Abstract
Articles in recent years have described two separate and distinct NF-κB activation pathways that result in the differential activation of p50- or p52-containing NF-κB complexes. Studies examining tumor-necrosis factor receptor-associated factors (TRAFs) have identified positive roles for TRAF2, TRAF5, and TRAF6, but not TRAF3, in canonical (p50-dependent) NF-κB activation. Conversely, it recently was reported that TRAF3 functions as an essential negative regulator of the noncanonical (p52-dependent) NF-κB pathway. In this article, we provide evidence that TRAF3 potently suppresses canonical NF-κB activation and gene expression in vitro and in vivo. We also demonstrate that deregulation of the canonical NF-κB pathway in TRAF3-deficient cells results from accumulation of NF-κB-inducing kinase (NIK), the essential kinase mediating noncanonical NF-κB activation. Thus, our data demonstrate that inhibition of TRAF3 results in coordinated activation of both NF-κB activation pathways.
Keywords: lymphotoxin-β receptor, p50, tumor-necrosis factor receptor, associated factor 3 (TRAF3)
The Rel/NF-κB family of transcription factors make essential contributions to a wide array of biological processes, including bone homeostasis, cellular proliferation and apoptosis, inflammation, and the initiation and propagation of innate and adaptive immune responses (1–3). Although the five NF-κB family members, p105 (which is constitutively processed to p50), RelA, cRel, p100 (which is processed to p52), and RelB can homodimerize and heterodimerize in numerous combinations, the predominant cellular species are p50:RelA, p50:cRel, and p100:RelB. Each of these dimers is bound in the cytoplasm by an inhibitor of κB (IκB), which prevents the nuclear translocation and transcriptional activation potential of the NF-κB complex (4, 5). Recent years have separated NF-κB signaling into two distinct activation pathways. The “classical,” or canonical, pathway requires the IκB kinase (IKK) complex, consisting of IKKα/β/γ (6). Activation of the IKK complex results in the phosphorylation and ubiquitin-dependent degradation of either IκBα or IκBβ and the nuclear translocation of p50-containing dimers. The “alternative,” or noncanonical, pathway requires the NF-κB-inducing kinase (NIK), which cooperates with IKKα to induce the processing of the p100 C terminus (termed IκBδ), which results in the nuclear translocation of p52:RelB complexes (7–10). Importantly, although activation of the canonical NF-κB pathway can occur within minutes and is independent of new protein synthesis, activation of the noncanonical NF-κB pathway requires several hours and the generation of new protein.
The tumor-necrosis factor receptor (TNFR) superfamily makes tremendous contributions to numerous biological processes, including orchestration of secondary lymphoid tissues and initiation and propagation of adaptive immune responses (11). TNFRs initiate biochemical pathway activation, in part, via recruitment of one or more of a family of adaptor molecules termed TNFR-associated factors (TRAFs) (12, 13). The TRAF family is composed of six members. Overexpression and genetic studies have identified positive roles for TRAF2, TRAF5, and TRAF6 in activation of the canonical NF-κB pathway (14–16). Among the TRAF molecules, TRAF3 has remained the most mysterious of family members. Unlike other TRAFs, overexpression of TRAF3 fails to activate the Jun N-terminal kinase (JNK) or canonical NF-κB pathways. Study of TRAF3 function was further complicated by the early postnatal lethality of TRAF3-null mice (17). Recent reports, however, have shed light on the function of TRAF3 in TNFR biology. In one study, Liao et al. (18) demonstrated that TRAF3 could promote the ubiquitination and degradation of NIK. In addition, Hauer et al. (19) established that any TNFR family member capable of binding TRAF3 could activate NIK-dependent processing of p100. Finally, we have recently shown (20) that the TRAF3-null phenotype results from constitutive activation of the noncanonical NF-κB pathway caused by the accumulation of NIK protein. Collectively, these data support a model in which NIK is constitutively degraded as a result of its association with TRAF3. Receptor recruitment of TRAF3 prevents this process, resulting in progressive accumulation of NIK and activation of the noncanonical NF-κB pathway.
Interestingly, initial characterization of NIK, which was first identified as a TRAF2 interacting protein by yeast-two hybrid assay, indicated that NIK was a potent inducer of the canonical NF-κB pathway (21). Later genetic studies, however, failed to identify a role for NIK in activation of the canonical NF-κB pathway but rather revealed a requisite role for NIK in activation of p100 to p52 processing and induction of noncanonical NF-κB activity (8, 10). Importantly, previous studies examining the contribution of NIK to activation of the canonical NF-κB pathway focused on immediate/early events after receptor ligation. An improved understanding of NIK biology, however, suggests that the potential role of NIK in activation of the canonical NF-κB pathway should be examined during periods of accumulated NIK protein.
Here, we show that TRAF3 is a potent suppressor of canonical NF-κB activity and gene induction in vitro and in vivo. In addition, we demonstrate that TRAF3 negatively regulates the IKK complex directly and that deregulated IKK complex activity in TRAF3−/− cells results from high levels of NIK. We also show that physiological accumulation of NIK, resulting from ligation of the TRAF3-binding lymphotoxin-β receptor (LTβR), results in long-term IKK complex activation and potent enhancement of NF-κB-mediated gene activation in a NIK-dependent manner. Consequently, induction of NIK protein provides a mechanism for the amplification of classical NF-κB-dependent processes. In summary, our findings indicate that receptor-mediated inhibition of TRAF3 inseparably tethers activation of the noncanonical NF-κB pathway to activation of the canonical NF-κB pathway as well.
Results
TRAF3 Deficiency Results in Increased Canonical NF-κB Activity In Vivo.
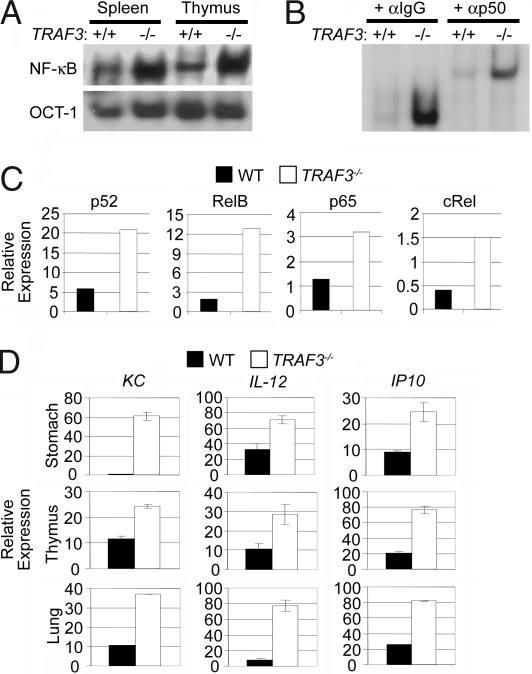
It recently was reported that targeted disruption of TRAF3 results in constitutive activation of the noncanonical NF-κB pathway (20). Previous studies examining TRAF3 function by overexpression suggested that TRAF3 also suppresses activation of the canonical NF-κB pathway. To test this finding, nuclear extract was harvested from the spleen and thymus of 8-day-old WT and TRAF3−/− mice. NF-κB activation status then was assessed by electrophoretic mobility shift analysis (EMSA) using a probe designed to preferentially bind canonical NF-κB subunits (22). As shown in Fig. 1A, TRAF3−/− organs displayed more NF-κB binding activity than WT tissues. To confirm that the increase in NF-κB binding activity was attributable to an increase in canonical pathway activation, we performed supershift analysis against the canonical NF-κB subunit p50 and proved that most of the elevated NF-κB binding activity in TRAF3−/− organs was p50-containing complexes (Fig. 1B). Next, nuclear extracts were prepared from thymus freshly harvested from 8-day-old mice and analyzed by quantitative ELISA. As expected, high binding of p52 and RelB was observed in TRAF3−/− extract compared with the WT control. In addition, a >2-fold increase in the DNA binding activity of both p65 and cRel was observed in the TRAF3−/− thymus extract (Fig. 1C), indicating that TRAF3, in addition to its role in suppressing the noncanonical NF-κB pathway, also suppresses the canonical NF-κB pathway in vivo. To assess the potential impact of heightened NF-κB activity, we harvested mRNA from the organs of WT and TRAF3−/− 8-day-old mice and analyzed the expression levels of multiple NF-κB target genes (23–25) by quantitative PCR (Q-PCR). Here, we observed that the expression levels of KC, IL-12, and IP-10 were elevated in multiple TRAF3−/− tissues, including the stomach, lung, and thymus (Fig. 1D). These data indicate that TRAF3 suppresses NF-κB target genes in vivo.
Fig. 1.
TRAF3-deficient tissues exhibit elevated canonical NF-κB activity and NF-κB target gene expression. (A) Nuclear extracts from WT and TRAF3−/− spleen and thymus harvested from 8-day-old pups were analyzed for NF-κB DNA binding activity by EMSA. Oct-1 EMSA served as a loading control. (B) Supershift assay against p50 was performed on nuclear extracts from WT and TRAF3−/− thymus. (C) Thymus nuclear extract harvested from WT and TRAF3−/− 8-day-old pups were analyzed for NF-κB DNA binding activity by quantitative ELISA. (D) RNA was isolated from WT and TRAF3−/− stomach, thymus, and lung of 8-day-old pups and assayed for levels of KC, IL-12, and IP10 mRNA by Q-PCR. Error bars are ± 1 SD between triplicate samples.
TRAF3−/− Mouse Embryonic Fibroblasts (MEFs) Exhibit Increased Basal and Inducible Canonical NF-κB Activity.
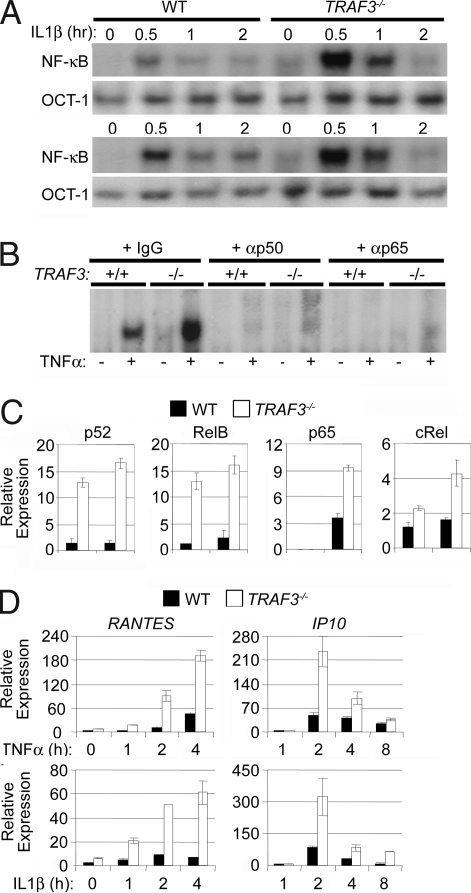
Although the increased expression of canonical NF-κB target genes in TRAF3−/− tissues was clear, it remained possible that these observations were indirect effects of TRAF3 deficiency. To address this possibility, WT and TRAF3−/− MEFs were treated with the proinflammatory cytokines IL-1β and TNFα, both of which solely induce the canonical NF-κB pathway (26). Here, EMSA analysis showed higher basal and cytokine-inducible NF-κB activity in TRAF3−/− MEFs compared with WT cells (Fig. 2A). Supershift analysis against p50 and p65 verified that this was an increase in the activation of the canonical NF-κB pathway (Fig. 2B). In addition, nuclear extract from TNFα-stimulated MEFs was analyzed by quantitative ELISA. As expected, the noncanonical NF-κB proteins, p52 and RelB, demonstrated constitutively high DNA binding activity in unstimulated nuclear extracts from TRAF3−/− MEFs and were not induced by TNFα in WT or TRAF3−/− MEFs. Alternatively, TNFα stimulation resulted in increased DNA binding activities of the canonical Rel proteins, p65 and cRel, in TRAF3−/− MEFs compared with the WT control (Fig. 2C). Importantly, the augmentation of canonical NF-κB activation resulting from TRAF3 deficiency was specific and not a result of global enhancement of TNFR/IL-1R signaling because TNFα and IL-1β-mediated ERK activation was similar in WT and TRAF3−/− MEFs [supporting information (SI) Fig. 6]. To assess the impact of heightened canonical NF-κB activation on gene induction patterns, we isolated mRNA from WT and TRAF3−/− MEFs stimulated with either TNFα or IL-1β. Again, TRAF3−/− MEFs showed increased production of both IP-10 and Rantes mRNA compared with WT cells (Fig. 2D). Collectively, these data indicate that TRAF3 suppresses the canonical NF-κB activation pathway.
Fig. 2.
TRAF3-deficient MEFs exhibit elevated canonical NF-κB activity. (A) Nuclear extracts from WT and TRAF3−/− MEFs stimulated with TNFα (20 ng/ml) or IL-1β (200 pg/ml) were analyzed for NF-κB DNA binding activity by EMSA. Nuclear extracts were incubated with the Oct-1 probe as a loading control. (B) Supershift assay against p50 and p65 was performed on nuclear extracts from WT and TRAF3−/− MEFs that were stimulated with TNFα for 30 min. (C) Nuclear extracts from WT and TRAF3−/− MEFs stimulated with TNFα for 30 min were analyzed for NF-κB DNA binding activity by using a quantitative ELISA kit. Error bars are ± 1 SD between duplicate samples. (D) WT and TRAF3−/− MEFs were stimulated with TNFα or IL-1β at the indicated times, and RNA was isolated and assayed for levels of Rantes and IP-10 mRNA by Q-PCR. Error bars are ± 1 SD between triplicate samples.
Increased IKK Complex Activity in TRAF3-Deficient MEFs.
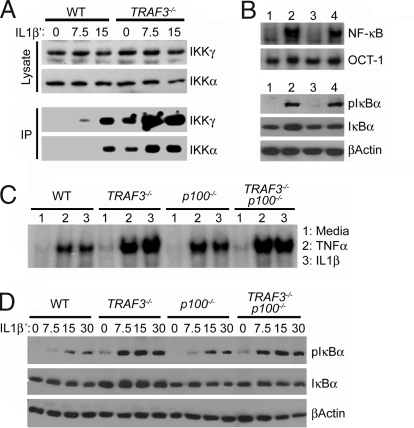
The mechanism of TRAF3 in suppression of canonical NF-κB activity could be multifaceted. First, overexpression of TRAF3 can inhibit the activation potential of other TRAFs, suggesting that loss of TRAF3 could augment the ability of TRAF2, TRAF5, or TRAF6 to activate the IKK complex. Second, TRAF3-deficient cells could exhibit an increase in basal and inducible canonical NF-κB independent of the IKK complex caused by constitutive deletion of the p100 C terminus (termed IκBδ), which also functions as an IκB against canonical Rel proteins (27, 28). To clarify these possibilities, WT and TRAF3−/− MEFs were stimulated with IL-1β, and activation of the IKK complex was then assessed by in vitro kinase assay using GST-IκBα1–54 as a substrate (29). As shown in Fig. 3A, TRAF3−/− MEFs displayed substantially higher basal and inducible activation of the IKK complex compared with WT cells. These data support a model in which TRAF3 suppression of canonical NF-κB activity is direct and not a result of secondary effects related to p100 to p52 processing. To show this, we examined basal canonical NF-κB activity, in WT, TRAF3−/−, p100−/−, and TRAF3−/−p100−/− MEFs. As shown by EMSA analysis (Fig. 3B Upper), only TRAF3−/− and TRAF3−/−p100−/− MEFs (Fig. 3B Upper, lanes 2 and 4) displayed high basal canonical NF-κB nuclear activity compared with the WT and p100−/− controls (Fig. 3B Upper, lanes 1 and 3). To assess basal IKK complex activity, we analyzed levels of the phosphorylated form of IκBα (pIκBα) by immunoblot (Fig. 3B Lower). Again, only TRAF3−/−p100−/− MEFs (Fig. 3B Lower, lanes 2 and 4) displayed high levels of pIκBα compared with the WT and p100−/− controls (Fig. 3B Lower, lanes 1 and 3). We next compared inducible canonical NF-κB activity in WT, TRAF3−/−, p100−/−, and TRAF3−/−p100−/− MEFs treated with either TNFα or IL-1β for 30 min by EMSA (Fig. 3C). Notably, the loss of p100 resulted in increased induction of canonical NF-κB activity compared with the WT control. TRAF3−/− and TRAF3−/−p100−/− MEFs, however, showed higher induction of canonical NF-κB activity compared with the WT and p100−/− controls. To see whether hyperinduction of the IKK complex was p100-independent, we compared inducible phosphorylation of IκBα in WT, TRAF3−/−, p100−/−, and TRAF3−/−p100−/− MEFs treated with IL-1β over a short time course (Fig. 3D). Here, only TRAF3−/− and TRAF3−/−p100−/− MEFs displayed earlier kinetics and increased magnitude of IκBα phosphorylation compared with the WT and p100−/− controls, indicating that TRAF3 suppresses basal and inducible IKK complex activity independent of the p100 gene.
Fig. 3.
TRAF3 negatively regulates activation of the IKK complex. (A) WT and TRAF3−/− MEFs were stimulated with IL-1β (200 pg/ml) for the indicated times. Equal amounts of whole-cell extract then were incubated with αIKKα or αIKKγ antibody, and immunoprecipitates were subjected to in vitro kinases assay with GST-IκBα1–54 as substrate. Input extracts were analyzed for total IKKα and IKKγ by immunoblot. (B) Nuclear and whole-cell extracts were isolated from WT (lane 1), TRAF3−/−p100+/+ (lane 2), TRAF3+/+p100−/− (lane 3), and TRAF3−/−p100−/− (lane 4) MEFs. Nuclear extracts were analyzed by EMSA for canonical NF-κB activity (Upper). Oct-1 EMSA is shown as a loading control. Whole extracts were analyzed for IKK complex activity by immunoblot analysis against phospho-IκBα (cells were incubated in the presence of the proteasome inhibitor MG132, 10 μM, to prevent pIκBα degradation) (Lower). (C) Nuclear extracts from WT, TRAF3−/−p100+/+, TRAF3+/+p100−/−, and TRAF3−/−p100−/− MEFs stimulated with TNFα (20 ng/ml) or IL-1β (200 pg/ml) were analyzed for NF-κB DNA binding activity by EMSA. (D) WT, TRAF3−/−p100+/+, TRAF3+/+p100−/−, and TRAF3−/−p100−/− MEFs were stimulated with IL-1β (200 pg/ml) for the indicated time points. All samples were incubated with MG132 for a total of 1 h. Whole-cell lysates then were analyzed by immunoblot using antibodies against phospho-IκBα, IκBα, and β-actin.
Accumulation of NIK Enhances Basal and Inducible IKK Complex Activity in TRAF3-Deficient and WT Cells.
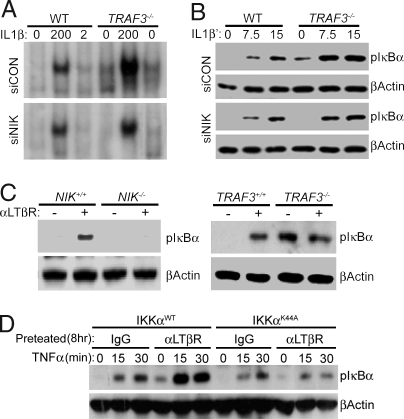
A recent report demonstrated that loss of TRAF3 results in marked accumulation of NIK protein (20). Importantly, initial characterization of NIK by overexpression demonstrated the capacity of NIK to activate the canonical NF-κB pathway (21). To examine whether NIK mediated the heightened basal and inducible canonical NF-κB activity in TRAF3−/− MEFs, we limited NIK expression with siRNAs in WT and TRAF3−/− MEFs. Cells then were stimulated with IL-1β for 30 min, and activation of the canonical NF-κB pathway was analyzed by EMSA. The specificity of the control and NIK siRNAs have been previously reported (20). TRAF3−/− MEFs treated with control siRNA displayed significantly heightened basal and inducible canonical NF-κB activity compared with the WT control (Fig. 4A Upper). In contrast, siRNA-NIK treatment of TRAF3−/− MEFs restored basal and inducible canonical NF-κB activity to WT levels (Fig. 4A Lower). This finding implicates NIK as the regulatory target of TRAF3 in suppression of the IKK complex. To test this, we next compared basal and IL-1β-inducible phosphorylation of IκBα in WT and TRAF3−/− MEFs treated with control siRNA or NIK siRNA. As expected, TRAF3−/− MEFs treated with control siRNA accumulated much more basal and inducible pIκBα compared with the WT control (Fig. 4B). In contrast, treatment of TRAF3−/− but not WT MEFs with NIK-siRNA reduced both basal and inducible levels of pIκBα. Together, these data suggest that regulation of NIK protein levels provides an additional mechanism for amplifying IKK complex activity.
Fig. 4.
Accumulation of NIK activates the IKK complex in TRAF3−/− and WT cells. (A) Nuclear extracts from IL-1β-stimulated (200 and 2 pg/ml) WT and TRAF3−/− MEFs transfected with control or NIK siRNA were analyzed for NF-κB DNA binding activity by EMSA. (B) WT and TRAF3−/− MEFs transfected with control or NIK siRNA were incubated in MG132 for a total of 1 h and stimulated with IL-1β for the indicated times. Whole-cell lysates were analyzed by immunoblot using antibodies against phospho-IκBα and IκBα. (C and D) WT and NIK−/− 3T3 cells (Left) and WT and TRAF3−/− MEFs (Right) were stimulated for 8 h with αLTβR antibody. Then, 10 μM MG132 was added to the cell cultures for the final hour. Whole-cell lysates then were analyzed for pIκBα and β-actin by immunoblot. (D) IKKα−/− 3T3 cells reconstituted with WT IKKα or the kinase dead mutant, IKKαK44A, were incubated 1 μg/ml of mouse IgG or αLTβR antibody for 8 h. Cells were stimulated with 20 ng/ml of TNFα for the indicated periods. Then, 10 μM MG132 was added to the media for the final hour. Whole-cell extracts were analyzed for pIκBα and β-actin.
Stimulation of WT MEFs with αLTβR antibody results in the slow accumulation of NIK to levels seen in TRAF3−/− MEFs (data not shown). We therefore speculated that accumulation of NIK in WT cells also would result in heightened IKK complex activation as seen in TRAF3−/− MEFs. To test this, NIK+/+ and NIK−/− 3T3 cells were stimulated with αLTβR antibody for 8 h and then analyzed for pIκBα by immunoblot analysis. Here, the capacity of LTβR to sustain long-term phosphorylation of IκBα required NIK (Fig. 4C). In addition, ligation of LTβR on TRAF3−/− MEFs did not result in any further increase in pIκBα levels (Fig. 4D). These data demonstrate that accumulation of NIK results in activation of the IKK complex. In addition, these data explain why previous analyses found no physiological role for NIK in IKK complex activation because these studies (10, 30) focused on immediate/early events after receptor ligation, i.e., at times before significant NIK accumulation.
A previous overexpression study indicated that NIK-mediated activation of the IKK complex required IKKα kinase activity (31). To determine whether this also was true at physiological levels, IKKα−/− 3T3 cells were reconstituted with WT IKKα or the kinase dead mutant, IKKαK44A. Immunoblot analysis of LTβR-induced p100 to p52 processing was performed to verify the IKKα kinase activity of the reconstituted cells (data not shown). To allow for NIK accumulation, IKKαWT and IKKαK44A reconstituted 3T3 cells were incubated for 8 h in the presence of IgG or αLTβR antibody and then stimulated with TNFα for the indicated time periods. Immunoblot analysis of phospho-IκBα levels demonstrated that IKKα kinase activity was required for the heightened basal and inducible IKK complex elicited by long-term stimulation of the LTβR. Together, these data demonstrate that TRAF3 modulates the IKK complex by regulation of the NIK–IKKα axis.
Synergy Between TNFR and LTβR in Inflammatory Gene Induction.
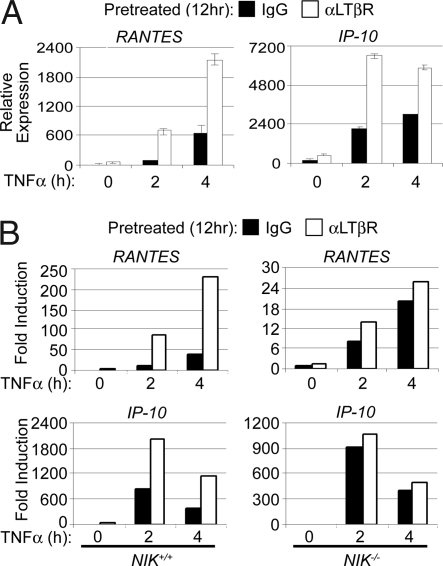
Several physiological contexts, including osteoclast differentiation and secondary lymphoid tissue development, involve integration of NIK and inflammatory cytokine signal transduction (2, 32, 33). Given our findings that accumulation of NIK heightened IKK complex activation, we speculated that accumulation of NIK also would augment NF-κB-mediated gene induction. To explore this possibility, we incubated WT MEFs in media alone or in the presence of αLTβR antibody for 12 h followed by a time course of TNFα stimulations. RNA then was harvested for analysis of gene induction by Q-PCR. As shown in Fig. 5B, cells receiving the combinatorial stimulation exhibited a significant increase in the induction of Rantes and IP-10 as seen with TNFα and IL-1β stimulations of TRAF3−/− MEFs (Fig. 2D). To determine whether NIK was required for the enhancement of gene induction, the same experiment was performed by using NIK+/+ and NIK−/− 3T3 cells. As shown in Fig. 5C, enhanced gene induction mediated by LTβR and TNFR1 engagement was lost in the absence of NIK. These results indicate that stabilization of NIK provides a mechanism for the amplification of canonical NF-κB-dependent cellular responses.
Fig. 5.
Accumulation of NIK mediates LTβR and TNFR synergistic gene activation. (A) WT MEFs were incubated in the presence of 1 μg/ml of mouse IgG or αLTβR antibody for 12 h. Cells then were stimulated with 20 ng/ml TNFα for the indicated times. RNA then was isolated and assayed for levels of Rantes and IP-10 mRNA by Q-PCR. Error bars are ± 1 SD between triplicate samples. (B) NIK+/+ and NIK−/− MEFs were stimulated as above, and RNA was isolated and assayed for levels of Rantes and IP-10 mRNA by Q-PCR. Error bars are ± 1 SD between triplicate samples.
Discussion
Numerous receptors can activate NF-κB transcription factors by either the canonical and/or noncanonical pathways. Although previous studies demonstrated the role of TRAF3 as a critical negative regulator of noncanonical NF-κB activities, our current work provides both in vitro and in vivo evidence that TRAF3 also suppresses canonical NF-κB activities. Our studies further revealed that TRAF3 modulates activity of the IKK complex through regulation of NIK protein, which is thought to be specifically involved in the activation of the noncanonical pathway. Importantly, our studies of LTβR and TNFR1 synergy suggest that, in addition to its essential role in activation of the noncanonical NF-κB pathway, NIK also functions as an amplifier of canonical NF-κB activities, which may play an important role in development of autoimmune and inflammatory diseases.
Our analysis of canonical NF-κB activation profiles in WT, TRAF3−/−, p100−/−, and TRAF3−/−p100−/− cells (review Fig. 4D) clearly demonstrates that hyperactivation of the IKK complex in TRAF3−/− cells was not a consequence of constitutive deletion of the p100 C terminus, which has the ability to bind and sequester RelA, cRel, and p50 in the cytoplasm. At the same time, EMSA analysis showed that p100 deficiency alone resulted in heightened canonical NF-κB nuclear activity in response to stress stimuli. Thus, the total increase in canonical NF-κB seen in TRAF3-deficient cells and tissues likely results from the combination of increased IKK complex activation and constitutive deletion of the p100 C terminus. In addition, a previous report demonstrated that overexpressed NIK-mediated activation of IKKα resulted in phosphorylation and activation of IKKβ (31). Because TRAF3 deficiency results in constitutive NIK–IKKα activity, there is likely some IKKα-mediated activation of IKKβ that would account for the higher basal and inducible canonical NF-κB in TRAF3−/− cells. This model is strongly supported by our finding that suppression of NIK protein levels reduced the high basal and inducible levels of phospho-IκBα in TRAF3−/− cells. In addition, we found that long-term ligation of LTβR resulted in activation of the IKK complex in a NIK- and IKKα-dependent manner (Fig. 4 C and D). Thus, it appears that TRAF3 suppresses both canonical and noncanonical NF-κB activation through inhibition of the NIK–IKKα axis.
In this article, we demonstrated that the TRAF3 phenotypes of hyper IKK complex activation and NF-κB-dependent gene induction in response to inflammatory cytokines also could be generated in WT MEFs with accumulated NIK, indicating that control of NIK protein levels provides an additional mechanism for regulating canonical NF-κB activity. There are several normal physiological contexts, including osteoclastogenesis, secondary lymphoid tissue development, and peripheral B cell activation, that involve the integration of sustained noncanonical NF-κB signaling with activation of the canonical NF-κB pathway (34–36). At the same time, the NIK-activating TNFR family members that mediate these processes—namely, RANK, BAFF, and CD40—also are implicated in many inflammatory diseases (37–40). In addition, recent work strongly indicates that elevated canonical and noncanonical NF-κB activity by deregulation of NIK directly contributes to the progression of primary multiple myelomas (41, 42). Collectively, these data suggest that TRAF3 functions as an important suppressor of inflammatory disease and cancer through the negative regulation of the canonical and noncanonical NF-κB pathways.
Materials and Methods
Mice Colony and Cell Culture.
C57BL/6 (The Jackson Laboratory) mice aged 6–12 weeks were used as recipients in fetal liver transplant experiments. Targeted disruption of the TRAF3 allele and the p100 allele has been described previously (17, 43). All mice were maintained and bred under specific pathogen-free conditions in the University of California Life Sciences mouse facility (Los Angeles), and experiments were conducted within the parameters of our approved protocol. MEFs and 3T3 cells were cultured in DMEM (Mediatech), supplemented with 5% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml).
Antibodies and Reagents.
The anti-IκBβ (C-20), anti-IκBα (C-21), and the anti-TRAF3 (C-20) antibodies for Western blotting and the p65 (C-20) and pSO H-119 antibodies were obtained from Santa Cruz Biotechnology. The antibodies to phospho- IκBα, phospho-ERK, and ERK were obtained from Cell Signaling. The anti-LTβR antibody was from Alexis Biochemicals and the anti-β-actin antibody, propidium iodide, and LPS were obtained from Sigma. Human IL-1β and mouse TNFα were purchased from R&D Systems.
Western Blot Analysis.
MEFs, B cells, or cells from tissues were lysed in RIPA buffer (50 mM Tris·Cl, pH 7.5/150 mM NaCl/1% Nonidet P-40/0.5% Na deoxycholate/0.1% SDS) for 30 min on ice. Equal amounts of whole-cell lysates were loaded onto 12% SDS/PAGE. Alternatively, equal volume of MEFs lysed in 50 mM Tris·Cl (pH 6.8), 20% 2-mercaptoethanol, 2% SDS, 10% glycerol, and 0.1% bromophenol blue was loaded onto 7% SDS/PAGE. Gels were transferred to PVDF membranes (Immobilon-P) and immunoblotted according to the manufacturer's recommended instructions.
EMSA and NF-κB ELISA.
Cells from tissues were isolated from 8-day-old mice with a syringe and cell strainer. To obtain nuclear fractions, cells were first lysed in buffer A (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, and 0.2% Nonidet P-40). Nuclei then were isolated and lysed in buffer C (25% glycerol, 20 mM Hepes, 0.6 M KCl, 1.5 mM MgCl2, and 0.2 mM EDTA). Nuclear extract was preincubated with a NF-κB probe or an Oct-1 probe for 15 min at room temperature in a final volume of 20 μl as described in ref. 44. The NF-κB probe used for EMSA contained the sequence 5′-GAGAGGGGATTCCCCGATTAGCTTTCGGGGAATCCCCTCT-3′, which was derived from the Igκ MHC H2 promoter. The Oct-1 probe contained the sequence 5′- TGTCGAATGCAAATCACTAGAA-3′. The NF-κB ELISA was performed according to manufacturer's recommended instructions (BD Biosciences). A total of 2.5 μg and 8 μg of nuclear extract from MEFs and thymus was used per reaction, respectively.
Real-Time Q-PCR.
RNA was isolated by using TRIzol Reagent (Invitrogen) according to the manufacturer's protocol. RNA was quantitated, and 1 μg of RNA was used to make cDNA templates by using iScript (Invitrogen) according to the manufacturer's instructions. Q-PCR analyses were performed by using the iCycler thermocycler (Bio-Rad Laboratories). Primer sequences for KC, VCAM-1, MCP-1, IL-12 p40, IP-10, Rantes, and L32 are available on request. L32 expression measurements were conducted in tandem with the gene of interest. All Q-PCR data are presented as relative expression units after normalization to the average L32 value to control for loading of total RNA.
siRNA.
siRNAs were transfected into MEFs by using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). NIK-specific and control siRNAs consisted of an equal molar mix of the following sequences (sense strands): siNIK, GGATTATGAGTATCGAGAA[dT][dT] and UCC ACA GAA UGA AGG ACA A[dT][dT]; siControl, (GGAGTATGACTAAGTTGAA[dT][dT] and TCCGAAAGTAAGGAACCAA [dT][dT]).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Robert Schreiber, Washington University, and Amgen for kindly providing NIK-deficient and control MEFs. We also thank Dr. Yangxin Fu (University of Chicago) for kindly proving agonistic αLTβR antibody. B.Z. is supported by the Warsaw Fellowship. J.Q.H. is supported by Clinical and Fundamental Immunology Training Grant AI07126-30. G.C. is a Lymphoma and Leukemia Society Scholar. Part of this work was supported by National Institutes of Health research Grants R01 AI056154, R01 CA87924, and R01 GM57559.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707959105/DC1.
References
- 1.Baeuerle PA, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 2.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Nat Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 3.Kopp EB, Ghosh S. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 4.Hayden MS, Ghosh S. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 6.Zandi E, Karin M. Mol Cell Biol. 1999;19:4547–4551. doi: 10.1128/mcb.19.7.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 8.Xiao G, Fong A, Sun SC. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 9.Xiao G, Harhaj EW, Sun SC. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 10.Yin L, Wu L, Wesche H, Arthur CD, White JM, Goeddel DV, Schreiber RD. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 11.Dempsey PW, Doyle SE, He JQ, Cheng G. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 12.Bradley JR, Pober JS. Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 13.Rothe M, Wong SC, Henzel WJ, Goeddel DV. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 14.Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothe M, Sarma V, Dixit VM, Goeddel DV. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 16.Saitoh T, Nakayama M, Nakano H, Yagita H, Yamamoto N, Yamaoka S. J Biol Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Cheng G, Baltimore D. Immunity. 1996;5:407–415. doi: 10.1016/s1074-7613(00)80497-5. [DOI] [PubMed] [Google Scholar]
- 18.Liao G, Zhang M, Harhaj EW, Sun SC. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 19.Hauer J, Puschner S, Ramakrishnan P, Simon U, Bongers M, Federle C, Engelmann H. Proc Natl Acad Sci USA. 2005;102:2874–2879. doi: 10.1073/pnas.0500187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, Dempsey PW, Cheng G. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malinin NL, Boldin MP, Kovalenko AV, Wallach D. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 22.Bonizzi G, Bebien M, Otero DC, Johnson-Vroom KE, Cao Y, Vu D, Jegga AG, Aronow BJ, Ghosh G, Rickert RC, Karin M. EMBO J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann A, Leung TH, Baltimore D. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy TL, Cleveland MG, Kulesza P, Magram J, Murphy KM. Mol Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohmori Y, Fukumoto S, Hamilton TA. J Immunol. 1995;155:3593–3600. [PubMed] [Google Scholar]
- 26.Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa H, Carrasco D, Claudio E, Ryseck RP, Bravo R. J Exp Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheinman RI, Beg AA, Baldwin AS., Jr Mol Cell Biol. 1993;13:6089–6101. doi: 10.1128/mcb.13.10.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 30.Matsushima A, Kaisho T, Rennert PD, Nakano H, Kurosawa K, Uchida D, Takeda K, Akira S, Matsumoto M. J Exp Med. 2001;193:631–636. doi: 10.1084/jem.193.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Mahony A, Lin X, Geleziunas R, Greene WC. Mol Cell Biol. 2000;20:1170–1178. doi: 10.1128/mcb.20.4.1170-1178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endres R, Alimzhanov MB, Plitz T, Futterer A, Kosco-Vilbois MH, Nedospasov SA, Rajewsky K, Pfeffer K. J Exp Med. 1999;189:159–168. doi: 10.1084/jem.189.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katakai T, Hara T, Sugai M, Gonda H, Shimizu A. J Exp Med. 2004;200:783–795. doi: 10.1084/jem.20040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aloisi F, Pujol-Borrell R. Nat Rev Immunol. 2006;6:205–217. doi: 10.1038/nri1786. [DOI] [PubMed] [Google Scholar]
- 35.Kalled SL. Immunol Rev. 2005;204:43–54. doi: 10.1111/j.0105-2896.2005.00219.x. [DOI] [PubMed] [Google Scholar]
- 36.Jones DH, Nakashima T, Sanchez OH, Kozieradzki I, Komarova SV, Sarosi I, Morony S, Rubin E, Sarao R, Hojilla CV, et al. Nature. 2006;440:692–696. doi: 10.1038/nature04524. [DOI] [PubMed] [Google Scholar]
- 37.Haynes DR. Inflamm Res. 2004;53:596–600. doi: 10.1007/s00011-004-1303-z. [DOI] [PubMed] [Google Scholar]
- 38.Mach F, Schonbeck U, Sukhova GK, Atkinson E, Libby P. Nature. 1998;394:200–203. doi: 10.1038/28204. [DOI] [PubMed] [Google Scholar]
- 39.Mackay F, Woodcock SA, Lawton P, Ambrose C, Baetscher M, Schneider P, Tschopp J, Browning JL. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toubi E, Shoenfeld Y. Autoimmunity. 2004;37:457–464. doi: 10.1080/08916930400002386. [DOI] [PubMed] [Google Scholar]
- 41.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarnegar B, He JQ, Oganesyan G, Hoffmann A, Baltimore D, Cheng G. Proc Natl Acad Sci USA. 2004;101:8108–8113. doi: 10.1073/pnas.0402629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beg AA, Sha WC, Bronson RT, Baltimore D. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.