Abstract
We demonstrate a role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. In particular, transient increased expression of Sirt1 is sufficient to stimulate basal rates of autophagy. In addition, we show that Sirt1−/− mouse embryonic fibroblasts do not fully activate autophagy under starved conditions. Reconstitution with wild-type but not a deacetylase-inactive mutant of Sirt1 restores autophagy in these cells. We further demonstrate that Sirt1 can form a molecular complex with several essential components of the autophagy machinery, including autophagy genes (Atg)5, Atg7, and Atg8. In vitro, Sirt1 can, in an NAD-dependent fashion, directly deacetylate these components. The absence of Sirt1 leads to markedly elevated acetylation of proteins known to be required for autophagy in both cultured cells and in embryonic and neonatal tissues. Finally, we show that Sirt1−/− mice partially resemble Atg5−/− mice, including the accumulation of damaged organelles, disruption of energy homeostasis, and early perinatal mortality. Furthermore, the in utero delivery of the metabolic substrate pyruvate extends the survival of Sirt1−/− pups. These results suggest that the Sirt1 deacetylase is an important in vivo regulator of autophagy and provide a link between sirtuin function and the overall cellular response to limited nutrients.
Keywords: mitochondria, starvation, acetylation, aging
Macroautophagy, referred to hereafter as autophagy, is an intracellular process that allows for the degradation of proteins and organelles (1–3). Morphologically, autophagy is characterized by the formation of a double-membrane structure termed the autophagosome. In yeast, the process of autophagy can be stimulated by the withdrawal of various nutrients, and evidence suggests that at least 16 separate autophagy genes (Atg) are ultimately required for the formation of the autophagosome and the subsequent induction of autophagy (1–3). Mammalian cells can also respond to nutrient withdrawal by inducing autophagy. Genetic studies have suggested that autophagy is required to maintain the animal's energetic needs during the small window of time immediately after birth but before neonates can get milk nutrients from their mothers (4). In addition, two recent studies have demonstrated that brain conditional knockouts of either Atg7 or Atg5 significantly accelerated the development of neuropathologies usually restricted to older animals (5, 6).
The sirtuins are a family of NAD-dependent deacetylases that have been linked to the regulation of life span. Increased expression of Sir2 can extend the life span of model organisms, and under certain experimental conditions, it appears that the activity of Sir2 is required for the life-extending benefits of caloric restriction in organisms such as yeast and flies (7). As opposed to these relatively simple organisms, mammalian species have seven different sirtuin family members (8, 9). The closest relative of yeast Sir2 is termed Sirt1 in mammalian cells. We have demonstrated that mice subjected to acute overnight starvation can increase their expression of Sirt1 in certain tissues (10). Similarly, it has been noted that chronic caloric restriction augments the level of Sirt1 (11). These results suggest that Sirt1 may participate in some fashion in protecting cells from nutrient stress. Based on the likely role of both Sirt1 and autophagy in nutrient adaptation and the putative role of both autophagy and Sirt1 in aging and age-related pathologies, we sought to understand whether there might be a connection between Sirt1 activity and the induction of autophagy.
Results
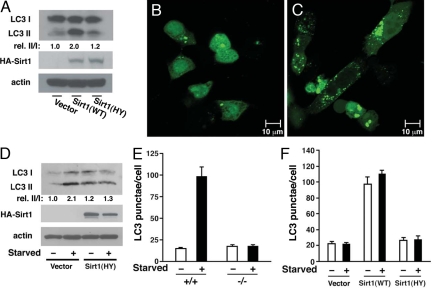
The formation of the autophagosome is conveniently assayed in living cells and tissues by the subcellular distribution of LC3, the mammalian homolog of the yeast Atg8 gene product. Previous studies have demonstrated that the conversion of LC3-I to LC3-II through proteolytic cleavage and lipidation is a hallmark of mammalian autophagy and that a fusion protein of green fluorescent protein and LC3 (GFP-LC3) provides a useful measurement of autophagy initiation through the assay of “LC3 dots or punctae” (4, 12). To assess the role of Sirt1 in autophagy, we transiently overexpressed wild-type Sirt1 or a deacetylase-inactive mutant of Sirt1(HY) in the HCT116 colon cancer cell line. As noted in Fig. 1A, wild-type Sirt1 but not the catalytically inactive mutant stimulated the conversion of endogenous LC3-I to LC3-II. Similarly, a transient increase of wild-type Sirt1 expression increased the formation of GFP-LC3 punctae, consistent with autophagosome formation [Fig. 1 B and C and supporting information (SI) Fig. 5]. In agreement with previous reports that the deacetylase-inactive Sirt1 mutant can act in a dominant-negative fashion (13, 14), expression of this mutant also partially blocked endogenous LC3-I to LC3-II conversion under starved conditions (Fig. 1D).
Fig. 1.
Sirt1 is necessary for autophagy. (A) Transient expression of wild-type (WT) Sirt1 but not a deacetylase-inactive (HY) point mutant of Sirt1 stimulates conversion of LC3-I to LC3-II in HCT116 cells. Quantification of the relative (rel.) levels of LC3-II/LC3-I is shown with the ratio of 1.0 being assigned to vector-transfected cells under fed conditions. (B and C) Distribution of GFP-LC3 in HCT116 cells under fed conditions when transfected along with either a control empty vector (B) or when cotransfected with wild-type Sirt1 (C). (D) The deacetylase-inactive mutant of Sirt1 acts in a dominant-negative fashion to inhibit starvation-induced conversion of LC3-I to LC3-II in HCT116 cells. (E) Quantification of GFP-LC3 punctae in wild-type (+/+) and Sirt1−/− MEFs under fed or starved conditions. (F) Level of autophagy under fed or starved conditions in Sirt1−/− MEFs transfected with GFP-LC3 along with an empty vector, wild-type Sirt1, or the described deacetylase-inactive mutant, Sirt1 (HY).
In an effort to extend these results, we quantified GFP-LC3 punctae formation in wild-type and Sirt1−/− mouse embryonic fibroblasts (MEFs). As noted in Fig. 1E, in wild-type MEFs, the switch to nutrient-depleted conditions resulted in an increase in the amount of GFP-LC3 punctae, whereas this response was markedly reduced in Sirt1−/− MEFs (Fig. 1E and SI Fig. 6). A similar reduction in autophagy was observed when the pharmacological agent rapamycin was used to stimulate autophagy in Sirt1−/− MEFs or when Sirt1 expression was knocked down with RNAi in HeLa cells (SI Figs. 7 and 8). We next asked whether transient reconstitution of Sirt1 could restore autophagy in Sirt1−/− cells. As shown in Fig. 1F, although expression of wild-type Sirt1 restored autophagy under starved conditions, expression of a deacetylase-inactive mutant of Sirt1 was unable to reconstitute activity. Transient overexpression of wild-type Sirt1 in MEFs, as was noted in HCT116 cells, was sufficient to stimulate basal autophagy under fed conditions (Fig. 1F).
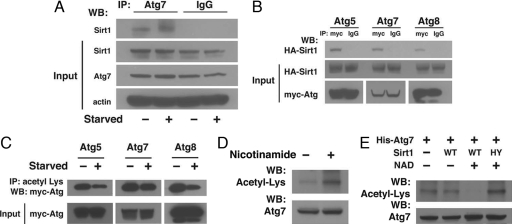
Given that Sirt1 appears to be required for both starvation- and rapamycin-stimulated autophagy, we thought it possible that Sirt1 might interact directly with components of the autophagy machinery. We therefore tested whether Sirt1 might coprecipitate with any of the 16 known autophagy gene products. Although there is limited availability of antibodies recognizing endogenous mammalian Atg gene products, we were able to find a commercially available antibody suitable for Atg7 immunoprecipitation. As noted in Fig. 2A, by using an antibody directed against endogenous Atg7, we could coprecipitate endogenous Sirt1 from HeLa cell lysates. The level of the Atg7–Sirt1 complex did not appear to change appreciably under fed or starved conditions. Similarly, Sirt1 and Atg7 appeared at least partially to colocalize within cells (SI Fig. 9). To extend these results further, we prepared epitope-tagged forms of several essential autophagy genes including Atg5, Atg7, and Atg8. As seen in Fig. 2B, when we immunoprecipitated Atg5, Atg7, or Atg8 we could readily detect the presence of coimmunoprecipitation Sirt1. Based on these observations, we next sought to understand whether autophagy-related proteins were potential substrate for Sirt1. As seen in Fig. 2C, by using an antibody directed against acetyllysine residues to immunoprecipitate Atg5, Atg7, and Atg8 we demonstrated that these proteins were acetylated under basal conditions and that acetylation levels were reduced under starved conditions. Levels of acetylation were also responsive to treatment with the broad-spectrum sirtuin inhibitor nicotinamide. In these experiments, acetylation was assessed by direct Western blotting with an antibody recognizing acetyllysine residues after purification of His-tagged Atg7 (Fig. 2D) or His-tagged Atg8 (SI Fig. 10) from transfected HeLa cell lysates. As demonstrated in Fig. 2E, purified wild-type Sirt1 could, in an NAD-dependent fashion, act as an effective in vitro deacetylase of His-tagged Atg7, whereas, as expected, addition of the deacetylase mutant was ineffective in this assay. Similar results were obtained when Atg8 was used as a substrate (SI Fig. 11).
Fig. 2.
Sirt1 interacts with components of the autophagy machinery. (A) Immunoprecipitation (IP) from 2 mg of HeLa cell protein lysate either with an Atg7-specific antibody or a control IgG serum demonstrating the endogenous interaction between Sirt1 and Atg7. Input represents Atg7 and Sirt1 expression in 50 μg of HeLa cell lysate. WB, Western blotting. (B) HeLa cells were transfected with Myc-tagged Atg5, Atg7, or Atg8 along with a HA epitope-tagged form of Sirt1. Lysates were subsequently immunoprecipitated with an antibody against the Myc epitope or with a matched IgG isotype control serum. Coimmunoprecipitated Sirt1 was assessed by Western blotting with a HA epitope-specific antibody. (C) Levels of acetylation in lysates prepared from transiently transfected HeLa cells with the indicated epitope-tagged Atg constructs. Acetylation was assessed under either fed or starved conditions and demonstrates reduced acetylation during starvation. (D) His-tagged Atg7 was purified from transfected HeLa cells treated or untreated with 10 mM nicotinamide for 14 h before harvest. Levels of Atg7 acetylation were detected by Western blotting with an acetyllysine antibody. Total levels of Atg7 were unaffected by nicotinamide treatment. (E) In vitro Sirt1 deacetylation reactions with purified acetylated His-Atg7 as a substrate in the presence or absence of 10 mM NAD and purified wild-type (WT) or deacetylase-inactive (HY) mutant Sirt1 protein.
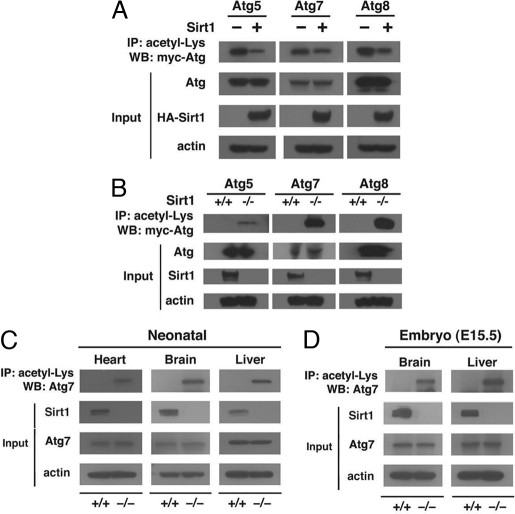
We next sought to characterize further the role of Sirt1 in regulating acetylation of the proteins controlling autophagy. As seen in Fig. 3A, in HeLa cells, increased expression of wild-type Sirt1 reduced the level of protein acetylation for all Atg proteins examined. These effects were specific for Sirt1 because similar expression of Sirt3, another sirtuin family member, did not alter autophagy protein acetylation (SI Fig. 12). To extend these results further, we assayed the level of acetylation in wild-type and Sirt1−/− MEFs. Consistent with a role for Sirt1 in regulating autophagy gene product acetylation, analysis of various independently isolated lines of MEFs revealed that in Sirt1−/− MEFs, the levels of each of the Atg proteins analyzed was markedly elevated (Fig. 3B). To confirm these results in vivo, we used our published line of Sirt1 knockout mice (15). As initially reported, these mice usually succumb shortly after birth, so tissues were harvested from wild-type and Sirt1−/− littermates within hours of delivery. Consistent with our analysis in MEFs, direct analysis of endogenous Atg7 in the liver, heart, and brain of wild-type and Sirt1−/− mice revealed substantial differences in acetylation in the absence of any differences in total Atg7 protein levels (Fig. 3C). These differences in acetylation were also present in utero because Sirt1−/− E15.5 embryos also demonstrated increased endogenous Atg7 acetylation (Fig. 3D).
Fig. 3.
Sirt1 regulates acetylation of autophagy gene products. (A) Transient increased expression of Sirt1 reduces acetylation. HeLa cells were transfected with the indicated epitope-tagged Atg construct along with, where indicated, wild-type Sirt1 (+). Measurements were done under fed conditions. IP, immunoprecipitation; WB, Western blotting. (B) Acetylation of autophagy proteins in wild-type or Sirt1−/− MEFs under fed conditions. Absence of Sirt1 dramatically increased acetylation. (C) Levels of endogenous Atg7 acetylation in various wild-type (+/+) and Sirt1−/− neonatal tissue. Tissues were harvested several hours after birth. (D) Levels of endogenous Atg7 acetylation in the brain and liver of approximately E15.5 embryos. Western blot data represent results obtained from one of two similar littermate pairs.
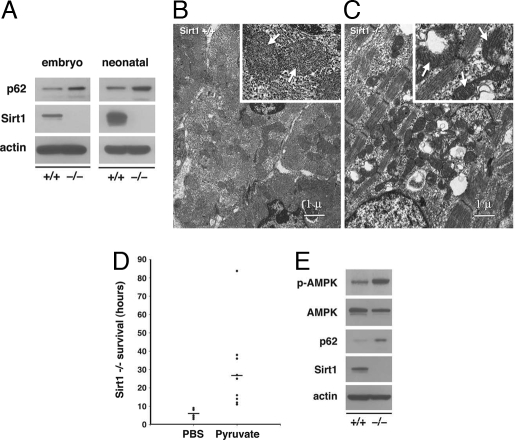
Recent evidence suggests that the accumulation of p62 represents a convenient in vivo marker for impaired autophagy (16, 17). We therefore assessed p62 levels in wild-type or Sirt1−/− littermates. As demonstrated in Fig. 4A, embryonic heart tissue of Sirt1−/− exhibited increased levels of p62. Similarly, when we analyzed neonatal tissues within the first 3 h after birth, levels of p62 were also elevated, especially within the myocardium (Fig. 4A). In addition to the accumulation of p62, previous studies have demonstrated that the absence of autophagy leads to the accumulation of damaged organelles (17, 18). In particular, accumulations of abnormally shaped mitochondria are a prominent feature in tissues that lack the normal autophagy clearance mechanisms (18). Electron micrographs of wild-type and Sirt1−/− myocardium demonstrated that mice lacking Sirt1 exhibited gross alterations in mitochondrial morphology (Fig. 4 B and C).
Fig. 4.
Absence of Sirt1 inhibits autophagy in vivo. (A) Accumulation of the autophagy marker p62 in the myocardium of embryonic and neonatal Sirt1−/− knockout tissue. Neonatal tissue was harvested in the first 3 h after birth. (B) Electron micrographs of neonatal wild-type myocardium. (Inset) Normal appearing mitochondria (white arrows). (C) Electron micrograph of the myocardium of a Sirt1−/− littermate. The tissue demonstrates prominent accumulation of abnormally shaped mitochondria, examples of which are seen with higher power in the Inset. (D) Survival of Sirt1−/− pups (in hours) for animals receiving either PBS injections or injections of ethyl pyruvate diluted in PBS while in utero. Median survival difference between groups was significant with P < 0.02. (E) Compared with wild-type littermates, phosphorylated AMPK (p-AMPK, Thr-172) levels are increased in the liver of unfed Sirt1−/− mice when assessed 15 h after birth. Western blot data are from one representative littermate pair with similar results obtained from at least four separate littermate pairs.
We were intrigued by the observations that Sirt1−/− animals die shortly after birth because a similar perinatal mortality has been observed in Atg5−/− mice (4). The perinatal period is believed to require autophagy as a means of providing energy in the absence of nutrient intake (4). Recently, it was demonstrated that energetic deficiencies caused by inadequate autophagy could be overcome, at least in part, by the delivery of exogenous pyruvate (19). Pyruvate is the end product of glycolysis, and it can readily enter cells where it can be used as a substrate for both aerobic and anaerobic metabolism. As such, delivery of pyruvate does not restore autophagy to Atg-deficient tissues but instead mitigates the overall energy deficit. We reasoned that if Sirt1−/− are unable to stimulate autophagy, the observed perinatal mortality of Sirt1−/− animals might be attenuated by pyruvate delivery. We therefore mated Sirt1+/− mice and injected pregnant mothers daily with either ethyl pyruvate or PBS. We next analyzed the survival of Sirt1−/− pups from ≈8–10 litters of either pyruvate-treated or PBS-treated animals. Similar to uninjected animals, PBS-treated mothers gave birth to Sirt1−/− pups that in our facility lived on average <8 h. In contrast, in animals treated with pyruvate in utero, the mean life span of Sirt1−/− pups was extended to >24 h (Fig. 4D). In some experiments, we attempted to continue to deliver pyruvate to Sirt1−/− pups, but for technical reasons we were unable to administer either PBS or pyruvate i.p. injections to these newborn animals. We do not believe this technical failure relates specifically to Sirt1 status because similar attempts in wild-type neonates was also technically difficult and led to high periinjection mortality.
Previous results indicated that unfed Atg5−/− mice usually live <12 h, and with artificial feedings, survival can only be extended to ≈40 h (4). Energy-responsive pathways, such as AMP-activated kinase (AMPK), indicative of an overall low-energetic state, have been shown to be activated in Atg5−/− mice (4). To assess such pathways in our Sirt1−/− animals, pyruvate-treated wild-type and Sirt1−/− littermates were separated from their mother after birth to avoid any potential differences in nutrient intake. We then assessed energy homeostasis in these unfed animals at ≈15 h after birth. As noted in Fig. 4E, the liver of Sirt1−/− mice demonstrated evidence for increased activation of AMPK. A similar induction of AMPK was seen in other tissues (SI Fig. 13). The neonatal tissues of Sirt1−/− mice also exhibited accumulation of p62, consistent with impaired autophagy (Fig. 4E).
Discussion
We demonstrate that Sirt1 activity is necessary for the induction of starvation-induced autophagy. In addition, transiently augmenting wild-type Sirt1 activity but not a deacetylase-inactive mutant of Sirt1 is sufficient to activate autophagy in cells even in the presence of nutrients. Essential components of the autophagy machinery interact with Sirt1, and the absence of Sirt1 dramatically increases the level of acetylation for multiple Atg-related gene products. Consistent with a requirement for Sirt1 in the execution of autophagy, embryos and neonatal mice lacking Sirt1 accumulate abnormal organelles, especially mitochondria, demonstrate increased levels of p62 and the activation of AMPK. In addition, the in utero delivery of pyruvate allows for prolonged survival of Sirt1−/− pups. The latter results need to be interpreted with caution because there is a growing body of data suggesting that ethyl pyruvate administration might be beneficial in a number of in vivo experimental stress conditions, and numerous modes of action besides the rescue of autophagy have been proposed (20, 21).
The simplest explanation of our results is that a starvation-induced increase in Sirt1 activity stimulates the deacetylation of the autophagy machinery. In this regard, one intriguing possibility is that the requisite covalent association between Atg5 and Atg12 that is mediated through a lysine residue on Atg5 might be a target of reversible acetylation (1). However, to date, we have been unable to prove definitively that this residue is acetylated by using mass spectrometry (perhaps caused by insufficient recovery of the corresponding peptide), and site-directed mutagenesis of this lysine did not appreciably alter the overall acetylation of Atg5 (SI Fig. 14). Nonetheless, one potentially attractive aspect of a Sirt1-dependent regulation of autophagy is that during starved conditions, Sirt1 activity, which is in turn dependent on NAD levels, could potentially tune the degree of autophagy to match current cellular needs with real-time metabolic status.
The accumulation of damaged proteins and organelles is believed to be an important aspect of the aging process (22). Indeed, as mentioned, in mice conditional deletion of autophagy genes leads to accelerated age-related pathologies (5, 6). Similarly, because the degree of autophagy both in vitro and in vivo is sensitive to nutrient availability (4, 12), it is tempting to speculate that basal levels of autophagy might be elevated during caloric restriction (CR). This speculation is indeed supported by previous observations (23). Our results would suggest that augmented Sirt1 expression could mimic CR conditions because both conditions would be expected to stimulate basal rates of autophagy. In this regard, it is of interest that in Caenorhabditis elegans, autophagy regulates life span (24, 25), whereas in yeast, Sir2 is required for the life-extending effects of CR (7, 26). Although to our knowledge there have been no genetic studies in yeast demonstrating a role for yeast Sir2 in autophagy, it is of interest to note that in Saccharomyces cerevisiae, Sir2 has been shown to be required for the proper segregation of damaged proteins between mother and daughter cells (27). In addition, in Drosophila, a recent study has linked the activity of another deacetylase, HDAC6, to autophagy (28). Future studies are needed to help clarify to what extent, if any, the life-extending effects of sirtuins relate to the ability of these proteins to regulate autophagy.
Because autophagy appears to contribute to a number of age-related mammalian pathologies, including diseases of mutant protein accumulation, our results suggest that augmenting Sirt1 activity might be clinically beneficial. Interestingly, treatment with a mammalian target of rapamycin (mTOR) inhibitor to stimulate autophagy has already been shown to be effective in animal models of Huntington's disease, a rare but particularly destructive neurodegenerative disease (29). Similarly, Sirt1 appears to protect against certain forms of neuronal degeneration (30), and the sirtuin activator resveratrol has also been shown to be effective in models of chronic neurological disease (31). Our results suggest that other neurodegenerative diseases and potentially a wide range of other age-related pathologies may also benefit from induction of basal autophagy through the augmentation of sirtuin activity.
Finally, it is interesting to note that we and others have demonstrated that Sirt1 can interact and regulate the activity the mitochondrial biogenesis regulator peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) (32–34). Most evidence suggests that Sirt1 can augment PGC-1α activity and thereby increase the supply of new mitochondria. In this report, our data would suggest that by regulating autophagy, Sirt1 may also be important for the clearance of old and damaged mitochondria. Taken together, these results would imply that Sirt1 may play a pivotal role in regulating the overall flux of cellular mitochondria. Given the central role of mitochondria in the aging process (35, 36), the role of Sirt1 in maintaining and potentially rejuvenating the pool of mitochondria may provide important insight into the overall biology of sirtuin function.
Materials and Methods
Cell Culture and DNA Constructs.
HeLa cells (obtained from American Type Culture Collection) were cultured at 37°C in DMEM (Invitrogen) supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). MEFs were prepared in the standard fashion and maintained in DMEM supplemented with 15% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Primary and immortalized MEFs gave similar results regarding acetylation; however, for autophagy assays only primary MEFs were used. HCT116 cells (a gift from P. Hwang, National Institutes of Health) were cultured in McCoy's 5A medium (Invitrogen) supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Epitope-tagged Myc-Atg8 was amplified by using the Atg8-specific primers 5′-AGC TTC GGA ATT CCG ACC ATG CCG-3′ and 5′-ATC CGG TGA GAT CTA CAC TGA CAA-3′ and then subcloned into pCMV-Myc (Clontech) or pcDNA4-HisMax (Invitrogen). Similar methods were used to clone Atg5 by using the primer set 5′-GCG TCG ACT CCT GGA AGA ATG ACA GAT-3′ and 5′-C GCC TCG AGT CCT TCA ATC TGT TGG CT-3′; Atg5 K130R using 5′-CA TGT ATG AGA GAA GCT GAT GCT-3′ and 5′-AGC ATC AGC TTC TCT CAT ACA TGA-3′ in addition to the primers for wild-type Atg5; and Atg7 using 5′-GCGA ATT CAA GAA ATA ATG GCG GCA GC-3′ and 5′-CAC AGC GGC CGC ATC TCA GAT GGT C-3′. All Myc and His-tagged constructs were confirmed by sequencing. The FLAG-tagged Sirt1 expression vector was obtained from Addgene.
Autophagy Assays.
The expression construct encoding the fusion protein GFP-LC3 was obtained by PCR amplification of a human brain cDNA library using the LC3–Atg8-specific primers 5′-CCG GAA TTC CCG ACC ATG CCG TCG-3′ and 5′-CGC GGA TCC TGC ACT GAC AAT TTC-3′. The fragment was subsequently cloned in-frame by using the pEGFP-C2 expression vector (Clontech) to create the chimeric molecule.
Autophagy was visualized in HCT116 and HeLa cells and MEFs by transfection of pGFP-LC3 followed by analysis with confocal microscopy (LSM 510 Meta; Zeiss). For most experiments, the number of GFP-LC3 punctae was assessed from at least six random high-power fields. A single random Z-section was used for each field, and a minimum of 30 cells per sample were counted with triplicate samples per condition per experiment. A LC3 punctae was considered to be a totally isolated GFP-positive structure greater than ≈1 μm in diameter. Unless stated otherwise, autophagy was induced by the replacement of full growth medium with Hanks' buffered saline solution (HBSS; Invitrogen) for 2 h in HeLa cells, 4 h in MEFs, or by treating cells with 0.2 μM rapamycin (Sigma) in the presence of complete growth medium for the same period. Results except where stated represent one experiment that is representative of at least three similar experiments. In general, although the absolute number of LC3 punctae varied between experiments, the switch to starvation medium resulted in an approximate 3-fold increase in HeLa cells and a 4- to 7-fold increase in LC3 punctae in wild-type MEFs.
Immunoprecipitation and Immunoblotting.
For immunoprecipitation analysis, HeLa or MEF cell lysates (2 mg) were mixed with antibodies (2 μg) at 4°C overnight followed by the addition of 60 μl of protein G–Sepharose (Amersham Biosciences) for 2 h at 4°C. Immune complexes were washed five times with lysis buffer [50 mM Tris (pH 7.4), 1% Triton X-100, 0.5% Nonidet P-40, 150 mM NaCl, protease, phosphatase inhibitor mixture (Sigma), and 10% (vol/vol) glycerol]. After boiling in 2× sample buffer, samples were subjected to SDS/PAGE. After transfer to nitrocellulose, membranes were immunoblotted with the indicated primary antibodies, including anti-HA (11867423001; Roche Applied Science), Sirt1 (07-131; Upstate), Atg7 (XW-7984; ProSci), GFP (632375; Clontech), actin (A2103; Sigma), c-myc (SC-40; Santa Cruz Biotechnology), Atg8–LC3 (NB100-2331; Novus), p62 (GP62; Progen), phospho-AMPKα (2535; Cell Signaling Technology), AMPKα (2532; Cell Signaling Technology), and acetyllysine (9441; Cell Signaling Technology) followed by the appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology or Pierce). Bands were visualized by enhanced chemiluminescence (Pierce). Where indicated, cells treated with 10 mM nicotinamide were harvested 14 h after exposure to the sirtuin inhibitor.
For analysis of in vivo endogenous Atg7 acetylation, embryonic tissues were harvested at E15–E16, and neonatal mouse tissues were harvested from wild-type or Sirt1−/− littermate mice within the first several hours after birth. Whole tissues were flash frozen and subsequently thawed in lysis buffer for 1 h at 4°C. These tissue lysates were then disrupted with a sonicator XL 2020 (Misonix, Inc.) for 5 s at 20% amplitude and subsequently clarified by centrifugation at 14,000 rpm (30 min at 4°C) before further processing as described above. Atg7 acetylation was assessed directly by immunoprecipitation of 1 mg of tissue protein lysate with an acetyllysine antibody (9441; Cell Signaling Technology) and subsequent Western blotting for Atg7. To observe the conversion of LC3-I into LC3-II, HCT116 cells were incubated in HBSS in the presence of 10 μg/ml pepstatin A (Sigma) and 10 μg/ml E-64d (Sigma) for 6 h as described in ref. 37.
Transfections and Immunofluorescence.
HeLa cells were routinely transfected with Effectene (Qiagen) and MEFs with FuGENE 6 (Roche Applied Science) or Effectene according to the manufacturer's recommendations. The HA-tagged Sirt1 wild-type or deacetylase-inactive mutant (HY) and myc-Sirt3 have been described (32). For the autophagy assays, cells were plated on chamber slides and then cotransfected with 0.8 μg of a Sirt1 expression construct or corresponding empty vector along with 0.4 μg of the pGFP-LC3 construct. Cells were visualized 24 h after transfection. For analysis of acetylation of autophagy proteins, 10-cm dishes of either HeLa cells or MEFs were transfected with 4 μg of the indicated epitope-tagged Atg constructs and analyzed 24 h after transfection in the presence of full medium or after starvation.
For Sirt1 knockdown experiments, HeLa cells were transfected with either 400 nM siRNA directed against SIRT1 (Dharmacon) or a corresponding control nontargeting RNAi along with 0.8 μg of pGFP-LC3. In these knockdown experiments, cells were transfected by using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Cells were analyzed 36 h after RNAi transfection.
For immunofluorescence, MEFs cells were grown on Lab-tek II (Nunc) dishes and transfected with GFP-LC3 with or without Sirt1 as above. Twenty-four hours later, cells were washed with ice-cold PBS and then fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were subsequently permeabilized in 0.5% Triton X-100. To detect transfected Sirt1, nonspecific sites were first blocked by incubating cells with a solution of 5% BSA and 0.05% Triton X-100 in PBS for 1 h. The cells were next incubated with the primary antibodies against the HA epitope (1:100 in 5% BSA) overnight at 4°C. Afterward, cells were washed with PBS and incubated with Alexa Fluor 594 goat anti-mouse IgG (1:250; Molecular Probes) for 1 h at room temperature. Images were recorded by using a confocal laser scanning microscope (510 Meta; Zeiss). For colocalization experiments, HeLa cells were grown on Lab-tek II and transfected with 0.5 μg of FLAG-SIRT1 and Myc-ATG7. Twenty-four hours later, cells were washed, fixed, and blocked as above. The cells were then incubated with primary antibodies against Sirt1 and Myc (1:100 in 5% BSA) overnight at 4°C followed by secondary Alexa Fluor 594 goat anti-mouse IgG and Alexa Fluor 488 goat anti-rabbit IgG (1:100; Molecular Probes) antibodies.
Assays for in Vitro Deacetylase Activity.
For the Sirt1-dependent in vitro deacetylase reaction, acetylated substrate was prepared by transfecting HeLa cells with expression constructs encoding either His-Atg8 or His-Atg7. We used 4 μg of each construct per 10-cm dish. Twenty-four hours after transfection, cells were harvested in lysis buffer, and 2 mg of transfected protein lysates was bound to Ni–nitrilotriacetic acid–agarose beads (Qiagen). In some experiments, to maximize levels of substrate acetylation, 10 mM nicotinamide was added for the last 14 h before cell harvest. Sirt1 protein was obtained by transfecting a FLAG-tagged wild-type or deacetylase-inactive Sirt1 expression construct (4 μg) into HeLa cells. The epitope product was purified by using an anti-FLAG M2 affinity gel (Sigma) and subsequently eluted competitively with five one-column volumes of a solution containing 100 μg/ml FLAG peptide (Sigma) in TBS [10 mM Tris·HCl, 150 mM NaCl (pH 7.4)].
The deacetylase reaction was performed in the presence or absence of Sirt1 or NAD (10 mM). Equal amounts of acetylated substrate were added to each tube, and the deacetylation reaction was performed for 2 h at 37°C in assay buffer [50 mM Tris·HCl, 1 mM MgCl2, and 150 mM NaCl (pH 8.0)] by using, where indicated, ≈300 ng of purified Sirt1 in a total reaction volume of 50 μl. The deacetylation of each protein substrate was subsequently detected after SDS/PAGE, transfer to nitrocellulose, and immunodetection with an anti-acetyllysine antibody as above.
Pyruvate Treatment and in Vivo Analysis.
Heterozygous mating pairs of Sirt1+/− mice were randomly assigned to either PBS or ethyl pyruvate (Sigma) injections. For the last 6–8 days before birth, pregnant mothers received either daily PBS or ethyl pyruvate (40 mg/kg) injection with a volume of 0.4 ml per dose. Ethyl pyruvate was initially dissolved in ethanol and then diluted in PBS. After birth, unless indicated, mice were allowed to feed if they were capable, and survival was assessed in hours after birth. Only live births whose timing of delivery could be accurately assessed were included in this analysis.
For electron micrographs, tissues were first washed several times in PBS, fixed in PBS-buffered 2.5% glutaraldehyde (Sigma), postfixed in 0.5% osmium tetroxide, and embedded into Spurr's epoxy resin (Ladd Research Industries). Ultrathin sections were stained with uranyl acetate–lead citrate and viewed with a Phillips CM10 transmission electron microscope.
Supplementary Material
ACKNOWLEDGMENTS.
We are grateful to Ilsa Rovira for help in the preparation of this manuscript and to C. Combs and D. Malide (National Institutes of Health) for the confocal images. This work was supported by National Institutes of Health Intramural Funds and a grant from the Ellison Medical Foundation (to T.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712145105/DC1.
References
- 1.Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 2.Shintani T, Klionsky DJ. Autophagy in health and disease: A double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: Cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 4.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 5.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 7.Guarente L, Picard F. Calorie restriction: The SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 8.Haigis MC, Guarente LP. Mammalian sirtuins: Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 9.Michan S, Sinclair D. Sirtuins in mammals: Insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 11.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 14.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 15.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 20.Das UN. Is pyruvate an endogenous anti-inflammatory molecule? Nutrition. 2006;22:965–972. doi: 10.1016/j.nut.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Fink MP. Ethyl pyruvate: A novel anti-inflammatory agent. J Intern Med. 2007;261:349–362. doi: 10.1111/j.1365-2796.2007.01789.x. [DOI] [PubMed] [Google Scholar]
- 22.Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Roso A, et al. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol. 2003;38:519–527. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- 24.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 25.Hars ES, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 26.Lin SJ, Defossez PA, Guarente L. Requirement of NAD, SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 27.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 28.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 29.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 30.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 31.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 32.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 33.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 34.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.