Abstract
The chromalveolates form a highly diverse and fascinating assemblage of organisms, ranging from obligatory parasites such as Plasmodium to free-living ciliates and algae such as kelps, diatoms, and dinoflagellates. Many of the species in this monophyletic grouping are of major medical, ecological, and economical importance. Nevertheless, their genome evolution is much less well studied than that of higher plants, animals, or fungi. In the current study, we have analyzed and compared 12 chromalveolate species for which whole-sequence information is available and provide a detailed picture on gene loss and gene gain in the different lineages. As expected, many gene loss and gain events can be directly correlated with the lifestyle and specific adaptations of the organisms studied. For instance, in the obligate intracellular Apicomplexa we observed massive loss of genes that play a role in general basic processes such as amino acid, carbohydrate, and lipid metabolism, reflecting the transition of a free-living to an obligate intracellular lifestyle. In contrast, many gene families show species-specific expansions, such as those in the plant pathogen oomycete Phytophthora that are involved in degrading the plant cell wall polysaccharides to facilitate the pathogen invasion process. In general, chromalveolates show a tremendous difference in genome structure and evolution and in the number of genes they have lost or gained either through duplication or horizontal gene transfer.
Keywords: gene gain, gene loss
The chromalveolates (1) account for about half of the recognized species of protists (e.g., apicomplexans and ciliates) and algae (e.g., kelps, diatoms, and dinoflagellates), and are of great ecological and economical importance. They are further subdivided into six diverse groups (2, 3). The Alveolata contain the parasitic apicomplexans, the plastid-less ciliates, and the dinoflagellate algae, whereas the Chromista include the cryptophyte, haptophyte, and stramenopile algae.
Despite general shared features, the alveolates have greatly diverged in many respects, including host specificities, tissue tropisms, and the requirement of multiple hosts (4). The obligate intracellular apicomplexans contain several protozoan pathogens that provoke severe diseases in mammals, including humans. Infections by Plasmodium falciparum, which causes human malaria, and Theileria parva and T. annulata, which are responsible for great economic losses in cattle in Africa, have profound medical, social, and economic effects (5, 6). Others such as Toxoplasma gondii, Cryptosporidium parvum, and C. hominis are primarily health threats in HIV+/AIDS and immunosuppressed populations (7). Ciliates are unique among unicellular organisms in that they separate germline and somatic functions (8).
The haptophytes and stramenopiles share characteristics such as tubular mitochondrial cristae, similar storage products, and fucoxanthin that suggest a specific relationship between these taxa (9). Stramenopiles include autotrophic and heterotrophic species that may differ enormously in their morphology and mode of life. Members of this group occupy key ecological niches in marine environments. For example, the diatoms constitute the most abundant component of marine plankton. Stramenopiles have also succeeded in occupying terrestrial environments as plant pathogens. For example, Phytophthora plant pathogens attack a wide range of agriculturally and ornamentally important plants (10). Phytophthora sojae costs the soybean industry millions of dollars each year (11). In California and Oregon, a newly emerged Phytophthora species, P. ramorum, is responsible for a disease called sudden oak death that affects not only the live oaks that are the keystone species of the ecosystem but also a large variety of woody shrubs that inhabit the oak ecosystems, such as bay laurel and viburnum (12).
All extant species of the chromist and alveolate groups are considered to have evolved from an ancestor that contained a red algal endosymbiont (the “chromalveolate” hypothesis), which later gave rise to the plastids found in most of the chromalveolates. Although there is ample molecular evidence that suggests that the alveolates and stramenopiles form a monophyletic group (13–17), the phylogenetic position of the cryptophytes and haptophytes, and their inclusion in the chromalveolate “superclade” is still debated. Recently, however, Hackett and colleagues (18) found additional support for the monophyly of the cryptophytes and haptophytes and their sister relationship to the chromalveolates based on a multigene dataset of nuclear genes.
Here, we have analyzed and compared 12 diverse chromalveolate species for which whole-sequence information is available. The aims of this analysis were to study the genome evolution for these organisms and to document gene loss and gene gain along the different chromalveolate lineages. Furthermore, we investigated the extent to which these events can be correlated with the differences in lifestyle between the various species.
Results and Discussion
A Parsimonious Scenario of Gene Loss and Gene Gain in the Chromalveolates.
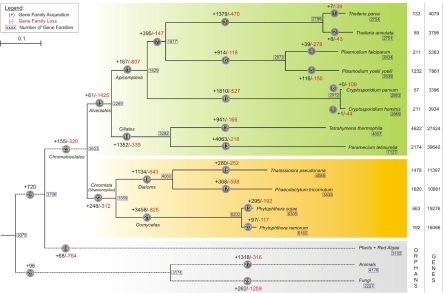
To study gene family evolution in this eukaryotic group of organisms, we assembled the publicly available protein sequences of 12 chromalveolate and eight outgroup species. This resulted in a dataset of 306,696 proteins that were grouped, based on sequence similarity and Markov clustering (see Methods), into 32,887 different multigene families and 58,331 orphans—i.e., genes without homology to others in the dataset. For all gene families, phylogenetic profiles were constructed that reflect the presence or absence of a particular gene family in a particular species. To make sure that an apparent gene family absence is not due to errors in annotation, or to increased evolutionary rates hiding “clear” homology, additional analyses were performed to see whether these two phenomena could have seriously biased our results [see supporting information (SI) Methods]. Based on these analyses, it seems that both annotation errors and increased evolutionary rates probably only had a very minor effect on our results (SI Fig. 4 and SI Table 1). To be able to study gene family evolution by means of phylogenetic profiles, information about the phylogenetic relationships between the different organisms in the dataset is required. Therefore, we have used a concatenated set of single copy core genes (20 families) to resolve the phylogenetic relationships between the 12 chromalveolates. After extracting these 20 families, multiple alignments were created and manually improved, and finally concatenated into one large multiple alignment of 5,360 aa (see SI Methods). The inferred phylogenetic tree is shown in Fig. 1.
Fig. 1.
A parsimonious scenario of gene loss and gene gain in the chromalveolates. At every time point (gray circles) in the chromalveolate tree (distance method, Poisson correction, NJ, all nodes 100% BS-supported), gene loss (red) and gene acquisition (black) was counted based on the Dollo parsimony principle. The inferred ancestral gene sets are shown in boxes. Next to every species, the number of orphans and number of predicted genes is indicated. The part of the tree drawn in solid black lines is drawn to scale; the part drawn in dashed lines is not. Detailed information on which genes and gene families have been gained or lost at certain points in chromalveolate evolution can be obtained from a clickable map of this figure at our web site (http://bioinformatics.psb.ugent.be).
Combining both the species tree and phylogenetic profiles, we were able to reconstruct a parsimonious scenario of chromalveolate genome evolution by mapping different gene loss and gain events onto the branches of the phylogenetic tree. Furthermore, to delineate the minimal gene set for the different ancestral nodes, the Dollo parsimony approach was applied (19), which assumes gene loss to be irreversible. In other words, a gene (family) can be lost independently in several evolutionary lineages but cannot re-evolve (except in a noticeably different form). By applying this methodology to all phylogenetic profiles (which were treated as strings of 0 and 1; i.e., absence resp. presence of the given gene family in the given species), we could associate every branch of the chromalveolate tree (defined as time points or TPs) with both gene family loss and gene family acquisition (Fig. 1). For more information regarding the performance of the Dollo parsimony principle on our data, see SI Methods and SI Fig. 5. To link the different evolutionary events (i.e., loss and gain) with information about the molecular function or biological process the corresponding gene families are involved in, the Gene Ontology (GO) vocabulary was used (20). Detailed information on which genes and gene families have been gained or lost during chromalveolate evolution can be found at http://bioinformatics.psb.ugent.be.
One of the most notable observations is the occurrence of massive gene loss in the branches leading to the obligate intracellular Apicomplexa [i.e., TP11 and TP15 (Fig. 1)]. This massive gene loss can be linked to the transition of a free-living to an obligate intracellular lifestyle (21). In addition, Phytophthora species seem to have lost a very large number of gene families (825 families, TP21). Although this observation seems surprising given their large number of genes [>16,000 (Fig. 1)], this massive loss is again explained by the lifestyle of these successful plant pathogens. When we consider the functions of these families in both lineages (SI Table 2, TP11, TP15, and TP21), we indeed observe that these organisms have lost many gene families involved in general processes such as amino acid metabolism, carbohydrate metabolism, and lipid metabolism.
Interestingly, the continuous loss of gene families (i.e., loss at different, successive TPs) in the Apicomplexa lineage is compensated by a similarly continuous acquisition of novel gene families in this lineage. As could be expected from the lifestyle of the Apicomplexa, we observed a significant gain (q-value <0.05) of gene families involved in pathogenesis in the ancestor of the Apicomplexa (SI Table 3, TP11). Similarly, gain of pathogenesis genes was also observed (q-value <0.05) at other TPs within the Apicomplexa lineage (TP4, TP6, and TP7). We also noted a significant gain of gene families with metallopeptidase activity at TP6, TP7, and TP11. For the malaria parasites (TP6), this can be explained by the fact that they hydrolyze host proteins (hemoglobin), using acid cysteine, aspartic, and metalloproteases. Hemoglobin is thereby processed into individual amino acids, which are subsequently used for their own protein synthesis (22). However, other Apicomplexa have also gained these metalloproteases, which suggests that they also are able to hydrolyze their host proteins and can afford loosing gene families involved in amino acid biosynthesis (SI Table 3, TP11).
Phytophthora, unlike the apicomplexans, seems to have acquired a huge number of novel gene families (3,458 families, TP21). We found no significant bias toward certain biological processes or functions for these newly acquired gene families, most probably because of the lack of functional annotation for many of these specific families. However, in Phytophthora ramorum, we observe a significant gain of gene families with potassium channel activity (SI Table 3, TP20), which can be explained by the fact that potassium ions play a key role in the locomotion and encystment of the Phytophthora zoospores (23). The process of encystment is directly correlated with the oomycete pathogenicity because this process allows the zoospore to perpetuate its life cycle at the expense of the infected organism.
The ciliates, which have by far the largest number of predicted genes (Fig. 1), have acquired many new gene families. Both ciliates show an enrichment of gene families with protein kinase activity and phosphotransferase activity (SI Table 3, TP12 and TP13), which is most likely correlated with the important process of exocytosis within the ciliates, because reversible protein phosphorylation is essential in controlling exocytosis (24–26). Compared with the large number of novel gene families, the number of gene families in ciliates that have been lost is quite limited (339 families, TP14).
All chromalveolates and plants are bikonts (i.e., ancestrally biciliate, or having two cilia), this in contrast to the Metazoa and Fungi, which are unikonts. However, during evolution, in all major eukaryote groups except the ciliates, different organisms have lost cilia and became secondary uniciliates (27). Our analyses indicate that among the chromalveolates, the diatoms, but also the Apicomplexa have also lost gene families responsible for cilium biogenesis (SI Table 2, TP18 resp. TP11), which is in concordance with the Apicomplexa lacking cilia and (at least the centric) diatoms (such as Thalassiosira) that became secondary uniciliates (27).
We also investigated the fraction of the lineage-specific gene families for the different major chromalveolate lineages—i.e., the alveolates (Fig. 1, TP15), Chromista (TP22), and Apicomplexa (TP11)—that has been maintained in all extant species (i.e., lineage-specific core gene families). These gene families probably contain the genes responsible for the shared, specific features of these lineages. However, for the alveolates, only 5 gene families of the 61 (or 8%) are still present in all extant species. These genes play a role in protein amino acid ADP-ribosylation, lipid metabolism, and electron transport. Within the Apicomplexa, 37 families (22%) of the 167 acquired families have been preserved. A significant fraction [q-value of 0.16, p-value <0.05 (data not shown)] of these gene families is involved in pathogenesis, which suggests that all Apicomplexa share a general set of pathogenicity genes. For the Chromista, 83 gene families (33%) are still present in all extant species, with a significant enrichment in prostaglandin metabolism and regulation of cell redox homeostasis [q = 0.37, p < 0.05 (data not shown)]. Finally, we observed a gain of 155 gene families in the ancestors of the chromalveolates (TP23) of which only two have survived in all extant species but, unfortunately, without informative gene descriptions available (i.e., hypothetical proteins).
The parsimony analysis also involves explicit reconstruction of the gene sets of ancestral chromalveolate genomes. Under this approach, 3,633 gene families were assigned to the last common ancestor of the chromalveolate supergroup (TP23). The last common ancestor of the Apicomplexa, in contrast, had the smallest number of gene families (i.e., 1,629), which implies that genome reduction had already occurred early in the evolution of these parasites.
Gene Family Expansions.
The results discussed so far focused on the presence or absence of gene families but not on the actual gene family sizes. Comparing the size of all gene families makes it possible to determine whether the variations in total gene number between the different species can, at least partially, be explained by gene amplification in certain gene families. Therefore, we also investigated lineage-specific gene family expansions during chromalveolate evolution. First, the 25% most variable multigene families were identified (see Methods). To compare these different phylogenetic profiles, the gene copy numbers were transferred into z-scores, which are expressed as standard deviations from their means (i.e., mean copy number over the species containing the gene family). This way, positive z-scores indicate gene copy numbers that are greater than the corresponding mean gene family size. To be able to observe different expansion patterns, these z-score profiles were hierarchically clustered. We removed the gene families that were only expanded in the outgroups and the gene families only expanded in one or both of the ciliates because their massive duplication events (28, 29) reduce the expansion signal of the other chromalveolates. This analysis yielded 119 gene families (see Methods) (Fig. 2). Again, detailed information on these gene families can be found at http://bioinformatics.psb.ugent.be.
Fig. 2.
Hierarchical clustering of species/lineage-specific gene family expansions. The gene family IDs are shown at the left, and the corresponding gene descriptions are shown at the right. The tree at the top reflects the chromalveolate phylogeny of Fig. 1. The blue and yellow scale (based on z-scores) shows that, given a certain gene family and organism, the gene family size is substantially smaller resp. larger than the mean gene family size. Hence, yellow little blocks reflect gene family expansions. The gene families that are described in more detail in the text are indicated by white (Plasmodium), yellow (Phytophthora), and blue (Theileria) arrows. This figure is also available as a clickable map at our web site (http://bioinformatics.psb.ugent.be).
As can be observed in Fig. 2, many of the z-score profiles show Phytophthora-specific expansions. Strikingly, many of these expanded families are involved in degrading the plant cell wall polysaccharides, which facilitates the pathogen invasion process. For example, the xylosidases (Fig. 2, families 259 and 842), which hydrolyze the xyloglucan network, help the physical penetration process by sufficient loosening of the host cell wall (30). The polygalacturonases (family 639), on the other hand, are involved in degrading the pectin backbone embedded in the plant cell wall. Similarly, the expanded endoglucanases (family 473) and exoglucanases (family 747) are involved in the degradation of cellulose. Moreover, the latter two enzymes are shown to be differentially expressed during the life cycle of the potato-pathogen Phytophthora infestans, as well as during the infection of potatoes (31). Additionally, the β-(1, 3)-endoglucanases are cell wall-associated in P. ramorum, suggesting an important role for this gene family in the interaction with the host plants (32). Finally, in P. sojae, these genes were found to be specifically expressed during infection (33).
Another group of gene families that are expanded in the Phytophthora genomes are families involved in the protection against oxidative stress, such as for example, the glutathione S-transferases (Fig. 2, families 3403 and 990), glutaredoxin (family 3430), and the catalase-peroxidase (family 220). These families were classified as putative pathogenicity genes by Torto-Alalibo and colleagues (33). Finally, the expansion of two other gene families can be explained by their role in the process of pathogenesis: first, the glucokinases (family 2414) that are expressed during the first 6 h of interaction with its host Glycine max (34), and second, the lipases (family 845), which are also shown to be potentially infection related (11).
In Plasmodium spp., three families are specifically expanded (Fig. 2, families 1526, 3, and 1997). One family, which is an Apicomplexa-specific gene family of trophozoite antigens R45, is only expanded in P. falciparum. This has also been observed by Schneider and Mercereau-Puijalon (35), who suggested that expansion of this family might contribute to P. falciparum virulence. The second gene family, a family of erythrocyte membrane-associated antigens, is expanded in both Plasmodium species and also plays a role in host–parasite interaction. Additionally, previous analyses by Hall et al. (36) demonstrated that these genes are among those with the highest rates of positive selection. Moreover, the set of genes that show the most positive selection includes many genes that one would expect to play a role in host–parasite interaction, which indicates that these signs of positive selection might be the result of the selective pressure from the host adaptive immune response. Therefore, we assume that these two families were expanded to augment the antigen variability and thereby escape recognition by the immune system of the host. Finally, a family of papain cysteine proteases seems also to be expanded in both Plasmodium species. These proteases, also known as falcipains, are hemoglobinases and potential drug targets (37). They hydrolyze hemoglobin during the erythrocytic stage of infection and thereby provide amino acids for parasite protein synthesis (38).
In Theileria spp., three families have specifically expanded (Fig. 2, families 111, 1573, and 1700). These two parasites are able to transform the cells they infect, which is unique among eukaryotic pathogens, and this capacity is also present in only some members of the genus (39). Proteins that function in lipid metabolism (e.g., the choline kinase genes, family 1700) were identified as one of the possible transformation candidates (40). In agreement with our results, Pain et al. (40) observed that these genes are present in high copy numbers compared with other apicomplexans.
To evaluate whether gene families have expanded mainly through tandem, block, or dispersed duplications, and to identify the number of duplicated blocks and tandem repeats in the chromalveolate genomes (SI Table 4), our previously developed i-ADHoRe software was used (see Methods). After analyzing the location of the genes of the above-mentioned expanded gene families in the genome, we observed that most of them are located in tandem clusters and thus are probably the result of more recent tandem duplications (SI Table 5). However, one family in Plasmodium, the trophozoite antigen family (SI Table 5, family 1526), has been expanded both by tandem and block duplications.
We also investigated the extent of gene family expansions at a genome-wide level. Therefore, the average gene family size was calculated for all organisms used in the current study and converted into a z-score. In Fig. 3, we observe that the ciliate Paramecium, which has the highest number of predicted genes (SI Fig. 6), also has the largest average gene family size. Running i-ADHoRe revealed that ≈95% (SI Table 4) of the genome is located in duplicated blocks. This is in agreement with previous analyses, which proved multiple rounds of whole-genome duplication in this ciliate (28). The other ciliate Tetrahymena has the second largest average gene family size. Eisen et al. (29) did not find any evidence for whole-genome duplications in this organism but reported extensive numbers of tandemly duplicated genes. However, our analysis indicates that, apart from those tandem duplications, also almost 40% of the genome is located within duplicated blocks (SI Table 4), which seems to point to large-scale gene duplication events in this organism as well.
Fig. 3.
Differences in average gene family size for different eukaryotes. For all organisms included in the current study, the average gene family size, converted into a z-score, is plotted on the ordinate. See text for details.
The second taxonomic group with large average gene family sizes is the group of the Viridiplantae containing Arabidopsis and rice, both having a history of genome duplications and family expansions as discussed in detail previously (41, 42). Next in line are the Metazoa, then the oomycetes (i.e., the Phytophthora spp.), Plasmodium spp., diatoms, Fungi, and finally the other Apicomplexa. This order is in accordance with the number of genes in the corresponding genomes. To our knowledge, so far no evidence was found for whole-genome duplications within the oomycetes, but our analyses indicate the existence of a large number of duplicated blocks (especially in P. sojae), as well as a high fraction of tandem duplications (SI Table 4). This observation will need some further study. In addition, the pathogen Plasmodium falciparum has a considerable part of its genome located in duplicated blocks [i.e., 21% (SI Table 4)]. However, whereas the duplicated blocks in the other genomes are randomly distributed over the whole genome, in P. falciparum they seem to be preferentially located at the telomeres and occasionally at the centromeres (data not shown).
Origin of the Chromalveolate Orphans.
Besides the set of genes that could be assigned to gene families, a substantial number of genes were uncovered that lack homologs in the eukaryotic data set. These so-called “orphans” represent de novo species-specific genes, falsely predicted gene models, or genes acquired through horizontal gene transfer (HGT) from other species (e.g., Bacteria, Viruses). To investigate the extent to which these genes have been obtained through HGT, all orphan genes and putative species-specific families (SSFs) were compared against a nonredundant protein database. After selection of highly similar sequences [using a relative bit score threshold (see SI Methods)], taxonomic information for all homologs was retrieved.
One first observation is that the orphans of four species (namely both Theileria species, Cryptosporidium parvum, and Plasmodium falciparum) do not show any homology with other genes at all. Whereas a small set of orphans (between 1 and 45) from Plasmodium yoelii yoelii, Cryptosporidium hominis, both Phytophthora species, and ciliates shows homology with bacterial species from a limited number of phyla, for both diatoms >100 orphan genes match a wide range of phyla (data not shown). In general, the fraction of orphans homologous to sequences from Archaea or Viruses is much smaller compared with genes showing homology with Bacterial genes (mostly Proteobacteria), suggesting that HGT between Bacteria and chromalveolates is most common. As for orphan genes, in most species only a minority of the SSFs have bacterial homologs, with Thalassiosira as an exception, where 42% (28/67) of all SSFs again match a wide range of phyla (data not shown).
We set out to document the genome evolution of a wide diversity of chromalveolate organisms. To this end, we inferred, by using a Dollo parsimony approach, both gene loss and gain events that have occurred along the chromalveolate tree. The great differences in genome content reflect the ancient history of the chromalveolate assemblage. Indeed, starting from an estimated 3,600 gene families in the ancestor of all chromalveolates, the number of gene families in extant chromalveolates ranges from <2,500 to >7,000, while the number of predicted genes ranges from ≈3,800 to almost 40,000. Extensive gene loss and, contrarily, gene family expansions explain the large differences in lifestyle but also many specific adaptations and the high degree of specialization some organisms within this eukaryotic “superkingdom” have undergone.
Methods
Gene Family Clustering.
To delineate gene families, a similarity search was performed (all-against-all BLASTP; E-value cutoff E-5) with all proteins from 12 different chromalveolate species, plus the protein sequences from eight outgroup organisms. For more information regarding the dataset, see SI Methods. Gene families were constructed with MCLBLASTLINE (inflation factor of 2.0) based on the BLASTP analyses. The performance of the MCLBLASTLINE protein clustering method was evaluated and compared with two other clustering methods (see SI Methods). We also evaluated the possible effects of errors in annotation and fast-evolving genes on gene family clustering (see SI Methods).
Functional Annotation of Genes and Gene Families.
Genes and gene families were functionally annotated by using Gene Ontology (GO) (20). In a first step, all proteins were annotated by using Blast2GO to assign the proteins to GO categories (43). Proteins mapped to a particular GO category were also explicitly included into all parental categories. GO annotation per family was obtained by listing the GO labels for all of the genes of that family. A weight, equal to the percentage of genes with GO annotation within the same subcategory (molecular function, cellular component, biological process) that carried this label, was attached to all of the GO labels. Only GO labels with a weight greater than 30% were considered as representative for the family. GO labels occurring in fewer than 10 gene families were discarded. The statistical significance of functional GO enrichment was evaluated by using the hypergeometric distribution, whereas multiple hypotheses testing was done by using FDR (44).
Loss and Acquisition of Gene Families.
The parsimonious evolutionary scenario, which included the loss and acquisition of gene families mapped onto the branches of the chromalveolate phylogenetic tree, was inferred by using the DOLLOP program of the PHYLIP package (45). The DOLLOP program is based on the Dollo parsimony principle, which assumes irreversibility of character loss (19) and has been used in similar studies describing the evolution of KOGs along the eukaryotic phylogenetic tree (46), and studying the intron gain and loss in paralogs (47).
Species/Lineage-Specific Gene Family Expansions.
For all gene families (orphans and SSFs excluded), the mean gene family size and standard deviation were calculated. The 25% most variable profiles, based on the standard deviations, were extracted. The matrix of these profiles was transformed into a matrix of z-scores to center and normalize the data. Subsequently, these profiles were hierarchically clustered (complete linkage clustering) by using Pearson correlation as a distance measure. The clustering and visualization was done by using Genesis (48). A description was added to all families based on the most frequently occurring gene descriptions in the corresponding gene family. Gene families only expanded in the outgroup were removed, in addition to those expanded only in one or both of the ciliates because of their massive gene duplication. Transposon-derived gene families were removed because the distribution of such families is likely to be a consequence of whether the gene models were derived from a repeat-masked genome sequence and therefore may be artifactual.
Detection of Block Duplications and Tandem Repeats.
The detection of tandem and block duplications within the 12 chromalveolate genomes was done with i-ADHoRe (49). After removal of transposable elements, the MCL pairs were used to define the homologous relations between the genes of every genome, which served as input for the i-ADHoRe algorithm. The following parameters were used in the i-ADHoRe analysis: gap size of 20 genes, q-value of 0.8, probability cutoff at 0.01, minimum of three homologs to define a block, and higher-level multiplicon detection disabled (level 2 only).
Origin of the Chromalveolate Orphans.
For the proteins that had no homologs in our dataset (i.e., the orphans or one gene representative per SSF), a similarity search (BLASTP, E-value cutoff E-5) was performed against the nonredundant protein database at NCBI (release 160.0). Valid hits were extracted by using a relative bit score (BS) threshold and taxonomic filtering of putative homologs (see SI Methods). Similar results were found by applying different relative BS thresholds [range 0.6–0.9 (data not shown)].
Supplementary Material
ACKNOWLEDGMENTS.
We thank Tine Blomme and Steven Maere for helpful discussions and Thomas Van Parys for technical assistance. C.M. is indebted to the Institute for the Promotion of Innovation by Science and Technology (Flanders, Belgium) for a predoctoral fellowship. K.V. is a postdoctoral fellow of the Research Foundation-Flanders. The sequence data of Phaeodactylum tricornutum were produced by the U.S. Department of Energy Joint Genome Institute www.jgi.doe.gov.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712248105/DC1.
References
- 1.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 2.Archibald JM, Keeling PJ. Recycled plastids: A ‘green movement’ in eukaryotic evolution. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- 3.Cavalier-Smith T. In: Organelles, Genomes and Eukaryotic Evolution. Hirt RP, Horner D, editors. London: Taylor & Francis; 2004. pp. 71–103. [Google Scholar]
- 4.Templeton TJ, et al. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billiouw M, et al. Theileria parva epidemics: A case study in eastern Zambia. Vet Parasitol. 2002;107:51–63. doi: 10.1016/s0304-4017(02)00089-4. [DOI] [PubMed] [Google Scholar]
- 6.Nahlen BL, Korenromp EL, Miller JM, Shibuya K. Malaria risk: Estimating clinical episodes of malaria. Nature. 2005;437:E3. doi: 10.1038/nature04178. discussion E4–E5. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan JE, Jones JL, Dykewicz CA. Protists as opportunistic pathogens: Public health impact in the 1990s and beyond. J Eukaryot Microbiol. 2000;47:15–20. doi: 10.1111/j.1550-7408.2000.tb00004.x. [DOI] [PubMed] [Google Scholar]
- 8.Jahn CL, Klobutcher LA. Genome remodeling in ciliated protozoa. Annu Rev Microbiol. 2002;56:489–520. doi: 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya D, et al. Algae containing chlorophylls a+c are polyphyletic: Molecular evolutionary analysis of the Chromophyta. Evolution (Lawrence, Kans) 1992;46:1801–1817. doi: 10.1111/j.1558-5646.1992.tb01170.x. and erratum (1993) 47:986. [DOI] [PubMed] [Google Scholar]
- 10.Erwin DC, Ribeiro OK. Phytophthora Diseases Worldwide. St Paul: Am Phytopathol Soc; 1996. [Google Scholar]
- 11.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 12.Rizzo DM, Garbelotto M, Hansen EM. Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu Rev Phytopathol. 2005;43:309–335. doi: 10.1146/annurev.phyto.42.040803.140418. [DOI] [PubMed] [Google Scholar]
- 13.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 14.Ben Ali A, De Baere R, Van der Auwera G, De Wachter R, Van de Peer Y. Phylogenetic relationships among algae based on complete large-subunit rRNA sequences. Int J Syst Evol Microbiol. 2001;51:737–749. doi: 10.1099/00207713-51-3-737. [DOI] [PubMed] [Google Scholar]
- 15.Dacks JB, Marinets A, Doolittle WF, Cavalier-Smith T, Logsdon JM., Jr Analyses of RNA polymerase II genes from free-living protists: Phylogeny, long branch attraction, and the eukaryotic big bang. Mol Biol Evol. 2002;19:830–840. doi: 10.1093/oxfordjournals.molbev.a004140. [DOI] [PubMed] [Google Scholar]
- 16.Stechmann A, Cavalier-Smith T. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol. 2003;57:408–419. doi: 10.1007/s00239-003-2490-x. [DOI] [PubMed] [Google Scholar]
- 17.Van de Peer Y, De Wachter R. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. J Mol Evol. 1997;45:619–630. doi: 10.1007/pl00006266. [DOI] [PubMed] [Google Scholar]
- 18.Hackett JD, et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 19.Farris J. Phylogenetic analysis under Dollo's law. Syst Zool. 1977;26:77–88. [Google Scholar]
- 20.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Green BR, Cavalier-Smith T. Phylogeny of ultra-rapidly evolving dinoflagellate chloroplast genes: A possible common origin for sporozoan and dinoflagellate plastids. J Mol Evol. 2000;51:26–40. doi: 10.1007/s002390010064. [DOI] [PubMed] [Google Scholar]
- 22.Rosenthal PJ. Hydrolysis of erythrocyte proteins by proteases of malaria parasites. Curr Opin Hematol. 2002;9:140–145. doi: 10.1097/00062752-200203000-00010. [DOI] [PubMed] [Google Scholar]
- 23.Appiah AA, van West P, Osborne MC, Gow NA. Potassium homeostasis influences the locomotion and encystment of zoospores of plant pathogenic oomycetes. Fungal Genet Biol. 2005;42:213–223. doi: 10.1016/j.fgb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Burgoyne RD, Morgan A. Regulated exocytosis. Biochem J. 1993;293:305–316. doi: 10.1042/bj2930305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plattner H. Regulation of membrane fusion during exocytosis. Int Rev Cytol. 1989;119:197–286. doi: 10.1016/s0074-7696(08)60652-x. [DOI] [PubMed] [Google Scholar]
- 26.Greengard P, Valtorta F, Czernik AJ, Benfenati F. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science. 1993;259:780–785. doi: 10.1126/science.8430330. [DOI] [PubMed] [Google Scholar]
- 27.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 28.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 29.Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costanzo S, Ospina-Giraldo MD, Deahl KL, Baker CJ, Jones RW. Gene duplication event in family 12 glycosyl hydrolase from Phytophthora spp. Fungal Genet Biol. 2006;43:707–714. doi: 10.1016/j.fgb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 31.McLeod A, Smart CD, Fry WE. Characterization of 1,3-β-glucanase and 1,3;1,4-β-glucanase genes from Phytophthora infestans. Fungal Genet Biol. 2003;38:250–263. doi: 10.1016/s1087-1845(02)00523-6. [DOI] [PubMed] [Google Scholar]
- 32.Meijer HJ, et al. Identification of cell wall-associated proteins from Phytophthora ramorum. Mol Plant–Microbe Interact. 2006;19:1348–1358. doi: 10.1094/MPMI-19-1348. [DOI] [PubMed] [Google Scholar]
- 33.Torto-Alalibo TA, et al. Expressed sequence tags from Phytophthora sojae reveal genes specific to development and infection. Mol Plant–Microbe Interact. 2007;20:781–793. doi: 10.1094/MPMI-20-7-0781. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Shen G, Wang Y, Zheng X, Wang Y. Identification of Phytophthora sojae genes upregulated during the early stage of soybean infection. FEMS Microbiol Lett. 2007;269:280–288. doi: 10.1111/j.1574-6968.2007.00639.x. [DOI] [PubMed] [Google Scholar]
- 35.Schneider AG, Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: Multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall N, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 37.Sijwali PS, Shenai BR, Rosenthal PJ. Folding of the Plasmodium falciparum cysteine protease falcipain-2 is mediated by a chaperone-like peptide and not the prodomain. J Biol Chem. 2002;277:14910–14915. doi: 10.1074/jbc.M109680200. [DOI] [PubMed] [Google Scholar]
- 38.Olliaro PL, Yuthavong Y. An overview of chemotherapeutic targets for antimalarial drug discovery. Pharmacol Ther. 1999;81:91–110. doi: 10.1016/s0163-7258(98)00036-9. [DOI] [PubMed] [Google Scholar]
- 39.Dobbelaere D, Heussler V. Transformation of leukocytes by Theileria parva and T. annulata. Annu Rev Microbiol. 1999;53:1–42. doi: 10.1146/annurev.micro.53.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Pain A, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- 41.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandepoele K, Van de Peer Y. Exploring the plant transcriptome through phylogenetic profiling. Plant Physiol. 2005;137:31–42. doi: 10.1104/pp.104.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 44.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 46.Koonin EV, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babenko VN, Rogozin IB, Mekhedov SL, Koonin EV. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 2004;32:3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sturn A, Quackenbush J, Trajanoski Z. Genesis: Cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 49.Simillion C, Vandepoele K, Saeys Y, Van de Peer Y. Building genomic profiles for uncovering segmental homology in the twilight zone. Genome Res. 2004;14:1095–1106. doi: 10.1101/gr.2179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.