserted genes coding for transcription factors.
Genes Implicated in Stress/Defense Signaling (Class A).
All of the differentially expressed genes found in the signal transduction category, and not related to the transgene's introduction or desired traits, could be related with stress/defense. Thus, in Table 1 we observe in this functional group a wall-associated kinase and a C2 domain-containing protein. In plants, many protein kinases and phosphatases are involved in environmental stress responses (8–10). The C2 domain is a Ca2+-dependent membrane-targeting module found in many cellular proteins involved in signal transduction or membrane trafficking and thought to be involved in binding calcium-dependent phospholipids (21). This domain has been correlated with stress signaling (22). In the stable mutagenized line we found two signal transduction-associated proteins, both also already characterized as stress/defense associated (Table 1): a receptor-like kinase (9, 10) and a phosphatidylinositol 3-kinase (17). Finally, concerning the unstable mutagenized line (Table 1) we found, in this category, an apyrase (23), a S receptor kinase (9, 10), a putative serine/threonine kinase (24), and a protein tyrosine kinase (25).
Genes Implicated in Stress/Defense/Apoptosis (Class C).
In this functional group we found four genes potentially involved in the ROS network, (one GST and three peroxidases) (12), three NB–ARC domain-containing proteins (26), one glyoxalase (27), one late embryogenesis abundant protein (28), one phosphoethanolamine methyltransferase (29), one β-glucosidase (30), one terpene synthase (31), and one putative thionin (32).
Genes Implicated in the Regulation of Transcription (Class E).
All of the differentially expressed genes found in this category, and not related with the transgenes' introduction or desired traits, could also be related with stress/defense. Thus, we found one AP2 domain, one zinc finger of the C2H2 type family, one WRKY DNA binding domain, a helix–loop–helix DNA-binding domain, a NB-ARC domain, and a Myb-like motif. All of these domain-containing proteins were previously associated with stress response (10, 14, 26, 33).
Transposons/Retrotransposons (Class F).
All of the tested plants showed detected alteration in the transcription of genes encoding transposons/retrotransposons. As stated above, these genes are sensitive markers of plant stress (15).
Other Genes That Could also Be Indirectly Related to Stress.
We could find in Table 1 some genes whose altered expression can be also indirectly related to stress. Thus, concerning the stable Estrela A mutagenized line (Table 1) the up-regulation of adenylyl-sulfate kinase can be related to gluthatione-based detoxification of methylglyoxal because this enzyme is involved in the sulfate assimilation pathway required for glutathione production (34). The up-regulation of a putative flavonol 3-O-glucosyltransferase could also be related to stress. This enzyme catalyzes the transfer of glucose from UDP-glucose to a flavonol, one of the last steps in anthocyanin pigment biosynthesis. Anthocyanins are produced by various plants as a result of stress and in senescing foliage as a consequence of the autumn hostile environment (35). Finally, the up-regulation of both pyruvate kinase and phosphomanose isomerase may also be related to stress. Pyruvate kinase is involved in glycolysis, and phosphomanose isomerase catalyzes the interconversion of mannose-6-phosphate and fructose-6-phosphate, also a component of the glycolytic pathway. The stress induction of glycolysis transcripts has been reported in other studies (36). Regarding the unstable Nipponbare 100-Gy line (Table 1), we also found, in the different functional groups, some genes already associated with stress/defense responses, specifically to NaCl-stress response: several aquaporin and lipid transfer proteins (37), one high-affinity nitrate transporter (38), one glycine-rich cell wall protein (24), and one endo-1,3–1,4-β-d-glucanase (39). The altered expression of the photosynthesis-associated genes encoding photosystem II protein D2 and chloroplast ATP synthase is consistent with the already known effect of γ-irradiation on the photosynthetic activity (40). Pyruvate phosphate dikinase up-regulation, NADH ubiquinone oxidoreductase down-regulation, and down-regulation of the photosynthesis-related genes may be a response to oxidative stress and a way of limiting mitochondrial ROS production while keeping the electron transport chain relatively oxidized (41).
The pie charts we obtained for the genetically unstable lines (SI Fig. 4) are strikingly similar to the one obtained for Arabidopsis under various stress conditions (9). This similarity also supports our statement about the relation between genetic modification and stress response.
In conclusion, we have demonstrated that:
(i) DNA microarray technology should be considered as a powerful profiling tool for studying altered gene expression induced by different breeding strategies. However, changes in transcriptome do not necessarily correlate with risk. Proteomic studies should thus be performed to provide data on the nature of proteins.
(ii) Transcript profile of the stable lines was less altered than that of unstable ones and tested GM plants showed fewer genetic alterations than mutagenized ones. This last difference remains well known for the tested stable lines despite the higher number of self-pollinations for the mutant stable line as compared with the transgenic (10 vs. 3). Although these results may be specific to the particular mutagenized and transgenic plants examined here, they show that transgenic plants may have fewer changes than mutagenized ones.
(iii) The improvement of a plant variety through the acquisition of a new desired trait or modification of a previous one (either by genetic engineering or mutagenesis) causes stress and thus has a broad impact on gene expression.
(iv) Even several generations after the breeding event, the plant still maintains the “memory” of that incident and responds accordingly.
(v) Similar phenotypes do not obligatorily mean similar transcript profiles, which was evident for the unstable mutant line (SI Fig. 5B). However, we cannot rule out that under certain environmental conditions different morphology would not become evident.
Finally, we believe that safety assessment of improved plant varieties should be carried out on a case-by-case basis and not simply restricted to foods obtained through genetic engineering.
Materials and Methods
Plant Materials.
Two genetically stable Oryza sativa L. ssp. japonica lines: a γ-irradiated rice mutant (cv. Estrela A) and a well characterized transgenic rice line (cv. Bengal) were used as well as controls (Fig. 1). The stable mutant was obtained in 1988 by γ-irradiation, had already gone >10 generations of self-pollination, and had a mature average height ≈45 cm lower than the wild type (SI Fig. 5A). The stable transgenic line, which was already in the third generation of self-pollination after transformation, expresses a ScFV antibody (ScFvT84.66) against carcinoembryonic antigen, a well characterized tumor-associated marker antigen (42).
We have also used two genetically unstable rice lines: the M1 generation of a 100-Gy γ-irradiated line (98% survival after mutagenesis) and the T1 generation of an Agrobacterium-transformed transgenic line (both cv. Nipponbare) containing one copy of the BCBF1 gene driven by the AtRD29A promoter from Arabidopsis and one copy of the hpt II gene (Fig. 1). We used seeds from the same self-pollinated panicle for control and irradiation/transgenesis. The nonstable mutant line chosen for this experiment was the one showing a phenotype more similar to that of the nonirradiated control (SI Fig. 5B).
In the case of the transgenic lines, stability was based on the stable inheritance of the introduced transgenes in the homozygous progeny. Regarding the mutagenized plants we have defined as genetically stable plants those that, after mutagenesis, had already gone through several cycles of self-pollination while maintaining the desirable traits.
Seed Treatment and Seedling Growth.
Seeds were manually peeled and immersed for 30 min at 50°C in 0.1% Benlate (fungicide). After washing in distilled sterilized water, seeds were surface-disinfected with 70% (vol/vol) ethanol for 1 min and then with a solution of 2% sodium hypochloride with traces of Tween 20, for 30 min, at room temperature. After thorough washing with distilled sterile water seeds were kept overnight in the final wash and then soaked in Yoshida's medium (43) for germination in the dark for 2 days at 28°C. Seedlings were further grown at 28°C for 10 days under a 12-h photoperiod regime. Yoshida's medium used for the transgenic lines was supplemented with 30 mg/liter of hygromicin B. Twelve-day-old seedlings were frozen in liquid nitrogen and kept at −80°C until RNA extraction.
RNA Extraction and Microarrays.
Two pools of six whole seedlings were prepared for each condition under test, and RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) following the manufacturer's instructions. Total RNA was kept at −80°C and sent to the Affymetrix core facility (Instituto Gulbenkian de Ciência, Oeiras, Portugal), where quality-control analysis was carried out before cDNA synthesis from the mRNA [with appropriate oligo(dT) primers], labeling (through synthesis of cRNA with incorporation of biotinylated ribonucleotide analogs), and hybridization to the GeneChip Rice Genome Array (Affymetrix). This array contains probes to query 51,279 transcripts representing two rice subspecies (48,564 japonica transcripts and 1,260 transcripts of indica subspecies).
Data Analysis.
Microarrays data analysis was performed with Partek Genomics Suite software. Affymetrix CEL files were imported by using the Robust Multichip Average method, which involves four steps: background correction of the perfect match values, quintile normalization across all of the chips in the experiment, Log2 transformation, and median polish summarization. The logged data were used for hierarchical cluster analysis and statistical analysis. Hierarchical cluster analysis was performed by using Pearson's dissimilarity product moment correlation coefficient and Ward's algorithm.
For the identification of differentially expressed genes we used ANOVA and a false discovery rate with a 0.05 threshold.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Eva Stöger (Institute of Molecular Biotechnology, Aachen University, Aachen, Germany) for providing the ScFv transgenic line and respective control, Estação Agronómica Nacional (Oeiras, Portugal) for the Estrela A mutant line, Prof. Jayamani Palaniappan for suggesting this line for our studies, Dr. Jorg Becker for permission to use Partek Genomics Suite software and help with its use, and Prof. Margarida O. Krause for the revision of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707881105/DC1.
References
- 1.McCouch S. Diversifying selection in plant breeding. PLoS Biol. 2004;2:1507–1512. doi: 10.1371/journal.pbio.0020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahloowalia BS, Maluszynski M. Induced mutations: A new paradigm in plant breeding. Euphytica. 2001;118:167–173. [Google Scholar]
- 3.Ahloowalia BS, Maluszynski M, Nichterlein K. Global impact of mutation-derived varieties. Euphytica. 2004;135:187–204. [Google Scholar]
- 4.Goodman RM, Hauptli H, Crossway A, Knauf VC. Gene transfer in crop improvement. Science. 1987;236:48–54. doi: 10.1126/science.236.4797.48. [DOI] [PubMed] [Google Scholar]
- 5.Malarkey T. Human health concerns with GM crops. Mut Res. 2003;544:217–221. doi: 10.1016/j.mrrev.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Kuiper HA, Kok EJ, Engel K-H. Exploitation of molecular profiling techniques for GM food safety assessment. Curr Opin Biotechnol. 2003;14:238–243. doi: 10.1016/s0958-1669(03)00021-1. [DOI] [PubMed] [Google Scholar]
- 7.Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- 8.Xiong L, Zhu J-K. Abiotic stress signal transduction in plants: Molecular and genetic perspectives. Physiol Plant. 2001;112:152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- 9.Mahalingam R, et al. Characterizing the stress/defense transcriptome of Arabidopsis. Gen Biol. 2003;4:R20.1–R20.14. doi: 10.1186/gb-2003-4-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schenk PM, et al. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 12.Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 13.Ryals J, Uknes S, Ward E. Systemic acquired resistance. Plant Physiol. 1994;104:1109–1112. doi: 10.1104/pp.104.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 15.Grandbastien M-A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. [Google Scholar]
- 16.Bartling D, Seedorf M, Schmidt RC, Weiler EW. Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: Key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc Natl Acad Sci USA. 1994;91:6021–6025. doi: 10.1073/pnas.91.13.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin WH, Ye R, Ma H, Xu ZH, Xue HW. DNA chip-based expression profile analysis indicates involvement of the phosphatidylinositol signaling pathway in multiple plant responses to hormone and abiotic treatments. Cell Res. 2004;14:34–45. doi: 10.1038/sj.cr.7290200. [DOI] [PubMed] [Google Scholar]
- 18.O'Mahony PJ, Oliver MJ. The involvement of ubiquitin in vegetative desiccation tolerance. Plant Mol Biol. 1999;41:657–667. doi: 10.1023/a:1006330623364. [DOI] [PubMed] [Google Scholar]
- 19.Parry G, Estelle M. Auxin receptors: A new role for F-box proteins. Curr Opin Cell Biol. 2006;18:152–156. doi: 10.1016/j.ceb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Dubouzet JG, et al. OsDREB genes in rice, Oryza sativa L, encode transcription activators that function in drought-, high-salt-, and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- 21.Davletov BA, Sudhof TC. A single C2 domain from Synaptotagmin I is sufficient for high-affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 22.Katagiri T, Takahashi S, Shinozaki K. Involvement of a novel Arabidopsis phospholipase D AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signaling. Plant J. 2001;26:595–605. doi: 10.1046/j.1365-313x.2001.01060.x. [DOI] [PubMed] [Google Scholar]
- 23.Navarro-Gochicoa M-T, Camut S, Niebel A, Cullimore JV. Expression of the Apyrase-Like APY1 genes in roots of Medicago truncatula is induced rapidly and transiently by stress and not by Sinorhizobium meliloti or Nod factors. Plant Physiol. 2003;131:1124–1136. doi: 10.1104/pp.102.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walia H, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Q, Fu H-H, Gupta R, Luan S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell. 1998;10:849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Biezen EA, Jones JDG. The NB-ARC domain: A novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998;8:R226–R227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 27.Singla-Pareek SL, Reddy MK, Sopory SK. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA. 2003;100:14672–14677. doi: 10.1073/pnas.2034667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali-Benali MA, Alary R, Joudrier P, Gautier M-F. Comparative expression of five Lea genes during wheat seed development and in response to abiotic stress by real-time quantitative RT-PCR. Biochim Biophys Acta. 2005;1730:56–65. doi: 10.1016/j.bbaexp.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Mou Z, et al. Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell. 2002;14:2031–2043. doi: 10.1105/tpc.001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulton JE. Cyanogenesis in plants. Plant Physiol. 1990;94:401–405. doi: 10.1104/pp.94.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnee C, Köllner TG, Gershenzon J, Degenhardt J. The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-β-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol. 2002;130:2049–2060. doi: 10.1104/pp.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina A, Ahl-Goy P, Frail A, Sanchez-Monge R, Garcia-Olmedo F. Inhibition of bacterial and fungal plant pathogens by thionins of type I, II. Plant Sci. 1993;92:169–177. [Google Scholar]
- 33.Zhou J, et al. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol. 2007;63:591–608. doi: 10.1007/s11103-006-9111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leustek T. Sulfate metabolism. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: Am Soc Plant Biologists; 2002. pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoch WA, Zeldin EL, McCown BH. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol. 2001;21:1–8. doi: 10.1093/treephys/21.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002;130:1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y, Deyholos MK. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006;6:25. doi: 10.1186/1471-2229-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Q, et al. Cloning and expression study of a putative high-affinity nitrate transporter gene from Dunaliella salina. J Appl Phycol. 2004;16:395–400. [Google Scholar]
- 39.Gong Z, et al. Genes that are uniquely regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakashita T, et al. Gamma-irradiation effect: Variation of photosynthetic activity of Euglena. Biomed Environ Sci. 2002;15:261–267. [PubMed] [Google Scholar]
- 41.Moller IM. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- 42.Stöger E, et al. Cereal crops as viable production and storage systems for pharmaceutical ScFv antibodies. Plant Mol Biol. 2000;42:583–590. doi: 10.1023/a:1006301519427. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory Manual for Physiological Studies of Rice. Manila, Philippines: International Rice Research Institute; 1976. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.