Abstract
Apis mellifera originated in Africa and extended its range into Eurasia in two or more ancient expansions. In 1956, honey bees of African origin were introduced into South America, their descendents admixing with previously introduced European bees, giving rise to the highly invasive and economically devastating “Africanized” honey bee. Here we ask whether the honey bee's out-of-Africa expansions, both ancient and recent (invasive), were associated with a genome-wide signature of positive selection, detected by contrasting genetic differentiation estimates (FST) between coding and noncoding SNPs. In native populations, SNPs in protein-coding regions had significantly higher FST estimates than those in noncoding regions, indicating adaptive evolution in the genome driven by positive selection. This signal of selection was associated with the expansion of honey bees from Africa into Western and Northern Europe, perhaps reflecting adaptation to temperate environments. We estimate that positive selection acted on a minimum of 852–1,371 genes or ≈10% of the bee's coding genome. We also detected positive selection associated with the invasion of African-derived honey bees in the New World. We found that introgression of European-derived alleles into Africanized bees was significantly greater for coding than noncoding regions. Our findings demonstrate that Africanized bees exploited the genetic diversity present from preexisting introductions in an adaptive way. Finally, we found a significant negative correlation between FST estimates and the local GC content surrounding coding SNPs, suggesting that AT-rich genes play an important role in adaptive evolution in the honey bee.
Keywords: adaptive evolution, FST, genetic differentiation
The relative contribution of selection and drift in shaping the observed patterns of genetic diversity within and between populations has been a subject of great debate in evolutionary biology. Recent studies have documented vast amounts of DNA polymorphism in both model and nonmodel organisms, yet we have very little understanding of the proportion of this diversity that is functional and its role in facilitating adaptation and phenotypic evolution (1–6). Such an understanding is needed to gain insights into the process of adaptive evolution and speciation at both the proximate and mechanistic levels (1–6). Furthermore, elucidating the role of molecular evolution associated with geographic radiations is likely to shed light on the genetics of range expansions in both native and invasive populations, a topic of immense theoretical and economic importance (6–8). The honey bee Apis mellifera provides an ideal system to study these longstanding questions.
The honey bee is an important pollinator in both managed and natural systems and has long served as a model organism for the study of behavior in insect societies (9). In its native range, A. mellifera is classified into approximately two dozen subspecies, which are further organized into four major geographically and genetically distinct groups: African, Western and Central Asian (hereafter referred to as Asian), Eastern European, and Western and Northern European (hereafter referred to as West European) (9–11). European honey bees were introduced by humans to the New World by European settlers as early as the 1600s. In Brazil in 1956, an intentional introduction of African honey bees (A. mellifera scutellata), which hybridized with previously introduced European bees, led to the establishment and spread of the highly invasive and economically devastating Africanized honey bees in North America and South America (12). Subsequent studies have shown that Africanized bees are predominantly African in ancestry with minor but consistent contribution from European genotypes (11, 12). Using recently developed SNP panels, Whitfield et al. (11) demonstrated that the honey bee originated in Africa and subsequently expanded into Eurasia in two or more independent ancient expansions. One expansion gave rise to Western European honey bees, and at least one other independent expansion gave rise to Asian and Eastern European honey bees. Honey bee subspecies vary in a host of phenotypic traits, such as morphology, behavior, physiology, and gene expression (9–11, 13, 14).
Adaptation in social insects involves selection on colony phenotypes mediated through social behaviors (15). For example, the expansion of honey bees into temperate regions was likely facilitated by selection for honey hoarding, ability to form a winter cluster, and differences in worker division of labor. The honey bee's multiple independent expansions out of Africa present a unique opportunity to examine how the ancestral African genome diverged in response to the different selective pressures experienced across its native and introduced ranges and to provide a population genetic context to the study of social regulation of behavior in the species. Our work provides the first step toward these goals by searching for genome-wide evidence for positive selection and adaptive evolution associated with the multiple ancient and recent (invasive) expansions of the honey bee.
Although there are several methods for detecting selection in population genetic data sets, methods based on multilocus comparisons of genetic differentiation provide a very useful framework for examining the extent of adaptive divergence between populations of the same species (1, 2, 4, 16–19). In subdivided populations, estimates of genetic differentiation derived from neutral loci should be concordant because the allele frequencies at such loci are controlled by drift and demographic processes that affect the entire genome. However, because selection acts on specific regions of the genome, allelic frequency at selected loci in subpopulations is expected to deviate from that of neutral loci, thereby creating deviations in the level of genetic differentiation estimated by the former versus the later (1, 17, 20–22). For example, heterogeneous selection at a locus can drive differences in allele frequencies between subpopulations, thereby increasing levels of genetic differentiation, commonly measured as FST (23, 24), when compared with neutral loci. On the other hand, balancing or purifying selection can maintain similar allele frequencies between subpopulations, reducing levels of FST when compared with neutral loci. As such, in surveys of genetic variation in subdivided populations, loci under selection are expected to provide outlier estimates of FST (1, 4, 16–18, 20, 21). Inferring selection based on these methods has recently been shown to be robust to demographic history (1, 18).
Tests of selection using genetic differentiation data can be used to indicate selection acting on specific loci, or across the genome as a whole. The former approach involves detecting outlier (i.e., selected) loci given an expected or observed neutral FST distribution. Many studies have successfully applied this approach to detect loci involved in adaptive population divergence in model and nonmodel organisms (25–30). Alternatively, the average effects of selection across the genome can be determined by comparing the FST distribution estimated from SNPs randomly distributed in coding and noncoding parts of the genome (20, 31). Noncoding SNPs are presumed to be mostly neutral, and genotype data at a large number of these loci provide an empirical (model-free) distribution of FST due to drift (20–22, 31). For example, higher FST in coding versus noncoding genomic regions indicates an overall signature of positive selection on the coding regions of the genome. To date, FST tests of selection have been applied only on the genome level in human populations (20, 31, 32). Given the recent development of genomic resources for the honey bee (11, 33) and the availability of geographically diverse genetic samples (11), it is now possible to examine the extent of selection in shaping genome-wide levels of genetic differentiation within native and invasive populations. In addition to its well characterized biogeography, the honey bee also possesses several useful properties that greatly facilitate studies of the genetic architecture of phenotypic traits (34, 35), such as haplodiploidy, high recombination rate, short generation time, extremely large family sizes, and the ability to control mating. The genetic properties and biogeographic history of the honey bee thus combine to provide an ideal system to examine the prevalence of genome-wide selection and the genetic basis of adaptive phenotypic evolution.
The goal of this study is to test for a signature of selection and adaptive evolution across the honey bee genome specifically in association with the bee's independent ancient and recent (invasive) expansions out of Africa. Using recently published genotypic data (11), we estimated FST among the four native honey bee groups and between native sub-Sahara A. m. scutellata and invasive A. m. scutellata-derived populations. We inferred selection in our data set as differences in the distribution of FST between coding and noncoding SNPs (20, 31, 32) and examined how such differences were associated with the evolutionary history of A. mellifera and its genomic properties.
Results and Discussion
We estimated FST at 444 SNPs that were randomly distributed across coding and noncoding genomic regions. The SNPs were typed in the four major honey bee population groups (each represented by 31–66 individuals from two or more subspecies) and an invasive Africanized population from South America (37 individuals) [see Materials and Methods and supporting information (SI) Fig. 5]. We classified SNPs into the following functional classes: exon, intron, and intergenic (SNPs <3 kb from exons were excluded from the latter two classes; see Materials and Methods). We gauged the overall effects of selection on the coding genome by comparing FST estimates of exon SNPs against those of the presumably neutral intron and intergenic SNPs. We examined the distribution (mean and variance) of allele frequencies in exon and noncoding (intergenic plus intron) SNPs to ensure that any observed difference in FST between the two classes is not confounded by differences in allele frequencies. In every population where we estimate FST, the distribution of allele frequencies did not significantly differ between exon and noncoding SNPs (SI Fig. 6) (means, two-tailed, P > 0.05 for all tests; variances, two-tailed, P > 0.05 for all tests), indicating that any observed difference in FST between the two classes must be caused by selection rather than nonadaptive causes (31).
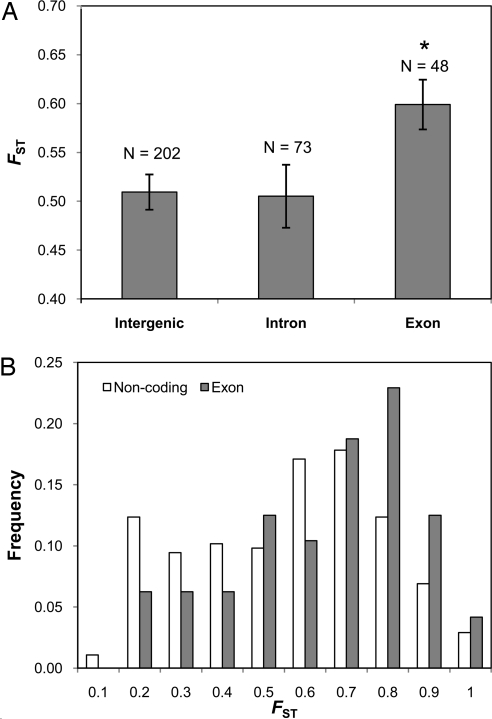
We found significant evidence for positive selection and adaptive evolution acting on the coding genome of the honey bee. In natural populations, exon SNPs had significantly higher FST estimates when compared with intergenic (P = 0.009), intronic (P = 0.009), and noncoding SNPs (P = 0.006) (Fig. 1). Because SNPs were randomly distributed throughout the honey bee's 16 chromosomes (SI Table 1 and SI Fig. 5), the observed differences in means between coding and noncoding SNPs are unlikely to be caused by selection on a small number of genes or a few selective sweeps affecting large chromosome portions. We predict that misclassification of SNPs (i.e., noncoding to coding and vice versa), a likely scenario given incomplete annotation of the honey bee genome (33), will reduce the signal for positive selection, suggesting that actual effects of selection may be stronger than those observed. We note that we do not interpret the results of the above analysis as direct selection on the actual SNPs used; indeed, the majority of exon SNPs were synonymous (SI Table 1) and most likely neutral. However, our results indicate that polymorphisms linked to these coding SNPs (i.e., in the coding regions themselves or nearby regulatory regions) are adaptively evolving in response to positive selection.
Fig. 1.
FST estimates varied between different SNP classes. (A) Exon SNPs had significantly higher FST when compared with intergenic, intronic, and noncoding (intergenic plus intron) SNPs, indicating the effects of positive selection acting on coding regions of the genome. Error bars represent SE. (B) Distribution of FST estimates in exon versus noncoding regions.
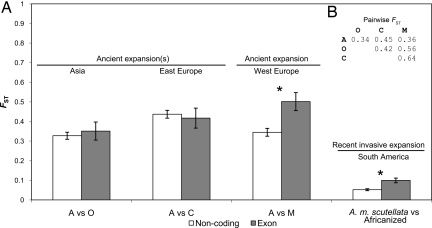
To further examine the causes of adaptive evolution in honey bee populations, we conducted the same analysis on pairs of populations that represent ancient and recent expansions of A. mellifera out of Africa (Fig. 2). The signature of selection found in the global analysis of bee population groups was driven primarily by adaptive evolution associated with the ancient expansion of A. mellifera out of Africa into Western and Northern Europe. Exon SNPs had significantly higher FST when compared with noncoding SNPs in the African versus West European comparison (P = 0.0032) (Fig. 2). It is worthwhile to note that West European honey bees, including the subspecies A. m. mellifera, represent the most ancient temperate expansion of A. mellifera out of Africa, extending into Northern Europe and West–Central Russia (10). We did not detect selection involved with the ancient expansion of A. mellifera in Asia and Eastern Europe (Fig. 2), which may be due to lack of power to detect the signature of adaptive evolution acting on a small number of genes, lack of actual adaptive population divergence, or subspecies heterogeneity within groups. The small number of coding SNPs that were polymorphic within the four main honey bee groups prevented us from examining the extent of adaptive divergence among subspecies within each of the four main groups.
Fig. 2.
Genetic differentiation in coding and noncoding SNPs in pairwise comparisons involving honey bee expansions out of Africa. (A) Significantly higher FST estimates in exon versus noncoding SNPs were observed in the ancient expansion of honey bees into Western Europe and the recent invasive expansion of Africanized honey bees in the New World. Error bars represent SE. (B) Pairwise estimates of FST between native honey bee groups by using all SNPs. A, O, C, and M refer to African, Asian, East European, and West European honey bee groups, respectively.
To explore the actual number of genes that experienced positive selection during A. mellifera's ancient expansion into Western and Northern Europe, we examined the number of outlier coding SNPs based on the observed distribution of noncoding SNPs. With cutoff criteria of >99% and >95% (FST ≥ 0.97 and 0.88, respectively), we observed three and five outlier coding SNPs, representing 8.1–13.5% of all coding SNPs surveyed in our study. Assuming that our randomly chosen SNPs represent the honey bee genome, our estimates imply that 852–1,371 genes (≈10% of the coding genome) experienced positive selection and adaptive evolution between the African and West European groups, on par with a recent study of adaptive evolution in human populations employing different methods (36). This is likely a conservative estimate because removal of outlier coding SNPs did not abolish the significant differences in FST between coding and noncoding SNPs. These results suggest that selection at a substantial number of genes may underlie adaptive evolution and phenotypic divergence in the ancient expansion of African bees into Western and Northern Europe.
We also detected a signal for positive selection associated with the invasion of Africanized bees in the New World. In a pairwise comparison between Africanized bees collected in South America and their African source, A. m. scutellata (12), we found that the coding genome was nearly twice as differentiated as the noncoding genome (coding FST = 0.10, nonoding FST = 0.05, P = 0.002), indicating positive selection (Fig. 2). Although A. m. scutellata experienced a bottleneck during its introduction to the New World, such an event would have affected both the coding and noncoding genomes equally and is thus not expected to generate the differences observed. Numerous studies have shown that introduced African bees hybridize with previously introduced European bees in the New World (12). The 37 Africanized bees analyzed here (from Brazil and northern Argentina) all exhibited a predominately African genome introgressed with Western European portions (11). It is possible that the signature of selection associated with the invasive expansion of Africanized bees may be related to hybridization of A. m. scutellata and West European bees in the New World.
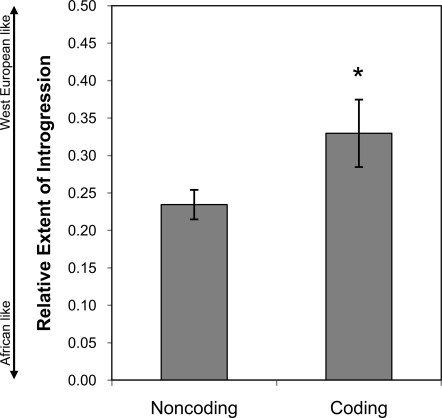
To investigate the role of introgression of African and West European genomes in the invasion of Africanized bees, we asked whether there were differences in the extent of introgression for coding and noncoding portions of the genome. Hybridization is a demographic process that involves the transfer of both coding and noncoding portions of the genome. In the absence of selection, we predict an equal extent of introgression for coding and noncoding portions of the West European genome in Africanized bees. Alternatively, if functional portions of the West European genome are either beneficial or detrimental to Africanized bees, we predict (respectively) greater or lesser introgression of coding relative to noncoding portions of the West European genome. For each variable SNP, we derived a relative measure of introgression of Africanized bees from 0 to 1 (A. m. scutellata-like to West European-like) as a function of pairwise FST estimates for Africanized bees versus A. m. scutellata and Africanized bees versus West European bees (Fig. 3). We found that SNPs in coding regions had significantly higher estimates of introgression (two-tailed, P = 0.032) when compared with SNPs in noncoding regions (Fig. 3), indicating that selection is acting to increase the average frequency of West European coding alleles over expectations for introgression without selection. This result is somewhat surprising given that West European bees are temperate, and the success of Africanized bees in tropical regions of the New World is clearly related to multiple morphological and behavioral traits derived from sub-Saharan A. m. scutellata (12). The adaptive value of functional (coding) portions of Western European genomes could be related to positive selection on novel variation in West European bees, to positive selection on novel hybrid gene combinations, and/or to selection for heterozygous genotypes (37). Our study thus provides direct evidence that invasive populations can exploit hybridization in an adaptive fashion—a finding of immense relevance to understanding the dynamics of biological invasions (37, 38).
Fig. 3.
Introgression of West European alleles in the genome of invasive Africanized bees. For each SNP we derived a relative measure of introgression as FST Africanized vs. A.m.scutellata/(FST Africanized vs. A.m.scutellata + FST Africanized vs. West European), which ranges from 0 to 1, indicating no to complete introgression of West European alleles into Africanized bees. Introgression was significantly greater in coding regions than in noncoding regions, indicating the effects of positive selection.
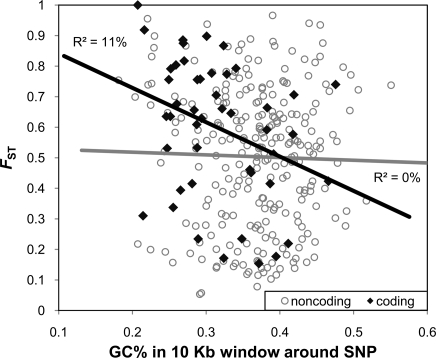
Finally, we found an interesting relationship between FST and the local GC content. In the native honey bee population, FST estimates were significantly negatively correlated with the GC content in a 10-kb window around exon SNPs (P = 0.025, r2 = 11%) but not around noncoding SNPs (P = 0.684, r2 ≈ 0%) (Fig. 4). The relationship between GC content and FST in exon SNPs was not mediated by differences in allele frequency because the correlation between an exon SNP's GC content and minor allele frequency was not significant (P = 0.35, r2 = 2%). Furthermore, GC-poor genes [defined as having average GC content at the third codon position (GC3) < 33%, following ref. 39] contained SNPs with significantly higher FST (P = 0.021) when compared with GC-rich genes (GC3 ≥ 33%). The negative relationship between FST and GC content implies that genes in GC-poor regions are contributing disproportionately to the adaptive evolution observed in honey bee populations. The honey bee genome is AT-rich in comparison to other sequenced invertebrates (33, 39). Furthermore, the honey bee genome is biomodal with respect to GC content, with both GC-poor and GC-rich regions, in stark contrast to the GC-rich genomes of both Drosophila melanogaster and Anopheles gambiae (33, 39). Jorgensen et al. (39) recently hypothesized that the isochore-like structure of the honey bee genome is caused by two markedly different mutational patterns, with a very strong bias toward A/T mutations acting in GC-poor regions, and an unbiased mutation pattern acting in GC-rich regions. Our results demonstrate that the derived GC-poor regions of the honey bee genome (33, 39) are of both functional and adaptive importance. Further work is needed to examine the mechanistic basis for this phenomenon.
Fig. 4.
Genetic differentiation and GC content. Genetic differentiation was negatively correlated with GC content in coding SNPs but not in noncoding SNPs, suggesting that AT-rich genes play an important role in facilitating adaptive evolution.
In summary, our study provides evidence for positive selection acting on the honey bee's protein-coding genome associated with the ancient radiation of honey bees into Western and Northern Europe and the recent invasion of African-derived honey bees in the New World. We estimate that positive selection acted on at least 10% of the genes in the honey bee genome during the former ancient expansion. We also show that the invasion of African-derived honey bees involved the preferential introgression of coding regions from previously introduced West European bees, suggesting that multiple introductions can provide functional genetic diversity that facilitates adaptive evolution in economically damaging invasive populations. Additionally, we show that the signal for selection in honey bee populations was biased toward the bee's unique AT-rich regions, suggesting that such regions play a major role in facilitating adaptive evolution. Further population genetic and genomic studies investigating selection at a finer scale in honey bees are likely to yield additional insights on theoretical and mechanistic aspects of adaptive evolution in native and invasive populations.
Materials and Methods
Samples.
Detailed information on sampling strategies, locations, and DNA extraction methods are provided elsewhere (11). The study population consisted of workers (females) from Africa (n = 66; 21 A. m. scutellata, 19 A. mellifera lamarckii, 19 A. mellifera intermissa, two A. mellifera litoria, three A. mellifera capensis, and two A. mellifera unicolor), Asia (n = 44; 18 A. mellifera anatoliaca, 14 A. mellifera caucasica, nine A. mellifera syriaca, and three A. mellifera pomonella), East Europe (n = 34; 18 A. mellifera ligustica and 16 A. mellifera carnica), and West Europe (n = 31; 20 A. mellifera mellifera and 11 A. mellifera iberiensis). We also used data from 37 Africanized honey bees from different regions of Brazil (n = 26) and northern Argentina (n = 11; north of the hybrid zone indicated in ref. 11).
SNP Panel and Typing.
Whitfield et al. (11) identified 1,536 putative SNPs in the honey bee genome based on (i) observed polymorphisms between the reference genome of A. mellifera (Assembly 3.0; sequenced from the North American DH4 strain, which was primarily A. m. ligustica) and genome sequence traces of Africanized honey bees (largely A. m. scutellata admixed with the genomes of both Western and Eastern European honey bees) and (ii) observed polymorphisms in ESTs. These SNPs were typed as described in ref. 11. In this study we restricted our analyses to 444 validated SNPs derived from the former set because they were identified from random genome traces without knowledge of their functional class. EST-derived SNPs were not analyzed here because they were derived from transcribed regions making them biased against neutrality when compared with the genome trace-derived SNPs.
Ascertainment Bias.
The same discovery panel and protocol were used to ascertain SNPs without knowledge of their functional class. Therefore, ascertainment bias is expected to affect coding and noncoding regions equally and not to systematically bias any particular class of SNPs. This is further supported by the finding that the distribution of allele frequencies does not significantly differ between coding and noncoding SNPs in our data set (SI Fig. 6). Evidence for selection in our study is thus not caused by variation in the discovery protocol among different genomic regions (5). Furthermore, although we acknowledge that ascertainment bias likely exists in our data set, two lines of evidence suggest that the effects of ascertainment bias are minor. First, the discovery panel for the SNPs used herein was relatively diverse (see SNP Panel and Typing), which is expected to reduce ascertainment bias in our data set (22, 40–42). Second, both genome sequence-derived and EST-derived SNPs (ascertained by using different discovery panels) yielded essentially the same results in analyses of population genetic structure (11).
Genetic Differentiation.
Previous analyses of population genetic structure among subspecies indicated that the majority of genetic differentiation was associated with the four major geographical honey bee groups (11). We thus treated each geographic group as a distinct subpopulation and estimated FST at each polymorphic SNP (defined as having a minor allele frequency ≥5%) in the global native honey bee population following standard methods (43, 44). We also conducted similar analyses involving specific subpopulation pairs as indicated in the text. Differences in the exclusion of rare alleles produced average FST estimates that differed slightly from ref. 11; however, relative differences between pairwise comparisons were essentially the same.
SNP Classification and Local GC Content.
Honey bee official gene predictions were generated by using Genome Assembly 2 (33), and SNPs were isolated by using Assembly 3. To classify SNPs, we mapped each SNP's 200-bp flanking sequence to Assembly 2 scaffolds using BLASTN. We retained SNPs that perfectly matched a unique position in Assembly 2. Using the coordinates of the honey bee gene set OGSv1 (33), we classified SNPs as belonging to exons, introns, or intergenic regions. Genetic diversity, measured as expected heterozygosity, in the global data set did not significantly differ between coding and noncoding SNPs (P = 0.49; also see SI Fig. 6). As a measure of local GC content, we estimated the GC% in a 10-kb window surrounding each SNP (33, 39). Windows containing >1,000 unknown bases were removed from analyses.
To provide the best possible benchmark for neutral SNPs, we used only data from intergenic and intronic SNPs that are ≥3 kb away from the nearest predicted exon in our analyses (note that introns are relatively large in honey bees compared with other sequenced insects). Noncoding regions near coding DNA are known to be both functional and under selection (45, 46). Intron SNPs close to exons had significantly higher mean FST estimates when compared with SNPs that were farther away (P = 0.021); intergenic SNPs showed a similar trend that was not significant (SI Fig. 7). Exclusion of near-exon SNPs had no effect on the outcome of our statistical tests and our conclusions.
Statistical Tests.
Because of the honey bee's extraordinary recombination rate of 19 cM/Mb (47) and large number of chromosomes (n = 16) and the low SNP density of the current data set (average 1.86 SNP/Mb), we expect little correlation between FST estimates because of physical distance between nearby SNPs, as found in dense human SNP data sets (48). In an earlier study using 1,136 SNPs (ref. 11; including the set used in this study) linkage disequilibrium rapidly declined over a distance of 5–10 kb. Given the above, we treated FST estimates from SNPs as independent with respect to physical distance. We used the nonparametric Wilcoxon rank-sums test (49) to examine differences in FST and other genomic parameters (e.g., GC content) between SNP classes. Because we predict stronger selection on functional SNPs a priori (1, 20, 31), we report one-tailed P values for all statistical tests, except where indicated and for tests of significance of regressions slopes (49). We compared the mean and variance of minor allele frequencies between coding and noncoding SNPs for all populations tested for selection, using the Wilcoxon rank-sums test (two-tailed) and Bartlett's test (two-tailed) for homogeneity of variances (49), respectively. Minor allele frequency represents the frequency of the less common SNP allele in any defined population where FST is estimated.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Andy Suarez, Kim Hughes, Stewart Berlocher, Jennifer Grixti, Gene Robinson, and anonymous reviewers for providing useful comments on the manuscript. This work was supported by the School of Integrative Biology and the Research Board at the University of Illinois (C.W.W.). A.Z. was partly supported by a National Science and Engineering Research Council of Canada Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800107105/DC1.
References
- 1.Beaumont MA. Adaptation and speciation: What can FST tell us? Trends Ecol Evol. 2005;20:435–440. doi: 10.1016/j.tree.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 3.Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 7.Allendorf FW, Lundquist LL. Introduction: Population biology, evolution, and control of invasive species. Conserv Biol. 2003;17:24–30. [Google Scholar]
- 8.Sakai AK, et al. The population biology of invasive species. Annu Rev Ecol Syst. 2001;32:305–332. [Google Scholar]
- 9.Winston ML. The Biology of the Honey Bee. Cambridge, MA: Harvard Univ Press; 1987. [Google Scholar]
- 10.Ruttner F. Biogeography and Taxonomy of Honeybees. New York: Springer; 1988. [Google Scholar]
- 11.Whitfield CW, et al. Thrice out of Africa: Ancient and recent expansions of the honey bee, Apis mellifera. Science. 2006;314:642–645. doi: 10.1126/science.1132772. [DOI] [PubMed] [Google Scholar]
- 12.Scott Schneider S, DeGrandi-Hoffman G, Smith DR. The African honey bee: Factors contributing to a successful biological invasion. Annu Rev Entomol. 2004;49:351–376. doi: 10.1146/annurev.ento.49.061802.123359. [DOI] [PubMed] [Google Scholar]
- 13.Whitfield CW, et al. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brillet C, Robinson GE, Bues R, LeConte Y. Racial differences in division of labor in colonies of the honey bee. Ethology. 2002;108:115–126. [Google Scholar]
- 15.Wilson EO. The sociogenesis of insect colonies. Science. 1985;228:1489–1495. doi: 10.1126/science.228.4707.1489. [DOI] [PubMed] [Google Scholar]
- 16.Cavalli-Sforza LL. Population structure and human evolution. Proc Biol Sci. 1966;164:362–379. doi: 10.1098/rspb.1966.0038. [DOI] [PubMed] [Google Scholar]
- 17.Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphism. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 19.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 20.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 22.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 24.Wright S. Inbreeding and homozygosis. Proc Natl Acad Sci USA. 1933;19:411–420. doi: 10.1073/pnas.19.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonin A, Taberlet P, Miaud C, Pompanon F. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frong (Rana temporaria). Mol Biol Evol. 2006;23:773–783. doi: 10.1093/molbev/msj087. [DOI] [PubMed] [Google Scholar]
- 26.Vasemagi A, Nilsson J, Primmer CR. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.). Mol Biol Evol. 2005;22:1067–1076. doi: 10.1093/molbev/msi093. [DOI] [PubMed] [Google Scholar]
- 27.Schlötterer C. A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics. 2002;160:753–763. doi: 10.1093/genetics/160.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blais J, et al. MHC adaptive divergence between closely related and sympatric African cichlids. PLoS ONE. 2007;2:e734. doi: 10.1371/journal.pone.0000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terai Y, et al. Divergent selection on opsins drives incipient speciation in Lake Victoria cichlids. PLoS Biol. 2006;4:e433. doi: 10.1371/journal.pbio.0040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rockman MV, et al. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 2005;3:e387. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 32.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt GJ, et al. Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften. 2007;94:247–267. doi: 10.1007/s00114-006-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page RE, Gadau J, Beye M. The emergence of hymenopteran genetics. Genetics. 2002;160:375–379. doi: 10.1093/genetics/160.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson SH, et al. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieseberg LH, et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambrinos JG. How interactions between ecology and evolution influence contemporary invasion dynamics. Ecology. 2004;85:2061–2070. [Google Scholar]
- 39.Jorgensen FG, Schierup MH, Clark AG. Heterogeneity in regional GC content and differential usage of codons and amino acids in GC-poor and GC-rich regions of the genome of Apis mellifera. Mol Biol Evol. 2007;24:611–619. doi: 10.1093/molbev/msl190. [DOI] [PubMed] [Google Scholar]
- 40.Akey JM, Zhang K, Xiong M, Jin L. The effect of single nucleotide polymorphism identification strategies on estimates of linkage disequilibrium. Mol Biol Evol. 2003;20:232–242. doi: 10.1093/molbev/msg032. [DOI] [PubMed] [Google Scholar]
- 41.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenblum EB, Novembre J. Ascertainment bias in spatially structured populations: A case study in the eastern fence lizard. J Hered. 2007;98:331–336. doi: 10.1093/jhered/esm031. [DOI] [PubMed] [Google Scholar]
- 43.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution (Lawrence, Kans) 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 44.Raymond M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 45.Kim SY, Pritchard JK. Adaptive evolution of conserved non-coding elements in mammals. PLoS Genet. 2007;3:e147. doi: 10.1371/journal.pgen.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 47.Beye M, et al. Exceptionally high levels of recombination across the honey bee genome. Genome Res. 2006;16:1339–1344. doi: 10.1101/gr.5680406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zar JH. Biostatistical Analysis. Upper Saddle River, NJ: Prentice-Hall; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.