Abstract
We previously reported the importance of the serum response factor (SRF) cofactor myocardin in controlling muscle gene expression as well as the fundamental role for the inflammatory transcription factor NF-κB in governing cellular fate. Inactivation of myocardin has been implicated in malignant tumor growth. However, the underlying mechanism of myocardin regulation of cellular growth remains unclear. Here we show that NF-κB(p65) represses myocardin activation of cardiac and smooth muscle genes in a CArG-box-dependent manner. Consistent with their functional interaction, p65 directly interacts with myocardin and inhibits the formation of the myocardin/SRF/CArG ternary complex in vitro and in vivo. Conversely, myocardin decreases p65-mediated target gene activation by interfering with p65 DNA binding and abrogates LPS-induced TNF-α expression. Importantly, myocardin inhibits cellular proliferation by interfering with NF-κB-dependent cell-cycle regulation. Cumulatively, these findings identify a function for myocardin as an SRF-independent transcriptional repressor and cell-cycle regulator and provide a molecular mechanism by which interaction between NF-κB and myocardin plays a central role in modulating cellular proliferation and differentiation.
Keywords: cell differentiation, serum response factor
Determining mechanisms that preferentially promote cardiomyocyte and vascular smooth muscle cell (VSMC) growth or differentiation remains an active challenge in cardiovascular biology (1). This process is regulated in part by serum response factor (SRF), a transcription factor that is widely expressed but enriched in muscle lineages during development (2). SRF binds to the CArG box (CC[A/T]6GG), a DNA consensus sequence located in the control regions of numerous growth factor-regulated and muscle-specific genes (3). Emerging evidence suggests that the regulation of SRF-driven gene transcription depends on its relationship with a wide variety of positive and negative cofactors (4, 5). Myocardin is expressed specifically in cardiac and smooth muscle cells and potently activates their gene expression by associating with SRF bound to CArG boxes (6). We have shown that the toggling of SRF toward growth-dependent or differentiation pathways is related to interplay between myocardin and another SRF-associated factor, Elk-1, in a manner where myocardin directly competes with Elk-1 for SRF binding and antagonizes proliferative signaling induced by PDGF (7). Although previous studies have shown that overexpression of myocardin can retard cell growth (8) and inactivation of myocardin promotes tumor growth (9), the molecular mechanism related to myocardin and cellular proliferation remains unknown.
The NF-κB family of transcription factors play key roles in regulating inflammatory and immune responses as well as cellular proliferation, differentiation, and survival (10, 11). Interactions between NF-κB dimers and other families of transcription factors have been widely reported, including that of SRF (10, 12). We have previously demonstrated that NF-κB(p65) can strongly induce cellular proliferation (13, 14). In light of our previous work detailing the effects of myocardin promoting cardiac and smooth muscle differentiation and p65 promoting cell growth, we hypothesized that p65 could inhibit myocardin-induced differentiation by interfering with its association with SRF. Conversely, we also wanted to determine the influence of myocardin as a transcriptional regulator of p65 and its downstream effect on cellular growth. Herein we describe a relationship between NF-κB and myocardin in regulating smooth muscle and cardiac gene expression and demonstrate a unique role for myocardin as a transcriptional repressor and inhibitor of cell proliferation.
Results
NF-κB Suppresses Myocardin Activation of Cardiac and Smooth Muscle Genes.
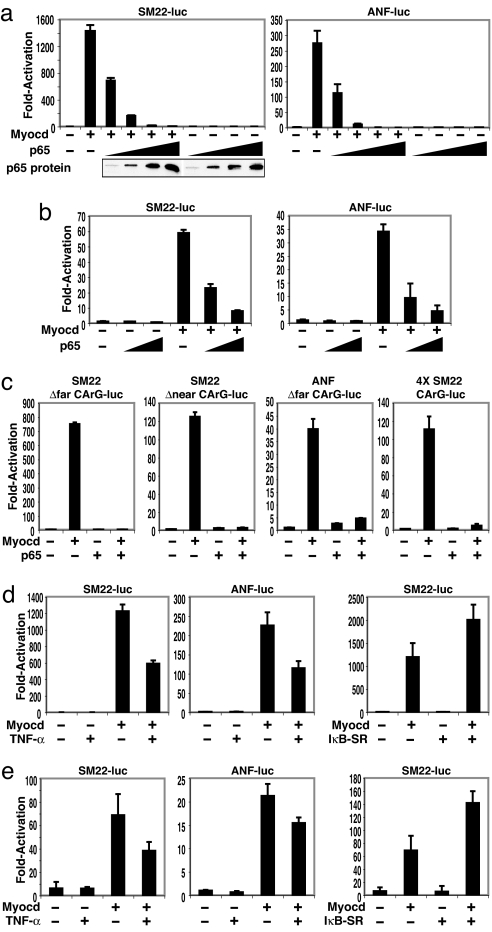
Smooth muscle 22 (SM22) and atrial natriuretic factor (ANF) promoters attached to luciferase reporters were transfected with myocardin and p65 expression plasmids in COS7 cells. Myocardin-induced transactivation of both SM22 and ANF was inhibited by cotransfection with p65 in a dose-dependent manner (Fig. 1a). Similar findings were also observed by using an α-MHC-luc reporter (data not shown) and corroborated in rat neonatal cardiomyocytes (Fig. 1b). Cotransfection of the p50 NF-κB subunit had no effect on myocardin transactivation (data not shown). Thus, p65 appears to functionally interfere with myocardin-dependent gene transcription.
Fig. 1.
Suppression of myocardin transcriptional activity by p65. Luciferase reporters controlled by SM22 and ANF were transfected into COS7 cells (a) or rat neonatal cardiomyocytes (b) and cotransfected with expression plasmids for myocardin (0.1 μg) and increasing amounts of p65 (COS7: 0.01, 0.05, 0.1, and 0.2 μg; cardiomyocytes: 0.02 and 0.1 μg). p65 protein assayed by Western blot. (c) COS7 cells were cotransfected with the indicated deleted CArG box within either the SM22 or ANF promoter. (d and e) COS7 cells (d) or cardiomyocytes (e) treated with TNF-α (10 ng/ml for 6 h) or cotransfected with an IκBα superrepressor. Values are the fold increase of luciferase activity relative to activation of the reporter alone of at least three experiments. Student's t test: P < 0.05, myocardin alone vs. myocardin plus p65, TNF-α, or IκBα-SR.
To investigate whether p65-mediated inhibition of myocardin transactivation is CArG-dependent, CArG-box mutation analysis of the SM22 promoter was performed. Although mutations of the distal CArG box (CArG-far) and the proximal CArG box (CArG-near) reduced responsiveness to myocardin (6), p65 nevertheless eliminated the ability of these mutant promoters to respond to myocardin (Fig. 1c). Similarly, p65 repressed myocardin activation on the ANF promoter with CArG-far box mutation. Next, we tested whether the CArG box is sufficient to mediate p65 repression of myocardin transactivity by using a luciferase reporter driven by four tandem CArG boxes derived from SM22 or c-fos promoters. Indeed, p65 inhibited myocardin activity on both CArG-box reporters (Fig. 1c and data not shown). Together, these data suggest that the CArG box is required for p65 to repress myocardin transactivation.
We have previously demonstrated that TNF-α is a potent mitogen for vascular smooth muscle and that this effect is NF-κB-dependent (13–16). Indeed, TNF-α repressed myocardin activation of SM22 and ANF reporters in both COS cells (Fig. 1d) and cardiomyocytes (Fig. 1e). Next, we examined the effect of blocking endogenous NF-κB on myocardin transactivity. Cotransfection with an inhibitory κBα superrepressor (IκBα-SR), a potent inhibitor of NF-κB activity (11), significantly enhanced myocardin activity for SM22 (Fig. 1 d and e) and ANF (data not shown). Together, these data demonstrate that TNF-α/NF-κB signaling is both necessary and sufficient to modulate myocardin transactivity in activation of cardiac and smooth muscle gene expression.
NF-κB Inhibits Myocardin Transactivity by Physically Interacting with Myocardin.
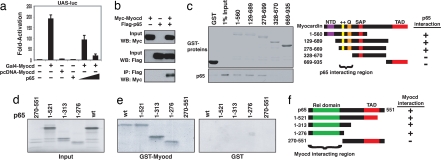
To characterize their mechanistic interaction, we tested whether p65 could still repress the activity of myocardin when it was fused to the DNA binding domain of GAL4, in such a way that its transcriptional activity is independent of SRF. Indeed, we observed that p65 repressed the myocardin-activated GAL4-dependent luciferase reporter (UAS-luciferase) in a dose-dependent manner (Fig. 2a), suggesting that p65 acts directly on myocardin to repress myocardin-dependent gene expression, rather than indirectly through SRF (12). Next, we used coimmunoprecipitation to demonstrate that myocardin and p65 had a direct physical association (Fig. 2b). With a series of myocardin deletion mutant proteins, we mapped the domains of myocardin responsible for its protein–protein interaction with p65. As shown (Fig. 2c), p65 interacts with GST fused to myocardin at amino acids 1–560, amino acids 129–689, and amino acids 78–669, but not with amino acids 328–670, amino acids 669–935, or GST alone. Deletion of N-terminal sequences of myocardin up to amino acid 328 abolished this interaction. Therefore, the N terminus of myocardin, including the basic and Q-rich domains, was necessary and sufficient for its interaction with p65. Interestingly, the p65-interaction region of myocardin protein was previously shown to be used for its interaction with SRF (7, 17), indicating that p65 and SRF might directly compete for myocardin binding.
Fig. 2.
Physical interaction between p65 and myocardin. (a) COS7 cells transfected with expression plasmids encoding full-length myocardin fused to GAL4 and UAS-luciferase reporter, with or without expression plasmid for p65. Values are the fold increase of luciferase activity relative to activation of the reporter alone in at least three experiments. Student's t test: P < 0.05, Gal4-myocardin vs. Gal4-myocardin plus p65. (b) Anti-Myc antibodies detected myocardin in immunoprecipitates prepared with anti-Flag antibodies and COS7 cells transfected with Flag-p65 and Myc-myocardin expression plasmids. (c) (Left) Coomassie-stained proteins corresponding to the amounts of GST and GST-myocardin protein in the pull-down assay are shown. (Right) Summary of p65 interaction with myocardin domains. (d) Autoradiogram indicating 1/20 input of radiolabeled p65 proteins used in pull-down assay. (e) Interaction between full-length and truncated p65 and GST-myocardin (Left) and GST alone (Right). (f) Summary of myocardin interaction with p65 domains.
Having identified the region on myocardin for p65 binding, we next sought to define the region of p65 that interacts with myocardin using truncated p65 proteins (Fig. 2d). Indeed, the N-terminal regions of p65 specifically interact with myocardin (Fig. 2e). Whereas the region including the Rel homology domain of p65 (amino acids 1–276) is sufficient to interact with myocardin, no interaction was detected in the C-terminal region of p65 (amino acids 270–551) (Fig. 2f). This finding compares favorably to a previous report demonstrating that SRF binds p65 between residues 234 and 245 (12).
NF-κB Prevents the Formation of the Myocardin/SRF/CArG Ternary Complex.
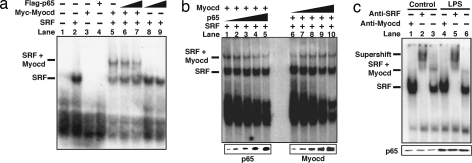
Myocardin does not directly bind to DNA; rather, it is recruited to target promoters by forming a stable complex with SRF bound to CArG box (6, 7). We therefore hypothesized that p65 could interfere with the myocardin/SRF protein complex bound to its associated CArG box. Gel shift assays were performed by using a radiolabeled consensus CArG box (derived from the c-fos promoter) in which SRF binds to the CArG box and SRF/myocardin associate with the CArG box to form a ternary complex (Fig. 3a). When p65 protein was added to the reaction, there was no additional complex formation and the SRF/myocardin/CArG ternary complex was not further shifted. However, the intensity of the ternary complex was decreased when increasing amounts of p65 were added (Fig. 3a, lanes 5–7). Although p65 is not a part of the stable component of the SRF/CArG or the myocardin/SRF/CArG complex, these results indicate that p65 does affect the formation of the ternary complex. This observation was further confirmed by using a series of increased amounts of p65 relative to the fixed amounts of SRF and myocardin (Fig. 3b Left). Conversely, addition of increased amounts of myocardin relative to fixed amounts of SRF and p65 resulted in enhancement of the ternary complex formation (Fig. 3b Right). Similarly, p65 interferes with the ternary complex formation of myocardin/SRF on the SM22 CArG box [supporting information (SI) Fig. 7]. Together, these data demonstrate that p65 and myocardin directly compete for their SRF association.
Fig. 3.
p65 inhibits the formation of myocardin/SRF/CArG ternary complex. (a) Myc-tagged myocardin, Flag-tagged p65, and SRF proteins were expressed by in vitro transcription and translation and incubated with radiolabeled probes, and gel mobility shift assays were performed. (b) Gel mobility shift assays were performed with increasing amounts of p65 (Left) and myocardin (Right). (c) Gel mobility shift assays were conducted from mouse hearts with or without LPS. Anti-SRF antibodies were applied for supershift. p65 protein was analyzed by Western blot.
We next sought to test whether p65 could inhibit formation of the SRF/myocardin/CArG complex under physiologic conditions in an in vivo sepsis model (18). LPS (15 μg/ml) was injected in mice, and hearts were later procured for myocardial protein. As expected, the p65 protein level was significantly induced in the LPS-treated hearts (Fig. 3c Lower). When nuclear protein extracts isolated from control mouse hearts were used in EMSAs, the formation of the SRF/CArG complex and the SRF/myocardin/CArG ternary complex was evident (Fig. 3c, lanes 1–3). Antibody supershift confirmed the identities of this complex (Fig. 3c, lane 2). Importantly, LPS treatment dramatically attenuated the SRF/myocardin/CArG ternary complex formation without affecting SRF CArG-box binding (Fig. 3c, compare lanes 6 and 3). These data demonstrate that p65 is a potent nuclear factor that inhibits myocardin transactivation by disrupting the formation of the SRF/myocardin/CArG complex both in vitro and in vivo.
Myocardin Inhibits NF-κB Signaling and Its Target Gene Expression.
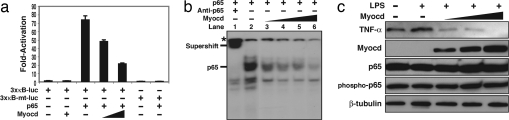
We next tested the effect of myocardin binding to p65 on the transcriptional activity of p65 itself. Luciferase constructs with three tandem κB-box promoters and its mutant were transfected with p65, and p65 appropriately induced luciferase activity of the wild-type κB promoters but not its mutant (Fig. 4a). When cotransfected with myocardin, p65-mediated luciferase activity was decreased in a dose-dependent manner (Fig. 4a). These data demonstrate that myocardin can inhibit p65 transcriptional activity.
Fig. 4.
Myocardin repression of p65 transcriptional activity. (a) COS7 cells transfected with a luciferase reporter construct of three tandem κB sites or its mutant, together with expression plasmids for p65 (0.1 μg), myocardin (0.1 and 0.2 μg), or both. Values are the fold increase in luciferase activity relative to activation of the reporter alone. Error bars represent SD of at least three experiments. Student's t test: P < 0.05, p65 alone vs. p65 plus myocardin. (b) Gel mobility shift assays were performed with a 32P-labeled κB probe with stable levels of in vitro translated p65 with or without increasing amounts of in vitro translated myocardin. (c) Aortic VSMCs infected with increasing amounts of adenoviral myocardin (multiplicity of infection: 20, 50, and 200) for 48 h and then stimulated with LPS (1 μg/ml) for 2 h. Western blots were performed on extracted proteins.
The functional inhibition of p65 transactivity by myocardin prompted us to determine whether myocardin interferes with DNA binding of p65. EMSAs were conducted by using a consensus κB box as a probe. Addition of p65 protein resulted in a specific shifted band (Fig. 4b, lane 2). Concomitant administration of anti-p65 antibody resulted in a supershift (Fig. 4b, lane 1). However, when the reaction was incubated with increasing amounts of myocardin protein, the p65 and κB-box complexes were gradually diminished (Fig. 4b, lanes 3–6). These results demonstrate that myocardin can indeed block p65 binding to its specific DNA element.
We next sought to determine whether myocardin could physiologically repress transcriptional activity of p65. Aortic smooth muscle cells were infected with Ad-myocardin (or Ad-GFP as a control) viruses for 48 h, and then 1 μg/ml LPS was added for 2 h. As expected, LPS increased TNF-α expression (Fig. 4c) that was essentially abolished with increased concentrations of myocardin. Interestingly, the expression levels of total p65 and phospho-Ser-536-p65 were not altered by myocardin (Fig. 4c). These data suggest that myocardin interferes with NF-κB signaling, not by its effect on the upstream phosphorylation cascade, but rather by directly inhibiting p65 transcriptional activity and expression of p65-dependent downstream target genes.
Myocardin Inhibits Cellular Proliferation.
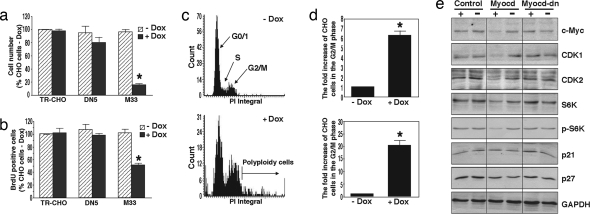
Because myocardin can functionally repress p65 transcriptional activity, and in particular decrease expression of TNF-α, we next sought to determine the influence of myocardin on cellular proliferation. We therefore stably overexpressed myocardin using a tetracycline-regulated expression system in CHO cells. Doxycycline (Dox) treatment of transfected cells that harbor wild-type myocardin (M33) successfully promoted a time-dependent overexpression of myocardin (SI Fig. 8a). When myocardin was induced, cellular growth and proliferation as measured by histology, cell numbers, and DNA synthesis was markedly reduced compared with cells that harbor either a control vector (TR-CHO) or the dominant negative myocardin (DN5) (Fig. 5 a and b and SI Fig. 8b). Myocardin overexpression was not associated with apoptosis because a sub-G0/G1 peak was not identified, nor did we detect DNA fragmentation or positive TUNEL staining (data not shown). The antiproliferative effect of myocardin was related to G2–M phase arrest and formation of polyploid cells (Fig. 5 c and d). To define transcriptional targets linking myocardin to cell-cycle regulation, we demonstrate that myocardin induction is associated with down-regulation of several important mediators of cell-cycle progression, including those previously known to be regulated by NF-κB such as c-myc (19) and cyclin-dependent kinase 2 (CDK2) (20), as well as ribosomal S6 kinase (S6K) and CDK1, but not p21 and p27 (Fig. 5e).
Fig. 5.
Myocardin inhibits CHO cell proliferation. (a) Numbers of cells harboring the myocardin-inducible system were counted 72 h after treatment with or without Dox (n = 7). *, P < 0.01. (b) Cells were treated for 3 days, pulse-labeled with BrdU for 60 min, and stained with BrdU (for both cell count and BrdU experiments (n = 7). *, P < 0.01. (c) Representative DNA histograms obtained through laser scanning cytometry in M33 CHO cells treated for 24 h. (d) Cell numbers in both G2–M phase and polyploid cell regions as detected by cytometry (n = 5). *, P < 0.01. (e) Western blot with corresponding antibodies for cell-cycle regulatory proteins in cells with (+) and without (−) Dox treatment for 3 days.
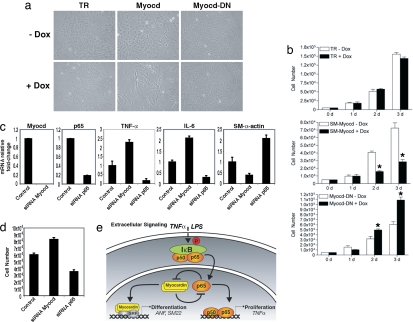
To investigate the endogenous influence of myocardin and p65 on cell proliferation and differentiation, we performed overexpression and loss-of-function studies in VSMC. Again, overexpression of myocardin using the tetracycline-regulated expression system decreased VSMC numbers, consistent with that seen in CHO cells (Fig. 6a). Interestingly, overexpression of the dominant negative myocardin increased numbers of VSMCs, whereas dominant negative myocardin in CHO cells had no such effect. We attribute this difference to interference of endogenous myocardin by dominant negative myocardin in VSMC, whereas CHO cells do not express myocardin. Quantitative analyses by direct cell counting over 3 successive days further support these results (Fig. 6b). Next, we knocked down myocardin or p65 expression using siRNAs in VSMC. Inhibition of myocardin resulted in increased expression of NF-κB-dependent and mitogenic cytokines TNF-α and IL-6. As expected, siRNA to p65 reduced both TNF-α and IL-6 levels (Fig. 6c). In contrast, myocardin inhibition decreased expression of SM-α-actin whereas p65-inhibition increased its expression, consistent with previous reports (Fig. 6c). Finally, we corroborated our overexpression studies in both CHO and VSMC by demonstrating that inhibition of myocardin increased VSMC numbers, whereas inhibition of p65 resulted in decreased VSMC growth (Fig. 6d). Taken together, these data establish that myocardin functions as an antiproliferative factor and show that this effect is coupled with its repression of NF-κB transcriptional activity.
Fig. 6.
Effect of endogenous myocardin on VSMC growth. (a) Photomicrographs of VSMC harboring the tetracycline-regulated (TR) inducible system for myocardin and its dominant negative treated with and without Dox for 24 h. (b) Cell numbers counted over 3 days in control (TR), myocardin (Myocd), and dominant negative (Myocd-DN) expressing VSMC (n = 5). *, P < 0.01. (c) Mouse aortic VSMC were transfected with siRNA to either myocardin or p65 for 24 h. Real-time PCR was performed to detect relative fold changes in mRNA expression (normalized to 18s rRNA). Values are the relative fold change in mRNA expression relative to control of at least three experiments. P < 0.05 for both siRNA vs. control and siRNA myocardin vs. siRNA p65. (d) VSMC directly counted 48 h after being transfected with either myocardin or p65 siRNA. Error bars represent SD of five experiments. P < 0.05 for both siRNA vs. control and siRNA myocardin vs. siRNA p65. (e) Working model balancing NF-κB(p65) and myocardin regulation of cellular growth and differentiation.
Discussion
This study reveals a relationship between two transcription factors, myocardin and NF-κB, in regulation of cellular proliferation and muscle differentiation. We found that p65 suppresses myocardin activation of CArG-dependent smooth muscle and cardiac genes and is mediated by physical interruption of the myocardin/SRF/CArG ternary complex. Conversely, myocardin inhibits p65 DNA binding, thereby repressing NF-κB-mediated target gene expression and function, including cellular proliferation. These findings add several layers to our current understanding of both NF-κB and myocardin biology. First, we offer evidence of the expanding complexity of myocardin-dependent signaling by demonstrating its unique regulation by p65. In addition, we establish additional DNA binding-independent effects of NF-κB. Finally, we demonstrate that myocardin can behave as a negative transcriptional cofactor in a SRF-independent manner to inhibit cellular proliferation.
Several reports have recently described mechanisms related to myocardin regulation of smooth muscle and cardiac gene expression and point to an intricate web of cofactors that control the influence of myocardin on SRF (21). Whereas regulation of myocardin activity is typically a function of its ability to physically associate with SRF (7), NF-κB regulation of gene transcription is classically a result of its binding to DNA. A functional interaction between p65 and SRF has been previously described and suggested that p65 complex formation with the SRF MADS domain may neutralize the inhibitory function of this region, thereby promoting SRF transcription (12, 22). In this article we offer an alternative mechanism of p65 regulation of SRF: disruption of the myocardin/SRF/CArG ternary complex, thus functionally inhibiting myocardin transactivation of SRF-regulated cardiac and smooth muscle genes.
In addition to demonstrating that p65 physically binds with myocardin and interferes with its transcriptional activation, we reveal an unexpected role of myocardin as a transcriptional repressor of p65 rather than as a SRF cofactor. Indeed, we demonstrate that myocardin has SRF-independent effects by directly interacting and preventing binding of p65 to its DNA target. Furthermore, myocardin completely abrogated TNF-α protein production after stimulation with LPS despite persistently high and stable levels of p65, suggesting that the influence of myocardin was not on upstream NF-κB activation but on its downstream activity. Our studies therefore assign a molecular mechanism for myocardin function.
We found that myocardin potently inhibits cell proliferation, at least in part by repressing NF-κB-dependent cell proliferation signals. This observation is consistent with a recent report in which inactivation of myocardin is associated with malignant tumor growth (9). In addition, we demonstrate the endogenous influence of myocardin to hinder cell growth. Our study specifically links myocardin to the control of G2–M transition of cell-cycle progression. Interestingly, a recent study also reported that NF-κB participates in regulation of G2–M cell-cycle progression through ERK5 (23). ERK5 activated NF-κB not through its classic upstream kinase but by S6K-dependent phosphorylation of IκBα. The observation that myocardin decreased expression of S6K and pS6K without associated changes in p65 levels is intriguing and may provide an alternative feedback mechanism to modulate cell-cycle progression. Similarly, activation of the cyclin-B–CDK1 complex is required for G2–M transition, and myocardin did effectively down-regulate CDK1 expression. In contrast, the cyclin-dependent kinase inhibitors p21 and p27, principally acting at the G1–S transition, were not influenced by myocardin induction, further supporting the view that myocardin specifically targets the G2–M phase. Taken together (Fig. 6), our results define a unique relationship between NF-κB and myocardin that govern signals balancing cardiac and smooth muscle cell growth and differentiation. We also reveal a function for myocardin in regulation of cell proliferation and provide a molecular mechanism by which myocardin can act as a transcriptional repressor to inhibit the NF-κB pathway.
Methods
Detailed materials and methods are in SI Methods. In brief, COS7, rat neonatal cardiomyocytes, and murine aortic VSMC were isolated and cultured (5). Luciferase reporter constructs were fused with the SM22 and ANF promoters (6). The myocardin adenoviral expression construct (Ad-MYCD) contained a cDNA encoding amino acids 129–935 of mouse myocardin (24).
Coimmunoprecipitation Assays.
COS7 cells were transiently transfected with plasmids encoding the epitope-tagged myocardin, SRF, and p65 proteins with FuGENE6. Epitope-tagged proteins were precipitated with antibodies and protein A/G beads and analyzed by Western blotting with directed antibodies.
GST Protein-Binding Assays.
Plasmids encoding a GST fusion with truncated myocardin were transformed into BL21 cells and added to the culture to induce protein expression. p65 full-length and truncated proteins translated in vitro were radiolabeled with [35S]methionine. Glutathione beads conjugated with GST fusion protein were incubated with radiolabeled proteins and analyzed by autoradiography.
EMSAs.
EMSAs were performed by using c-fos CArG probe (6) or NF-κB-specific probe (AGT TGA GGG GAC TTT CCC AGGC). For mouse studies, mice were killed, the left ventricular muscles were isolated, and cytoplasmic and nuclear proteins were extracted and subsequently incubated with labeled probes. Antibody supershift experiments were conducted with anti-SRF (Santa Cruz Biotechnology), anti-FLAG, or anti-Myc (Sigma).
Myocardin Tetracycline-Regulated System and Cellular Proliferation.
A tetracycline-regulated system in CHO cells or human aortic VSMCs (American Type Culture Collection) harboring wild-type myocardin (M33) and dominant negative (DN5) myocardin were analyzed by Western blot using anti-myocardin and anti-Flag M2 monoclonal antibody, respectively. Cell counting, BrdU staining, and laser scanning cytometry were performed and compared with control cells (TR) with or without Dox.
siRNA Transfection and Real-Time PCR.
Primary aortic VSMCs were transfected with predesigned siRNAs of myocardin and p65 (Ambion) by applying program D-33 and using Basic Nucleofector Kit for primary smooth muscle cells (VPI-1004; Amaxa). TaqMan polymerase, as well as TaqMan matched probes and primers, were applied for real-time PCR (Applied Biosystems).
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the American College of Surgeons (C.H.S.), the National Institutes of Health (A.S.B. and D.-Z.W.), the Canadian Institutes of Health Research (X.-L.Z.), the March of Dimes Birth Defects Foundation (D.-Z.W.), the Muscular Dystrophy Association (D.-Z.W.), and the American Heart Association (D.-Z.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705842105/DC1.
References
- 1.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miano JM. Serum response factor: Toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 3.Posern G, Treisman R. Actin' together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Callis TE, Cao D, Wang DZ. Bone morphogenetic protein signaling modulates myocardin transactivation of cardiac genes. Circ Res. 2005;97:992–1000. doi: 10.1161/01.RES.0000190670.92879.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, et al. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: A component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 9.Milyavsky M, et al. Inactivation of myocardin and p16 during malignant transformation contributes to a differentiation defect. Cancer Cell. 2007;11:133–146. doi: 10.1016/j.ccr.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin AS., Jr Series introduction: The transcription factor NF-kB and human disease. J Clin Invest. 2001;107:3–6. doi: 10.1172/JCI11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: Possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 12.Franzoso G, et al. Activation of the serum response factor by p65/NF-kappaB. EMBO J. 1996;15:3403–3412. [PMC free article] [PubMed] [Google Scholar]
- 13.Selzman CH, et al. Monocyte chemotactic protein-1 directly induces human vascular smooth muscle proliferation. Am J Physiol. 2002;283:H1455–H1461. doi: 10.1152/ajpheart.00188.2002. [DOI] [PubMed] [Google Scholar]
- 14.Selzman CH, et al. Liposomal delivery of purified inhibitory-kappaBalpha inhibits tumor necrosis factor-alpha-induced human vascular smooth muscle proliferation. Circ Res. 1999;84:867–875. doi: 10.1161/01.res.84.8.867. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerman MA, Reznikov LL, Sorensen AC, Selzman CH. Relative contribution of the TNF-alpha receptors to murine intimal hyperplasia. Am J Physiol. 2003;284:R1213–R1218. doi: 10.1152/ajpregu.00434.2002. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman MA, et al. Lack of TNF-alpha attenuates intimal hyperplasia after mouse carotid artery injury. Am J Physiol. 2002;283:R505–R512. doi: 10.1152/ajpregu.00033.2002. [DOI] [PubMed] [Google Scholar]
- 17.Wang DZ, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shames BD, et al. Increased levels of myocardial IkappaB-alpha protein promote tolerance to endotoxin. Am J Physiol. 1998;275:H1084–H1091. doi: 10.1152/ajpheart.1998.275.3.H1084. [DOI] [PubMed] [Google Scholar]
- 19.Duyao MP, Buckler AJ, Sonenshein GE. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc Natl Acad Sci USA. 1990;87:4727–4731. doi: 10.1073/pnas.87.12.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahradka P, et al. NF-kappaB activation is essential for angiotensin II-dependent proliferation and migration of vascular smooth muscle cells. J Mol Cell Cardiol. 2002;34:1609–1621. doi: 10.1006/jmcc.2002.2111. [DOI] [PubMed] [Google Scholar]
- 21.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 22.Montaner S, Perona R, Saniger L, Lacal JC. Activation of serum response factor by RhoA is mediated by the nuclear factor-kappaB and C/EBP transcription factors. J Biol Chem. 1999;274:8506–8515. doi: 10.1074/jbc.274.13.8506. [DOI] [PubMed] [Google Scholar]
- 23.Cude K, et al. Regulation of the G2-M cell cycle progression by the ERK5-NF{kappa}B signaling pathway. J Cell Biol. 2007;177:253–264. doi: 10.1083/jcb.200609166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing W, et al. Myocardin induces cardiomyocyte hypertrophy. Circ Res. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.