Abstract
The numerous steps in protein gene expression are extensively coupled to one another through complex networks of physical and functional interactions. Indeed, >25 coupled reactions, often reciprocal, have been documented among such steps as transcription, capping, splicing, and polyadenylation. Coupling is usually not essential for gene expression, but instead enhances the rate and/or efficiency of reactions and, physiologically, may serve to increase the fidelity of gene expression. Despite numerous examples of coupling in gene expression, whether splicing enhances mRNA export still remains controversial. Although splicing was originally reported to promote export in both mammalian cells and Xenopus oocytes, it was subsequently concluded that this was not the case. These newer conclusions were surprising in light of the observations that the mRNA export machinery colocalizes with splicing factors in the nucleus and that splicing promotes recruitment of the export machinery to mRNA. We therefore reexamined the relationship between splicing and mRNA export in mammalian cells by using FISH, in combination with either transfection or nuclear microinjection of plasmid DNA. Together, these analyses indicate that both the kinetics and efficiency of mRNA export are enhanced 6- to 10-fold (depending on the construct) for spliced mRNAs relative to their cDNA counterparts. We conclude that splicing promotes mRNA export in mammalian cells and that the functional coupling between splicing and mRNA export is a conserved and general feature of gene expression in higher eukaryotes.
During gene expression, pre-mRNA undergoes several processing steps in the nucleus, including capping, splicing, and polyadenylation, and the mature mRNA is then exported to the cytoplasm for translation. All of the steps in gene expression are carried out by distinct multicomponent machines, but it is now clear that there is extensive physical and functional coupling among them (1).
Initial studies on the role of splicing in gene expression revealed that splicing of the simian virus 40 (SV40) intron was required for expression of late viral genes (2, 3). Similarly, splicing was required for efficient β-globin expression from SV40 expression vectors (4, 5). Since then, numerous studies have reported a splicing-dependent enhancement of gene expression for multiple genes (6–15). Furthermore, this effect has been observed in Arabidopsis, Drosophila, maize cells, transgenic mice, and a variety of cell lines, indicating that it is a conserved and important feature of gene expression (7, 9, 11, 13, 15). The specific mechanisms by which splicing promotes gene expression have also been extensively investigated, revealing that splicing affects several steps, including transcription, total mRNA levels, and 3′ end processing (1, 16–20). In addition, splicing enhances both translation and localization of mRNA (13, 14, 21).
Although splicing has also been reported to promote mRNA export, this functional connection has been controversial. In early studies, Ryu and Mertz (18) used transfection of mammalian cells followed by biochemical fractionation of the nucleus and cytoplasm to show that SV40 mRNAs generated by splicing were efficiently exported to the cytoplasm, whereas their cDNA counterparts were largely retained in the nucleus. Subsequently, two studies (13, 14) used this approach but instead concluded that splicing is of little or no consequence for the nuclear export of most mRNAs. The issue was addressed again by Tokunaga et al. (22) who used microinjection of RNA into mammalian cell nuclei together with FISH to show that spliced mRNAs were efficiently exported, whereas the corresponding cDNA transcripts were largely degraded. In their study, Tokunaga et al. were unable to distinguish between nuclear retention of the cDNA transcripts followed by degradation or export of the cDNA transcripts followed by degradation in the cytoplasm. The relationship between splicing and mRNA export has also been addressed in Xenopus oocyte microinjection studies. One study (23) indicated that spliced mRNAs were exported more rapidly and efficiently than mRNAs generated from their cDNA counterparts, whereas other studies (24, 25) have led to the view that this is not the case.
In contrast to the conflicting reports on the functional relationship between splicing and mRNA export, biochemical and subcellular localization studies are less controversial and do support a role for splicing in mRNA export. In particular, the conserved TREX complex, which functions in mRNA export, colocalizes with the splicing machinery in nuclear speckle domains (24, 26, 27). Moreover, the TREX complex, which contains the proteins Aly/REF and UAP56, and the multisubunit THO complex, is recruited to mRNA during the splicing reaction (27, 28). Finally, splicing promotes efficient recruitment of the TREX complex to the 5′ end of mRNA, which occurs via an interaction between the cap-binding protein CBP80 and Aly (28). Although Aly and the rest of the TREX complex can be recruited to cDNA transcripts, this recruitment is less efficient than to spliced mRNA and is also cap-independent (28). Thus, it is possible that cDNA transcripts interact with the export machinery via nonspecific binding to the RNA. In addition, a recent study (29) reported that the nucleotide sequence that encodes the signal sequence coding region can function as a nuclear export signal for cDNA transcripts and spliced mRNAs.
In light of the clear connections between the machineries that function in splicing and mRNA export, we sought to reinvestigate whether splicing promotes mRNA export in mammalian cells. To address this question, DNA constructs containing or lacking introns were introduced into mammalian cell nuclei by transfection or microinjection. All constructs contained the CMV promoter and the BGH polyadenylation site. FISH was used to analyze the nucleocytoplasmic distribution of the mRNAs. Significantly, these data revealed that mRNAs generated by splicing were mostly cytoplasmic, whereas their cDNA counterparts were mostly nuclear. Moreover, these effects appear to be general, as we obtained similar results with three different genes and in both HeLa cells and SV40-transformed mouse embryonic fibroblasts (SV40-MEFs). Analysis of the total FISH fluorescence signal in our microinjection study revealed that the overall levels of spliced mRNA and cDNA transcript were similar, indicating that the difference in the nucleocytoplamic distribution of mRNAs is a consequence of splicing promoting mRNA export. We conclude that splicing enhances mRNA export in mammalian cells and that the functional coupling of these two steps is a conserved and general feature of gene expression in higher eukaryotes.
Results
Splicing Increases the Cytoplasmic-to-Nuclear (C/N) Ratio of mRNAs 6- to 10-Fold.
During the last 30 years, the β-globin gene has been used extensively in gene expression studies (3–5, 13, 16, 17, 22, 24, 30–32) and was therefore used as our first model gene. To investigate the possibility that splicing promotes mRNA export in mammalian cells, HeLa cells were transiently transfected with β-globin DNA containing or lacking its natural introns (Fig. 1A). Steady-state β-globin mRNAs were visualized 30 h posttransfection by FISH, with the same probe for both the cDNA transcript and spliced mRNA (Fig. 1B). Quantitation of FISH images for 70 individual cells per construct revealed that the C/N ratio of the FISH signal was 5.44 for the spliced mRNA versus only 0.59 for the cDNA transcript (Fig. 1C). Thus, spliced β-globin mRNA was largely cytoplasmic, whereas the cDNA transcript was mostly nuclear.
Fig. 1.
Splicing results in a 10-fold increase in the C/N ratio of β-globin mRNA. (A) Schematic of β-globin constructs. The CMV promoter and BGH poly(A) site are indicated. The WT and cDNA constructs are isogenic except for the presence or absence of introns. The positions of the FISH and RPA probes are shown. (B) HeLa cells were transiently transfected with 900 ng of β-globin WT or cDNA constructs. FISH was performed to visualize β-globin mRNA, and DAPI staining was used to identify the cell nucleus. (Magnification: ×630.) (C) The C/N ratio of β-globin mRNA was determined for a minimum of 70 cells per construct. The graph shows the average C/N ratio for spliced mRNAs and cDNA transcripts, and error bars indicate standard error.
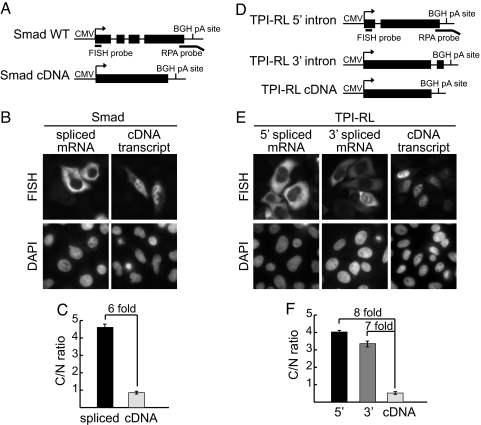
To investigate whether the effect of splicing on the nucleocytoplasmic distribution of mRNA was general, we transfected a Smad construct and two TPI-RL constructs, the latter of which were previously used to examine the effect of splicing on mRNA export (14). The Smad construct contains three introns, and the TPI-RL constructs contain an intron either near the 5′ end (TPI-RL5′) or near the 3′ end (TPI-RL3′) of the gene (Fig. 2 A and D). Consistent with our β-globin results, the FISH data showed that spliced Smad, TPI-RL5′ and TPI-RL3′ mRNAs were all largely cytoplasmic, whereas their cDNA counterparts were mostly nuclear (Fig. 2 B, C, E, and F).
Fig. 2.
Splicing enhances the C/N ratio for Smad and TPI-RL mRNAs. (A) Schematic of Smad constructs. The positions of the FISH and RPA probes are indicated. (B) HeLa cells were transiently transfected with 900 ng of Smad WT or Smad cDNA constructs. FISH was performed to visualize Smad mRNA, and DAPI staining was used to identify the cell nucleus. (C) The C/N ratio of Smad mRNA was determined for a minimum of 90 cells per construct. The graph shows the average C/N ratio for spliced mRNAs and cDNA transcripts, and error bars indicate standard error. (D, E, and F) The same as A, B, and C, respectively, except TPI-RL was used. (Magnification: B and E, ×630.)
Together, the data with all of our constructs show that splicing enhances the C/N ratio of mRNA in the range of 6- to 10-fold, depending on the construct. Considering the direct association between the mRNA export machinery and the splicing machinery (see Introduction), a likely explanation for this enhancement is that splicing promotes efficient mRNA export. However, other possible reasons for the enhancement include differences in transcription, 3′ end formation, and RNA degradation for the intron-containing versus intron-lacking constructs. Thus, we next carried out studies to address these possibilities.
Splicing-Dependent Differences in Nucleocytoplasmic Ratios of mRNAs Are Not Caused by Differences in 3′ End Formation.
Previous work showed that splicing promotes efficient 3′ end formation (13, 33, 34). Additional studies indicated that defective 3′ end formation results in retention of nascent transcripts at the site of transcription (32, 35), raising this as a possible explanation for our data. To address this possibility, we carried out RNase protection assays (RPAs) of total RNA from the transfected cells. As shown in Fig. 3A [and see supporting information (SI) Fig. 5], read-through products were not detected for any of the cDNA transcripts or spliced mRNAs (β-globin, Smad, TPI-RL). In the previous work in which cDNA transcripts were found to be defective in 3′ end formation, the constructs contained a genomic polyadenylation signal (13, 34). In our studies, we used the strong BGH polyadenylation signal (36), which may explain why 3′ end formation defects were not observed when this site was used in cDNA constructs. Together, the data indicate that defective 3′ end formation is not a likely explanation for the splicing-dependent differences that we observed in nucleocytoplasmic distribution of mRNAs.
Fig. 3.
RPA analysis of β-globin spliced mRNAs versus cDNA transcripts. (A) HeLa cells were transiently transfected with 900 ng of β-globin WT or β-globin cDNA constructs and 900 ng of a transfection control plasmid, xpSER, which encodes the human tRNASER gene with a tag at the 5′ end. Total RNA was extracted 24 h after transfection and RPA was performed. Reactions in lanes 1 and 3 contained 250 ng of cellular RNA, and reactions in lanes 2 and 4 contained 1 μg of cellular RNA. β-globin probe (24 fmol) and 2 fmol of xpSER probe were used. Lane 5 was carried out in the absence of target RNA. Lane 6 shows the probe in the absence of target RNA and RNase. (B) Graph of the fold difference in mRNA levels for each gene. mRNA levels were normalized to the levels of xpSER, and the ratio of mRNA generated from intron-containing genes to mRNA from intronless genes was calculated. Data represent the average of three experiments, and error bars indicate standard errors.
Another variable that could potentially affect the nucleocytoplasmic distribution of mRNAs is differential transcription and/or degradation of cDNA transcripts versus spliced mRNAs. Indeed, previous studies have shown that significantly greater levels of spliced mRNAs accumulate in transfected cells relative to the corresponding cDNA transcripts (refs. 14 and 15 and references therein). Consistent with this observation, our RPAs showed that levels of spliced mRNAs were approximately four to nine times greater (depending on the construct) than the corresponding cDNA transcripts (Fig. 3A and see SI Fig. 5). In contrast, the transfection control, a tagged serine tRNA, was present at the same levels in cells cotransfected with either the intron-lacking or intron-containing constructs (Fig. 3A and see SI Fig. 5). Additional studies are required to determine whether differential transcription and/or degradation accounts for the greater levels of spliced mRNAs in previous studies and in our work. For the purposes of the present study, we sought to determine whether the differences in mRNA levels of cDNA transcripts versus spliced mRNAs might have any bearing on the nucleocytoplasmic distribution of these mRNAs. To approach this question, we used a nuclear microinjection assay in combination with FISH, as described below.
Evidence That Splicing Enhances mRNA Export in Mammalian Cells.
To further investigate the basis for the splicing-dependent enhancement of the C/N ratio of mRNAs, we microinjected our DNA constructs into HeLa cell nuclei. After incubation for 15 min to allow transcription, α-amanitin was added to block further transcription, and the C/N ratios were determined after 1, 2, 3, or 4 h of continued incubation. Consistent with the transfection data (Figs. 1 and 2), the microinjection data show the same splicing-dependent enhancement of the C/N ratios for β-globin mRNA (Fig. 4A) and Smad and TPI-RL mRNAs (SI Figs. 6A and 7A). Microinjection studies using SV40-MEF cells also revealed similar splicing-dependent enhancements of the C/N ratio for all of the mRNAs (SI Fig. 8). We conclude that the splicing-dependent enhancement not only occurs in HeLa cells but also in SV40-MEFs, a cell type that more closely resembles a primary cell.
Fig. 4.
Evidence that β-globin-spliced mRNAs are exported more rapidly and efficiently than cDNA transcripts. (A) β-Globin WT and cDNA constructs (50 ng/μl) shown in Fig. 1A were microinjected into HeLa cell nuclei along with FITC-conjugated dextran as an injection marker. α-Amanitin (50 μg/ml) was added 15 min after injection to inhibit transcription. Cells were incubated for the indicated times before fixation. Representative pictures are shown for each time point. Large pictures show FISH signal; inset pictures show the injection marker. (Magnification: ×630.) (B) The ln-total FISH florescence was determined for a minimum of 20 cells per construct per time point. The graph shows the average ln-total fluorescence for spliced mRNAs and cDNA transcripts at each time point. (C) The same as B, except C/N ratios were calculated.
We next determined whether there was a difference in total mRNA levels in the microinjected HeLa cells by quantitating total fluorescence over the time course (Fig. 4 B and SI Figs. 6B and 7B). Significantly, these data showed similar levels of spliced mRNA and cDNA transcript throughout the time course, indicating no significant differences in transcription/degradation before α-amanitin was added and no difference in degradation after transcription was inhibited by α-amanitin. Thus, the microinjection data provide strong evidence that the splicing-dependent enhancement of C/N ratios is caused by increased export efficiency of the spliced mRNAs, rather than any major transcription/degradation differences.
Finally, quantitation of the C/N ratios over the time course fits a linear model for both the cDNA transcripts and spliced mRNAs. Thus, the differences in C/N ratios are reflected as differences in the slopes of the lines. These data indicate that β-globin-spliced mRNA was exported significantly faster (four times) than the cDNA transcript (Fig. 4 A and C). Similar results were obtained for Smad and TPI-RL, indicating that spliced Smad was exported about five times faster than its cDNA counterpart, and spliced TPI-RL5′ and TPI-RL3′ were exported faster (seven and six times, respectively) than the cDNA (see SI Figs. 6 and 7). We conclude that splicing enhances the kinetics of mRNA export in mammalian cells. Furthermore, we note that the export kinetics of TPI-RL5′ and TPI-RL3′ spliced mRNA, while faster than those of TPI-RL cDNA, are slower (three to five times) than those of β-globin- and Smad-spliced mRNAs. This difference in export kinetics could reflect a gene-specific difference in the rate of mRNA export.
Discussion
In this study, we have reinvestigated the debated issue regarding the role of splicing in promoting mRNA export. To do this, we used quantitative FISH analysis in combination with both transient transfection and nuclear microinjecion of DNA constructs containing or lacking introns. Multiple constructs were analyzed, and two different cell types were used. In all cases, our data showed that the ratio of C/N mRNA was enhanced in a splicing-dependent manner, with the enhancement ranging between 6- and 10-fold depending on the construct. As previous studies showed that the conserved TREX export complex colocalizes with splicing factors in nuclear speckle domains and is recruited to the 5′ end of mRNAs in a splicing-dependent manner during the splicing reaction (24, 26–28), our data are most consistent with the conclusion that splicing promotes mRNA export. However, we also considered potential contributions from other steps in gene expression, including 3′ end formation, transcription, and mRNA degradation.
In previous studies, 3′ end formation was found to be defective for cDNA transcripts, resulting in retention of mRNAs at the site of transcription (13, 32, 34, 35). However, in our studies, we used the strong BGH polyadenylation signal (36) and did not detect any 3′ end formation defects for any of our constructs, suggesting that this was not the reason for the splicing-dependent enhancement of the C/N ratio of our mRNAs. Consistent with previous work, we did observe significantly greater levels of spliced mRNAs relative to cDNA transcripts by using RPA analysis of transfected cells, raising the possibility that differential transcription and/or degradation could affect the C/N ratios. To address this possibility, we microinjected our DNA constructs, allowed transcription for an initial period, and then blocked further transcription with α-amanitin. We then determined the total FISH florescence signal. This analysis showed that both spliced mRNA and cDNA transcripts were present at similar levels throughout the time course, indicating no significant differences in transcription or degradation. Based on all of these observations, we conclude that the splicing-dependent enhancement of the C/N ratios of our mRNAs can be attributed to splicing promoting mRNA export. Moreover, of the low levels of cDNA transcripts that were exported, the kinetics were significantly slower than the corresponding spliced mRNAs.
A potentially contradictory observation that we made was that the level of spliced mRNA was greater than that of the cDNA transcript when the total population of transfected cells was measured by RPA (Fig. 3), yet the levels of these two mRNAs were similar when total FISH fluorescence was measured in the microinjection assay (Fig. 4B). However, a likely reconciliation of these data comes from our observation that the total number of transfected/microinjected cells containing FISH signal was significantly lower for the cDNA transcripts compared with the spliced mRNAs (unpublished work). Thus, cDNA transcripts failed to accumulate to detectable levels in a subset of the cells, which in turn, resulted in the lower RPA signals for the cDNA transcripts. This lack of cDNA transcripts in a subset of cells may be a consequence of rapid degradation of cDNA transcripts and requires further investigation. Of importance for the present study, our data showed that of the subset of cells that did contain FISH signal for the cDNA transcripts, the levels of FISH signal were similar to those of spliced mRNAs. Taken together, the data lead to a model in which cDNA transcripts are degraded in a subset of cells, and those that escape this pathway are largely retained in the nucleus. Based on previous data (27, 28), this nuclear retention most likely occurs because of inefficient recruitment of the mRNA export machinery to cDNA transcripts.
Our results with mRNA export in mammalian cells are consistent with our previous conclusions using the Xenopus oocyte export assay system (23). Thus, we conclude that splicing and mRNA export are functionally coupled and that this coupling is a general and conserved feature of gene expression in higher eukaryotes. In addition, the intron-containing β-globin, Smad, and TPI-RL differ in their number of introns, but the spliced mRNAs are exported with similar efficiencies. These data indicate that splicing of a single intron is sufficient to promote mRNA export. In contrast to our conclusions and previous work (18, 37), recent studies (13, 14) reported that splicing had no significant effect on mRNA export with TPI-RL and other constructs. However, we observed the effect of splicing with all of our constructs and their TPI-RL (14). A possible reason for the discrepancy between our conclusion and the other studies (13, 14) is that we used FISH to directly visualize and quantitate the nucelocytoplasmic distribution of mRNAs rather than biochemical fractionation of the nucleus and cytoplasm. TPI-RL5′ mRNA levels were also found to be higher than those of TPI-RL3′ in the previous work (14). We did not observe a significant difference between the levels of these mRNAs, and the reason for this difference is unknown.
Our results with splicing and export typify those of most coupled reactions. Specifically, coupling between different steps in gene expression is usually not essential, but instead enhances the rate/efficiency of the coupled reactions. For example, transcription can occur when uncoupled from splicing and vice versa. Likewise, polyadenylation can occur when uncoupled from splicing and the reverse is also true. Indeed, similar conclusions can be drawn for the >25 different coupled interactions that have been described so far. The physiological explanation for the rate/efficiency enhancement that occurs with coupling is that there is cross-talk between the different machineries involved in each of the reactions. The coupling between splicing and export provides one of the few examples in which this cross-talk has been characterized, as the export machinery has been shown to associate with the spliceosome, colocalize with it in the nucleus, and load onto the mRNA during the splicing reaction (24, 26, 27, 38). Thus, the functional, biochemical, and cytological data all are consistent with the model that there is functional cross-talk between the splicing and mRNA export machineries, and this cross-talk results in a significant splicing-dependent enhancement of mRNA export. The magnitude of the enhancement varies from gene to gene, possibly providing a means for gene-specific regulation at this step in the gene expression pathway.
Materials and Methods
Plasmids.
PCR was used to insert the AdML intron into three exon–exon junctions of Xenopus laevis Smad (39). A 5′ Myc tag and a 3′ HA tag were added to both intron-containing and intron-lacking Smad and β-globin by PCR. Smad genes were cloned into EcoRI and EcoRV sites in pcDNA 3.1/myc-His, and β-globin genes were cloned into KpnI and EcoRV sites in pcDNA3. TPI-RL constructs were as described (14). The transfection control, xpSER, was created by adding a 30-nt sequence to the 5′ end of the human gene encoding tRNASer. A PCR product was then cloned into the BglII and HindIII sites of pSUPER.
Cell Culture and Transfections.
SV40-MEFs were derived from control mice that have been transformed with the SV40 large T antigen. HeLa and SV40-MEF cells were cultured in DMEM supplemented with 10% FBS. HeLa cells were transfected on 12-well plates at 80–90% confluency with Lipofectamine 2000.
FISH.
FISH probes were 70-nt oligos labeled at the 5′ end with Alexa Fluor 546 NHS Ester and then HPLC-purified. Probe sequences were as follows: Smad probe, cacttgcagacttcttccatcctagcaggcgcttcactactggcggggtgaaagg c a a g a t g gacgacat; β-globin probe, cttcatccacgttcaccttgccccacagggcagtaacggcaga c t t c t c ctcaggagtcaggtgcaccat; and TPI-RL probe, gggtgctctgagccaccgcatcagagacgttggacttcagccatcctcggagcttctcgtgtacttccat. Transiently transfected HeLa cells were plated on fibronectin-coated 35-mm dishes with glass coverslip bottoms (MatTek) 24 h posttransfection. Cells were allowed to attach for 6 h, and FISH was performed (29). Transfected cells were then stained with 5 μg of DAPI in FISH hybridization buffer for 25 min. Cells were then washed with 50% formamide in 1× SSC. Images were captured with an EM-CCD camera, model C9100–12 (Hamamatsu) on an inverted microscope (200M; Zeiss) using Metamorph software (Molecular Devices). Equal exposure times were used to capture FISH images for cDNA transcripts and spliced mRNAs. Figures were produced by adjusting the contrast and brightness of the 14-bit images, which were then converted to 8-bit images.
FISH quantitation was carried out by using ImageJ 1.33u software (National Institutes of Health). Measurements were obtained for background fluorescence (Sb), fluorescence in the nucleus for FISH probe (Sn), total fluorescence of the cell for FISH probe (Sc), area of the nucleus (An), and area of the cell (Ac). The cytoplasmic (C′) amount of mRNA was calculated as: C′ = 1 − [An (Sn-Sb)/Ac (Sc-Sb)]. C/N ratios were calculated as C/N = C′/(1 − C′).
DNA Microinjections.
HeLa and SV40-MEF cells were plated on fibronectin-coated 35-mm dishes with glass coverslip bottoms (MatTek). For microinjections (29), plasmid DNA was microinjected at 50 ng/μl along with FITC-conjugated 70-kDa dextran (Molecular Probes). In each experiment 60–70 cells were microinjected and incubated in a 37°C incubator with constant (5%) CO2 levels. Fifteen minutes after microinjection, 50 ng/ml α-amanitin was added, and the cells were incubated for the indicated times before fixation.
RPA.
RPA probes were transcribed from linearized plasmids by using T7 RNA polymerase and gel purified from 6.5% denaturing polyacrylamide gels. Total RNA was extracted from transiently transfected HeLa cells by using Stratagene's Absolutely RNA Miniprep Kit. The RNA was then retreated with DNase I. RNA (250 ng or 1 μg) was hybridized with 24 fmol of β-globin, Smad, or TPI-RL probe and 2 fmol of xpSER probe. RPAs were performed by using the RPA III kit from Ambion. Protected RNA fragments were analyzed on denaturing polyacrylamide gels and quantitated by PhosphorImager. φX174 end-labeled DNA was used as a size marker.
Supplementary Material
ACKNOWLEDGMENTS.
We thank A. Palazzo for sharing his expertise with mammalian microinjections, Y. Chretien for assistance with statistical analyses, M. J. Moore (University of Massachusetts Medical School, Worcester) for TPI-RL constructs, T. Maniatis (Harvard University, Cambridge) for SV40-MEFs, the National Cell Culture Center (Minneapolis) for HeLa cells, and J. Hsu, K. Dufu, H. Cheng, and M. Bühler for useful discussions and comments on the manuscript. This work was supported by a National Institutes of Health grant supplement (to P.V.) and a National Institutes of Health grant (to R.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800250105/DC1.
References
- 1.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 2.Lai CJ, Khoury G. Deletion mutants of simian virus 40 defective in biosynthesis of late viral mRNA. Proc Natl Acad Sci USA. 1979;76:71–75. doi: 10.1073/pnas.76.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruss P, Khoury G. Rescue of a splicing defective mutant by insertion of an heterologous intron. Nature. 1980;286:634–637. doi: 10.1038/286634a0. [DOI] [PubMed] [Google Scholar]
- 4.Hamer DH, Leder P. Splicing and the formation of stable RNA. Cell. 1979;18:1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- 5.Hamer DH, Smith KD, Boyer SH, Leder P. SV40 recombinants carrying rabbit β-globin gene coding sequences. Cell. 1979;17:725–735. doi: 10.1016/0092-8674(79)90279-4. [DOI] [PubMed] [Google Scholar]
- 6.Gasser CS, Simonsen CC, Schilling JW, Schimke RT. Expression of abbreviated mouse dihydrofolate reductase genes in cultured hamster cells. Proc Natl Acad Sci USA. 1982;79:6522–6526. doi: 10.1073/pnas.79.21.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 8.Deng TL, Li Y, Johnson LF. Thymidylate synthase gene expression is stimulated by some (but not all) introns. Nucleic Acids Res. 1989;17:645–658. doi: 10.1093/nar/17.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA. 1991;88:478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson JJ, Foresman MD, Wilson N, McIvor RS. Intron requirement for expression of the human purine nucleoside phosphorylase gene. Nucleic Acids Res. 1992;20:3191–3198. doi: 10.1093/nar/20.12.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncker BP, Davies PL, Walker VK. Introns boost transgene expression in Drosophila melanogaster. Mol Gen Genet. 1997;254:291–296. doi: 10.1007/s004380050418. [DOI] [PubMed] [Google Scholar]
- 12.Zieler H, Huynh CQ. Intron-dependent stimulation of marker gene expression in cultured insect cells. Insect Mol Biol. 2002;11:87–95. doi: 10.1046/j.0962-1075.2001.00312.x. [DOI] [PubMed] [Google Scholar]
- 13.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose AB. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 2004;40:744–751. doi: 10.1111/j.1365-313X.2004.02247.x. [DOI] [PubMed] [Google Scholar]
- 16.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu WS, Mertz JE. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang MT, Gorman CM. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiou HC, Dabrowski C, Alwine JC. Simian virus 40 late mRNA leader sequences involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J Virol. 1991;65:6677–6685. doi: 10.1128/jvi.65.12.6677-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 22.Tokunaga K, et al. Nucleocytoplasmic transport of fluorescent mRNA in living mammalian cells: Nuclear mRNA export is coupled to ongoing gene transcription. Genes Cells. 2006;11:305–317. doi: 10.1111/j.1365-2443.2006.00936.x. [DOI] [PubMed] [Google Scholar]
- 23.Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues JP, et al. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno M, Segref A, Kuersten S, Mattaj IW. Identity elements used in export of mRNAs. Mol Cell. 2002;9:659–671. doi: 10.1016/s1097-2765(02)00454-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, et al. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 27.Masuda S, et al. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng H, et al. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 29.Palazzo AF, et al. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamer DH, Leder P. SV40 recombinants carrying a functional RNA splice junction and polyadenylation site from the chromosomal mouse βmaj globin gene. Cell. 1979;17:737–747. doi: 10.1016/0092-8674(79)90280-0. [DOI] [PubMed] [Google Scholar]
- 31.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol Cell. 2005;20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Custodio N, et al. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfarr DS, et al. Differential effects of polyadenylation regions on gene expression in mammalian cells. DNA. 1986;5:115–122. doi: 10.1089/dna.1986.5.115. [DOI] [PubMed] [Google Scholar]
- 37.Rafiq M, et al. Expression of recombinant human ceruloplasmin: An absolute requirement for splicing signals in the expression cassette. FEBS Lett. 1997;407:132–136. doi: 10.1016/s0014-5793(97)00325-6. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 39.Das R, et al. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.