Abstract
Deletion of phenylalanine-508 (Phe-508) from the N-terminal nucleotide-binding domain (NBD1) of the cystic fibrosis transmembrane conductance regulator (CFTR), a member of the ATP-binding cassette (ABC) transporter family, disrupts both its folding and function and causes most cystic fibrosis. Most mutant nascent chains do not pass quality control in the ER, and those that do remain thermally unstable, only partially functional, and are rapidly endocytosed and degraded. Although the lack of the Phe-508 peptide backbone diminishes the NBD1 folding yield, the absence of the aromatic side chain is primarily responsible for defective CFTR assembly and channel gating. However, the site of interdomain contact by the side chain is unknown as is the high-resolution 3D structure of the complete protein. Here we present a 3D structure of CFTR, constructed by molecular modeling and supported biochemically, in which Phe-508 mediates a tertiary interaction between the surface of NBD1 and a cytoplasmic loop (CL4) in the C-terminal membrane-spanning domain (MSD2). This crucial cytoplasmic membrane interface, which is dynamically involved in regulation of channel gating, explains the known sensitivity of CFTR assembly to many disease-associated mutations in CL4 as well as NBD1 and provides a sharply focused target for small molecules to treat CF. In addition to identifying a key intramolecular site to be repaired therapeutically, our findings advance understanding of CFTR structure and function and provide a platform for focused biochemical studies of other features of this unique ABC ion channel.
Keywords: ABC transporter, cystic fibrosis, domain interactions, modeling, protein misfolding
The cystic fibrosis transmembrane conductance regulator (CFTR) plays a fundamental role in metazoan epithelial ion transport, providing a rate-limiting step in the regulation of salt secretion and reabsorption (1). Secretory diarrhea results from persistent activation of CFTR by enterotoxins (2). When CFTR is absent or defective in humans, normal salt and water homeostasis in epithelial tissues cannot be maintained, resulting in the accumulation of macromolecular secretions and chronic infection and inflammation of the airways (3). At the biochemical and cell biological levels, CFTR is distinguished primarily by two characteristics. First, belonging to the human C subfamily of ATP-binding cassette (ABC) transporters, CFTR is unique as the only member of the entire family known to function as an ion channel (4). Progress toward understanding how fundamentally similar structures accomplish rapid bidirectional ion permeation in the case of an ion channel, and much slower vectorial active transport by transporters, is beginning to shed new light on the mechanism of each (5). The second feature of CFTR that is of fundamental importance is the major CF-causing mutation, deletion of Phe-508 (ΔF508), which results in misfolding and misassembly of the complex multidomain protein (6).
The in-frame deletion of the single Phe-508 codon present in ≈90% of CF patients prevents conformational maturation in the endoplasmic reticulum (ER). Any mutant protein that reaches the cell surface remains thermally unstable, capable of only partial chloride channel activity, and is rapidly endocytosed and degraded (7). In vitro experiments with the isolated first nucleotide-binding domain (NBD1) reveal that the ΔF508 mutation decreases folding yield (8). However, other than the absence of the Phe-508 residue from its surface, the overall 3D structure of the domain is little altered (9–11). This observation led to the hypothesis, for which there is some support (12), that Phe-508 may mediate a critical interaction between NBD1 and a site elsewhere in the protein. As yet, the hypothesis has not been confirmed nor has the site been identified. There are indications that it is probably not in NBD2 (13, 14). However, gaining a fuller understanding of both the impact of the ΔF508 and the utilization of the ABC architecture to generate ion channel activity has been hampered by the lack of 3D structural information. Although the crystal structure for the isolated NBD1 is available (9), there is only a low resolution structure of the whole protein (15). High-resolution structures of several bacterial ABC transporters have recently been determined by x-ray crystallography (16–19). These structures provided important new insights into the mechanisms by which these proteins accomplish transmembrane translocation of their substrates (20). Although functioning as a channel rather than either an importer or an exporter, CFTR is a member of the exporter class of ABC proteins. Sequence and biochemical similarities between several of these exporters, and those for which atomic structures have been determined, Sav1866 (16) and MsbA (21), suggest that there are likely to be strong structural similarities as well.
Results
Molecular Modeling.
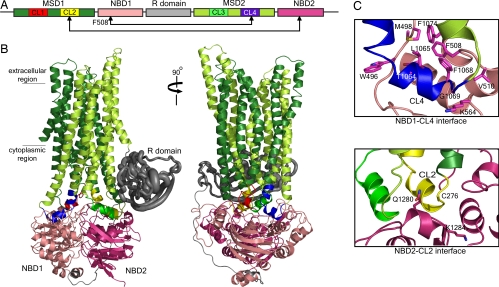
We constructed a 3D structure of CFTR by molecular modeling [for detailed methods, see supporting information (SI) Text]. CFTR consists of nucleotide-binding domains NBD1 and NBD2, membrane-spanning domains MSD1 and MSD2, and a regulatory region called the R domain (Fig. 1). There are existing 3D structures of NBD1 and also a homology model of NBD2 derived from structures of NBDs of other ABC transporters (22). To arrive at a structural model of the complete CFTR, we built models of the membrane-spanning domains from the known structure of the full-length ABC exporter, Sav1866 (SI Fig. 5). Full-length ABC proteins can be grouped into two classes according to the number and conformation of their transmembrane helices. Bacterial importers have variable numbers of helices that are short, positioning their NBDs close to the membrane plane. The exporters such as Sav1866 possess 12 transmembrane helices that are longer than those of the importers, thus their NBDs are farther from the membrane plane. CFTR contains 12 transmembrane helices, and its intracellular loops are of a length similar to those of Sav1866 (16), which suggested that CFTR MSDs can be modeled from those of Sav1866 (See SI Text). To organize the different domains of CFTR, we followed the tertiary organization of the Sav1866 domains. The structural model is consistent with available experimental data on the orientation and packing of CFTR transmembrane helices (see SI Text and SI Fig. 6). At least some of these data derive from measurements of open-channel currents, whereas the Sav1866 structure is believed to represent the closed state of the translocation pathway of that transporter (16). Nevertheless, the interdomain contacts in the modeled CFTR structure, which are our primary focus here, are strongly confirmed by biochemical experiments (see below). Significantly, analogous contacts have been shown to be maintained in both open and closed conformations of the MsbA exporter (21).
Fig. 1.
Theoretical model of CFTR structure. (A) Schema of CFTR primary structure containing two nucleotide-binding domains (NBD1 and NBD2), two membrane-spanning domains (MSD1 and MSD2), and a regulatory region (R domain). Each MSD contains two cytoplasmic loops (CL) that form interfaces with the NBDs. (B) Homology model of CFTR constructed from Sav1866 exporter (16) (see Results), where the domains are colored as in the schema. The unique-to-CFTR R domain, which is largely unstructured (23), was approximated by constructing an ensemble of dynamically accessible conformations derived from ab initio folding (see SI Text). R domain backbone size is rendered in proportion to variations of Cα atoms. (C) Close-up view of the interfaces formed between NBD1/CL4 and NBD2/CL2. Cross-linking of Cys pairs F508C/L1065C, F508C/F1068C, F508C/G1069C, and F508C/F1074C confirms that Phe-508 in NBD1 associates with CL4 in MSD2 (Fig. 3 and SI Fig. 7). Cross-linking of C276/Q1280C and C276/K1284C confirms interaction of CL2 and NBD2.
The R domain unique to CFTR is largely unstructured (23). Secondary structure prediction algorithms as well as our ab initio folding simulations (see SI Text) suggested the formation of persistent secondary structural elements and tertiary arrangement of the chain. We approximated the R domain by constructing an ensemble of observed conformations and representing it by the centroid of the most dominant structure in this ensemble (Fig. 1B). The placement of the R domain in the structure is consistent with preliminary EM observations of purified CFTR, with nanogold labeling of polyhistidine sequences inserted into the R domain and localized dynamic changes on phosphorylation by protein kinase A (PKA) (R. C. Ford and J.R.R., unpublished observations).
Cytoplasmic–Membrane Domain Interfaces.
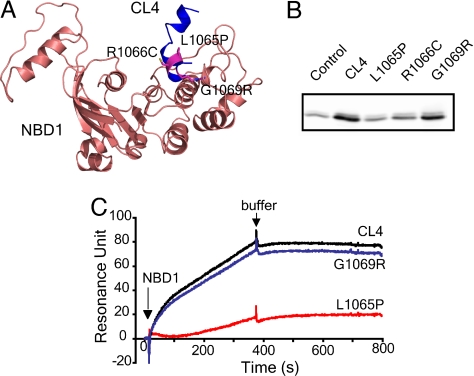
Most of the electrophysiological and biochemical constraints accommodated by the structure relate to the interactions of the two NBDs or MSDs (SI Fig. 6) with each other but not those between NBDs and MSDs. The NBD/MSD interfaces are likely to be crucial in both assembly during biogenesis and in mediating conformational signals that influence channel activity. Therefore, we focused on verification of these interfaces. Because Sav1866 was the primary template for building the structural model, these interfaces comprise contacts between the NBD in one half of the molecule and the MSD in the other. Earlier studies had shown many disease-associated missense mutations in CL4, including several within a short sequence at its center, corresponding to a so-called coupling helix in Sav1866 (24, 25). The positions of three of these mutated residues in proximity to NBD1 are shown in Fig. 2A. The abilities of synthetic 18-mer peptides containing the wild type or each of the mutated CL4 sequences to bind NBD1 were tested in pull-down experiments. The wild-type peptide bound strongly, whereas the two mutations (L1065P and R1066C) that prevent conformational maturation (24) reduced this binding to control levels (Fig. 2B). The third mutation (G1069R), which does not prevent maturation but impairs channel function (25), causes a lesser reduction in binding. Surface plasmon resonance analysis (Fig. 2C) of the binding yielded essentially the same result in which interaction of purified NBD1 with the wild-type CL4 peptide was nearly completely ablated by a folding mutation (L1065P) but only slightly diminished by one influencing channel function (G1069R).
Fig. 2.
CL4 peptide binding to NBD1. (A) Location of disease-associated mutations (L1065P, R1066C, and G1069R) at the NBD1/CL4 interface. (B) Disease-causing mutations in CL4 abolish or diminish the CL4 and NBD1 interaction. Biotinylated CL4 or its mutant peptides immobilized on NeutrAvidin beads were incubated with purified recombinant human NBD1 (see SI Text). Bound proteins were eluted with sample buffer and detected by Western blotting with CFTR antibody 660. NeutrAvidin beads without bound peptide were used as control. (C) CL4 binds to NBD1 as detected with surface plasmon resonance. Biotinylated peptides were immobilized on a BIAcore streptavidin sensor chip to 200 resonance units. NBD1 was injected, and the binding was detected by surface plasmon resonance and BIAcore 2000. The binding of NBD1 to the chip without peptide was subtracted from NBD1 binding to the peptides.
Cysteine Cross-Linking.
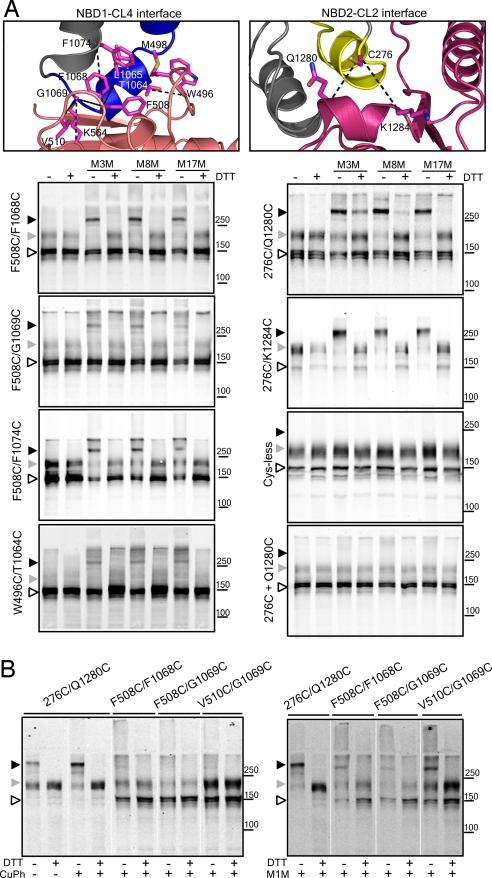
As a second means of assessing the NBD1/CL4 interface and to locate specifically the contact sites between CL4 and NBD1, pairs of cysteine residues were introduced into a functional Cys-less CFTR construct (26) at positions in CL4 and NBD1 that are in close proximity according to our modeled structure (Fig. 3A, Top Left). As seen in the Left column of Fig. 3A, several such Cys pairs could be cross-linked by bifunctional methane-thiosulfonate (MTS) reagents. In control experiments where separate constructs containing individual rather than both members of these pairs were coexpressed, cross-linking did not occur (SI Fig. 7). Strikingly, Phe-508 apparently plays a central role in this interface because it can be cross-linked to cysteines introduced at many positions in CL4. These positions include Leu-1065, Phe-1068, Gly-1069, and Phe-1074 (Fig. 3A and SI Fig. 8). Cross-linking also occurs between cysteines substituted for another hydrophobic residue (Val-510) near Phe-508 on the NBD1 surface and G1069C in CL4 (SI Fig. 8). In addition to the Phe-508-containing NBD1 surface patch, residues in other regions of the domain also interact with CL4 residues as evidenced by cross-linking of Cys pairs involving amino acids closer to the Q loop (Gln-493), including W496C/T1064C and M498C/L1065C as well as nearer the Walker B motif (Asp-572) such as K564C/G1069C (Fig. 3A and SI Fig. 8). These data confirm the extensive NBD1/CL4 contact apparent in the structure.
Fig. 3.
Cross-linking of interfacial cysteine pairs. (A) Confirmation of the interaction, in cells, between NBD1 and CL4 (MSD2) and between NBD2 and CL2 (MSD1). Phe-508 participates in an apparent aromatic cluster with residues from CL4 (see also SI Fig. 10). CL4 also interacts with other regions in NBD1 as suggested by cross-linking of residues close to the Q loop (W496C/T1064C and M498C/L1065C) and a residue near the Walker B motif (K564C/G1069C). HEK293 cells transiently transfected with Cys-less CFTR containing Cys pairs were harvested and incubated in the presence of 200 μM MTS cross-linkers of different spacer arm lengths for 15 min at room temperature. Samples with or without DTT were subjected to SDS/PAGE and Western blotting with mAb596. Cross-linked species migrate above 250 kDa. (Right, Bottom two) Controls showing the lack of effects of cross-linking of the Cys-less construct and individually expressed and mixed single cysteine constructs (labeled 276C + Q1280C) under the same conditions (see also SI Fig. 8). (B) (Right) Cross-linking by the shortest reagent 1,1-methanediyl bismethanethiosulfonate (M1M), in isolated membranes, indicates close contact and probable mobility of residues across both interfaces. (Left) There is greater propensity for disulfide bond formation at the CL2/NBD2 interface (276C/Q1280C) than at the CL4/NBD1 interface. Open arrowhead, immature core-glycosylated CFTR (band B); gray arrowhead, mature complex-glycosylated CFTR (band C); black arrowhead, cross-linked mature protein.
Most of the specific Cys pair cross-links were mediated by MTS reagents with spacer arms ranging from 3.9 to 24.7 Å in length, which initially seemed somewhat surprising but must indicate considerable flexibility in the positioning of each member of the pair with respect to each other (Fig. 3 A and B, Right and SI Fig. 8). Nevertheless, the proximity or relative orientation of the F508C/F1068C, F508C/G1069C, and V510C/G1069C pairs permitted very little disulfide bond formation on oxidation catalyzed by copper phenanthroline, i.e., only a very small proportion of mature band was converted to cross-linked band (Fig. 3B, Left). In contrast, in all cases where strong cross-linking occurred, only the mature form of CFTR but not the immature one was converted to the cross-linked species of slower mobility (Fig. 3 and SI Fig. 8). This finding provides strong evidence that residue associations occur only in the mature native form of the protein. There is indirect evidence that the Phe-508 side chain is involved in both domain assembly (11) and channel gating (26). Our present findings identify the site of the interdomain association and in so doing confirm that the so-called domain swapping described by Dawson and Locher in the Sav1866 exporter (16) and also observed in the P-glycoprotein multidrug exporter (27) and the MsbA lipid exporter (21) occurs in CFTR.
Unlike Sav1866, which is a homodimer containing two identical NBDs and MSDs, CFTR contains two distinct domains of each type. The structural model predicts that domain swapping occurs symmetrically between NBDs and MSDs in opposite halves of the molecule. Cysteine cross-linking confirms that domain swapping occurs between CL2 and NBD2 (Fig. 3). In fact, the Q1280C and K1284C substitutions in NBD2 of Cys-less CFTR containing the native Cys-276 residue in CL2 (illustrated in Fig. 3A, Top Right) enabled cross-linking by all MTS reagents tested. Furthermore, disulfide bond formation between cysteines at positions 276 and 1280 occurred spontaneously because of oxidation during membrane isolation and was further promoted by copper phenanthroline (Fig. 3B, Left). This observation indicates close contact between the native Cys-276 residue in CL2 and Gln-1280 in NBD2. Thus, when NBD2 is synthesized to complete the CFTR structure, it integrates with CL2 in MSD1. However, although there are many disease-associated mutations in both NBD1 and CL4 that compromise assembly, few have been identified in either NBD2 or CL2 (www.genet.sickkids.on.ca/cftr). This finding now can be understood in view of the realization that incorporation of NBD2 is not required for the correct assembly of the other domains into a structure that satisfies ER quality control (13, 14).
Role of Interfaces in Channel Gating.
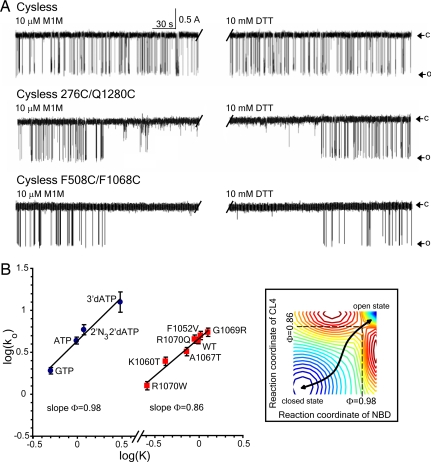
It is likely that both of these interfaces mediate conformational signals elicited by ATP binding and hydrolysis to the channel gate. If this is the case, it is unclear whether the channel-activating stimuli, ATP binding and phosphorylation, should influence the formation of cross-links between cysteines on either side of an interface or whether their cross-linking should influence channel gating. Treatment of cells with cAMP-elevating stimuli, which caused CFTR phosphorylation by PKA, did not alter cross-linking of cysteines between either CL4 and NBD1 or CL2 and NBD2 (SI Fig. 9). With membranes isolated from these cells, high concentrations of nucleotides [MgATP, adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP), or ADP + vanadate] with or without phosphorylation by PKA also did not strongly influence formation of the cross-links. These results would imply that if conformational signals were being transmitted, structural elements on either side of the interface may move in concert. If so, channel gating might or might not be expected to be influenced by covalent cross-linking between cysteines on either side of the interface. In fact, we found that single-channel gating, which persists after the introduction of a Cys pair at each interface, was completely inhibited by cross-linking (Fig. 4A). In both cases, this inhibition was completely reversed on reduction with DTT. Hence, these points of contact are integral elements of the structure, and covalent coupling between residues on either side restricts channel activity. This restriction is unlikely to be caused by prevention of signal transmission per se but probably reflects the restriction of dynamics at the interfaces, which also is suggested by cross-linking by reagents of different lengths. The lack of effect of gating stimuli (PKA + ATP) on the interactions between the NBDs and the CLs (SI Fig. 9) also suggests that there may be coordinated movements of the two sides of the interfaces.
Fig. 4.
Role of domain–domain interactions in CFTR channel gating. (A) Inhibition of CFTR channel gating by cross-linking. Single-channel recording after exposure to 10 μM M1M from the cis side of the bilayer followed by 10 mM DTT. For Cys-less CFTR, the last 4 min from the total 20 min of M1M treatment is shown in the first half of the trace before the interruption. The last 4 min of the total 20 min of DTT treatment is shown in the second half. O, open state; c, closed state. (Top) Cys-less CFTR (n = 4). (Middle) Cys-less CFTR with 276C/Q1280C (n = 3). (Bottom) Cys-less CFTR with F508C/F1068C (n = 4). (Middle and Bottom) Change in functional state in the middle of a 4-min portion of the total 20-min recording. No effect of MTS reagents was observed in constructs containing single cysteines that contribute to cross-linkable pairs (SI Fig. 11). (B) NBD1 and CL4 participate early in the gating cycle. Brønsted plots for wild-type CFTR gated by 2 mM nucleotide ligands shown by each experimental point (Left) and substitutions of CL4 residues (Right). Both graphs are linear with the slope Φ values indicated. Points on both graphs are shown as mean values ± SEM of at least four different experiments. (Inset) Hypothetical free energy landscape of CFTR gating where troughs (stable states) are colored blue and crests (unstable states) are red. At the transition state (saddle point), NBD1 has proceeded 98% toward the open state, and CL4 has proceeded 86%.
As an independent means of comparing the stages in the gating reaction at which NBD1 and CL4 participate, rate equilibrium–free energy relationship (REFER) analysis (28) was applied (Fig. 4B). The slopes (Φ values) of the linear log/log plots relating the rate constants of single-channel opening and the equilibrium constants approach 1 (Φ = 0.98) for different channel-activating nucleotide ligands and only a slightly lesser value for multiple amino acid substitutions in CL4 (Φ = 0.86). These values indicate that ligand binding is essentially complete before the transition state for channel opening is reached, whereas CL4 has proceeded 86% of the way to the open state at the transition state. Thus, both parts of the structure appear to participate early in the gating cycle, consistent with a dynamic functional role of the NBD1/CL4 interface. Significantly, a Φ value near 1 for NBD1 was also obtained by similar analysis of different chimeric constructs of the domain (29).
Discussion
Thus, a 3D structure of CFTR derived by molecular modeling reveals important aspects of the interfaces between the NBDs and MSDs of CFTR. The most critical contact in conformational maturation during biosynthesis, which is precluded by the ΔF508 mutation, is between a site containing that residue on the surface of the N-terminal NBD and a cytoplasmic loop in the C-terminal MSD. This finding is consistent with and helps to explain previous observations of disruption of CFTR membrane domains by the ΔF508 mutation (13, 14). The mutation is known to act at two levels: the first to decrease the folding yield and transitions between intermediate states in the folding of NBD1 itself, and the second, to prevent an interaction of the NBD1 surface elsewhere in the protein. We have identified the site of this interaction, dependent on the phenylalanine aromatic side chain. It appears to participate in an aromatic cluster with residues from CL4 (Fig. 3A and SI Fig. 10), which may contribute to the stability of this vital tertiary interaction as in other proteins (30, 31). The confirmed 3D structure of this interfacial site provides precise information on what has to be restored or mimicked to counteract the defect caused by the mutation. The corresponding crossing-over between NBD2 and MSD1 is less crucial to maturation of the protein because NBD2 is the last portion of the polypeptide to be translated and may be incorporated posttranslationally (12).
In addition to establishing the precise intramolecular contact formed by the Phe-508 side chain in the assembly of the N- and C-terminal portions of the protein, our findings provide some insight into the role of this interface in the regulation of the CFTR channel. The formation of covalent cross-links between cysteines on either side of this interface arrests channel gating, indicating a requirement for dynamic contact at these sites. This finding applies to both the Phe-508-containing NBD1/CL4 interface and the counterpart between NBD2 and CL2. These interactions between NBDs and MSDs in opposite halves of the molecule, which have been referred to as domain swapping (16) or intertwining (21) between subunits of homodimer ABC transporters, are probably also important in the transmission of regulatory signals. The fact that the ability of these interfaces to be cross-linked is not perturbed by the stimuli that activate channel gating is consistent with their action as connecting joints between other portions of the molecule that may undergo larger ranges of motion. This interpretation is supported by the REFER analysis of single-channel kinetics, which suggests that conformational movements in ligand-binding sites and cytoplasmic loops between transmembrane helices are tightly coupled.
Methods
Structural Modeling.
The Homology suite from INSIGHTII (Accelrys) was used to construct an atomic model of CFTR MSDs, The x-ray structure of NBD1 (9) and an existing homology model of NBD2 (22) were used. The R domain was approximated by an ensemble of conformations derived by ab initio folding.
CL4-NBD1 Binding.
Biotinylated synthetic peptides corresponding to wild-type and mutant versions of a CL4 peptide were immobilized on NeutrAvidin beads and used to pull down purified human NBD1 protein kindly provided by P. J. Thomas (University of Texas Southwestern Medical Center, Dallas, TX). Binding also was assayed by surface plasmon resonance (BIAcore 2000).
Cysteine Cross-Linking.
HEK293 or BHK cells or membranes expressing Cys-less CFTR containing specific Cys pairs were incubated with MTS reagents of different chain lengths and analyzed by SDS/PAGE with or without reduction with 30 mM DTT and Western blotting.
Single-Channel Recording.
Measurements were made in planar lipid bilayers as described (26), and REFER analysis of gating kinetics was performed according to Mitra et al. (28).
Details of all of these procedures are in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grant DK051619 (to J.R.R.) and Cystic Fibrosis Foundation Grant DOKHOL07I0 (to N.V.D.). A.W.R.S. is a Predoctoral Fellow of the American Heart Association, Grant 0715215U.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800254105/DC1.
References
- 1.Quinton PM. Cystic fibrosis: Lessons from the sweat gland. Physiology. 2007;22:212–225. doi: 10.1152/physiol.00041.2006. [DOI] [PubMed] [Google Scholar]
- 2.Al Awqati Q. Alternative treatment for secretory diarrhea revealed in a new class of CFTR inhibitors. J Clin Invest. 2002;110:1599–1601. doi: 10.1172/JCI17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis PB. Cystic fibrosis since 1938. Am J Respir Crit Care Med. 2006;173:475–482. doi: 10.1164/rccm.200505-840OE. [DOI] [PubMed] [Google Scholar]
- 4.Bear CE, et al. Purification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR). Cell. 1992;68:809–818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]
- 5.Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riordan JR. Assembly of functional CFTR chloride channels. Annu Rev Physiol. 2005;67:701–718. doi: 10.1146/annurev.physiol.67.032003.154107. [DOI] [PubMed] [Google Scholar]
- 7.Sharma M, Benharouga M, Hu W, Lukacs GL. Conformational and temperature-sensitive stability defects of the ΔF508 cystic fibrosis transmembrane conductance regulator in postendoplasmic reticulum compartments. J Biol Chem. 2001;276:8942–8950. doi: 10.1074/jbc.M009172200. [DOI] [PubMed] [Google Scholar]
- 8.Qu BH, Strickland EH, Thomas PJ. Cystic fibrosis: A disease of altered protein folding. J Bionenerg Biomembr. 1997;29:483–490. doi: 10.1023/a:1022439108101. [DOI] [PubMed] [Google Scholar]
- 9.Lewis HA, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis HA, et al. Impact of the ΔF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 11.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe-508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du K, Sharma M, Lukacs GL. The F508 cystic fibrosis mutation impairs domain–domain interactions and arrests posttranslational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 13.Younger JM, et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 14.Cui L, et al. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- 16.Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 17.Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 18.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 19.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal chelate-type ABC transporter. Science. 2007;315:373–377. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 20.Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callebaut I, Eudes R, Mornon JP, Lehn P. Nucleotide-binding domains of human cystic fibrosis transmembrane conductance regulator: Detailed sequence analysis and three-dimensional modeling of the heterodimer. Cell Mol Life Sci. 2004;61:230–242. doi: 10.1007/s00018-003-3386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker JMR, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seibert FS, et al. Disease-associated mutations in the fourth cytoplasmic loop of cystic fibrosis transmembrane conductance regulator compromise biosynthetic processing and chloride channel activity. J Biol Chem. 1996;271:15139–15145. doi: 10.1074/jbc.271.25.15139. [DOI] [PubMed] [Google Scholar]
- 25.Cotten JF, Ostedgaard LS, Carson MR, Welsh MJ. Effect of cystic fibrosis-associated mutations in the fourth intracellular loop of cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1996;271:21279–21284. doi: 10.1074/jbc.271.35.21279. [DOI] [PubMed] [Google Scholar]
- 26.Cui L, et al. The role of cystic fibrosis transmembrane conductance regulator phenylalanine-508 side chain in ion channel gating. J Physiol (Lond) 2006;572:347–358. doi: 10.1113/jphysiol.2005.099457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zolnerciks JK, Wooding C, Linton KJ. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. FASEB J. 2007;21:3939–3948. doi: 10.1096/fj.07-8610com. [DOI] [PubMed] [Google Scholar]
- 28.Mitra A, Tascione R, Auerbach A, Licht S. Plasticity of acetylcholine receptor gating motions via rate–energy relationships. Biophys J. 2005;89:3071–3078. doi: 10.1529/biophysj.105.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott-Ward TS, et al. Chimeric constructs endow the human CFTR Cl(−) channel with the gating behavior of murine CFTR. Proc Natl Acad Sci USA. 2007;104:16365–16370. doi: 10.1073/pnas.0701562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano L, Bycroft M, Fersht AR. Aromatic–aromatic interactions and protein stability. Investigation by double-mutant cycles. J Mol Biol. 1991;218:465–475. doi: 10.1016/0022-2836(91)90725-l. [DOI] [PubMed] [Google Scholar]
- 31.McGaughey GB, Gagne M, Rappe AK. pi–stacking interactions: Alive and well in proteins. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.