Abstract
Developmental commitment involves activation of lineage-specific genes, stabilization of a lineage-specific gene expression program, and permanent inhibition of inappropriate characteristics. To determine how these processes are coordinated in early T cell development, the expression of T and B lineage-specific genes was assessed in staged subsets of immature thymocytes. T lineage characteristics are acquired sequentially, with germ-line T cell antigen receptor-β transcripts detected very early, followed by CD3ɛ and terminal deoxynucleotidyl transferase, then pTα, and finally RAG1. Only RAG1 expression coincides with commitment. Thus, much T lineage gene expression precedes commitment and does not depend on it. Early in the course of commitment to the T lineage, thymocytes lose the ability to develop into B cells. To understand how this occurs, we also examined expression of well defined B lineage-specific genes. Although λ5 and Ig-α are not expressed, the μ0 and Iμ transcripts from the unrearranged IgH locus are expressed early, in distinct patterns, then repressed just before RAG1 expression. By contrast, RNA encoding the B cell receptor component Ig-β was found to be transcribed in all immature thymocyte subpopulations and throughout most thymocyte differentiation. Ig-β expression is down-regulated only during positive selection of CD4+CD8– cells. Thus several key participants in the B cell developmental program are expressed in non-B lineage-committed cells, and one is maintained even through commitment to an alternative lineage, and repressed only after extensive T lineage differentiation. The results show that transcriptional activation of “lymphocyte-specific” genes can occur in uncommitted precursors, and that T lineage commitment is a composite of distinct positive and negative regulatory events.
In the mammalian hematopoietic system, cells differentiating from the same precursor can adopt any of multiple distinct developmental fates. Choice of a developmental lineage involves the activation of lineage-appropriate genes and the repression, or blocking of induction, of inappropriate genes. Many specific genes have been identified that characterize particular hematopoietic cell types, and the expression of these genes has begun to provide useful landmarks for hematolymphoid differentiation (1–10). However, the mechanism of lineage specification and the “commitment” events that make it irreversible still are not well understood at the level of the control of lineage-specific gene expression. One major question is whether “master switch” events exist, in which cellular identity is established by turning on a set of lineage-specific genes coordinately. Another major question is whether cells can activate such lineage-specific genes, yet still keep the option of following other developmental pathways. These unanswered questions cast shadows over attempts to infer the timing and mechanism of developmental commitment from the timing of activation of particular lineage-specific genes.
To address these issues we have focused on early murine T cell differentiation. In this system there are substantial data to establish a succession of lineage-restriction events, when developmental potential is narrowed, and to define their timing relative to defined phenotypic changes (11, 12). We have used this system to investigate, first, which T cell-specific genes are activated at key stages of commitment, and second, when the cells lose the ability to express “inappropriate” genes. Our results show that T lineage genes are activated noncoordinately, that many of these genes are activated before commitment, that at least one B lineage gene is also activated during T lineage differentiation, and that repression of such inappropriate genes can occur long after the cells are committed.
MATERIALS AND METHODS
Cell Preparation and Reverse Transcriptase–PCR (RT-PCR) Conditions.
All thymocytes were obtained from animals between 3.5 and 5.5 weeks old and stained as described (13). Stained cells, 5–20 × 104 per selected population, were sorted into chilled tubes containing Hanks’ balanced salt solution without phenol red, with 0.25% BSA, 5% fetal bovine serum, and azide. A portion of each sorted sample was reserved for reanalysis, and the rest of the cells were spun down, carrier tRNA was added, and first-strand cDNA was synthesized from each sample in a 40-μl reaction as described (13).
Primer pairs and annealing temperatures for the RT-PCRs were taken from the following references: HPRT (14); RAG-1, Ig-α, λ5, and terminal deoxynucleotidyltransferase (TdT) (1); CD3ɛ (15); T cell antigen receptor-Cβ (TCR-Cβ) (16); pTα (17); Iμ (18); μ0 and Ig-β (7). All amplification products cross an exon boundary. Different sample inputs were normalized to obtain matched signals with hypoxanthine phosphoribosyltransferase primers at a limiting number of cycles. Amplification was for 29–35 cycles (usually 35) in the experiments shown, and each lane shows the product amplified from ≈1% of the cells in the initial sample. Repeated analyses with various primers at lower and higher PCR cycle numbers were routinely used to verify relative levels of expression in different populations. The identity of the Ig-β PCR product (nucleotides 194–688), was confirmed by hybridization with an end-labeled internal oligonucleotide (nucleotides 571–591). Hybridization (at 20–25 cycles) was quantitated by PhosphorImager (Molecular Dynamics).
Fetal Liver and Spleen Cell Fractionation.
C57BL/6 fetal liver samples were obtained at E14.5 and stained with antibodies against B220 (CD45R, RA3–6B2), Sca-1 (Ly-6A, E13–161.7), and Gr-1 (Ly-6G, RB6–8C5), all from PharMingen. Preparative cell sorting with three-color flow cytometry was used to enrich four populations: (i) the B220− Sca-1− Gr-1− majority population (≈92% of the population was in triple negative quadrants; 66% within sorting gate); (ii) the B220+ Sca-1+ Gr-1− population (0.4% of cells); (iii) the B220− Sca-1+ Gr-1− population (1.0%); and (iv) the Sca-1− Gr-1+ population (2.5%). T enriched splenocytes were obtained by magnetic cell sorting with streptavidin magnetic beads (MACS, Miltenyi Biotec) and biotinylated antibodies against B220, Gr-1, and Ter-119 (PharMingen) to deplete cells of other lineages. CD4+ and CD8+ subsets, obtained from the T enriched cells by fluorescence-activated cell sorting, were each >98.5% pure.
Fractionation of Thymocytes Undergoing Positive Selection.
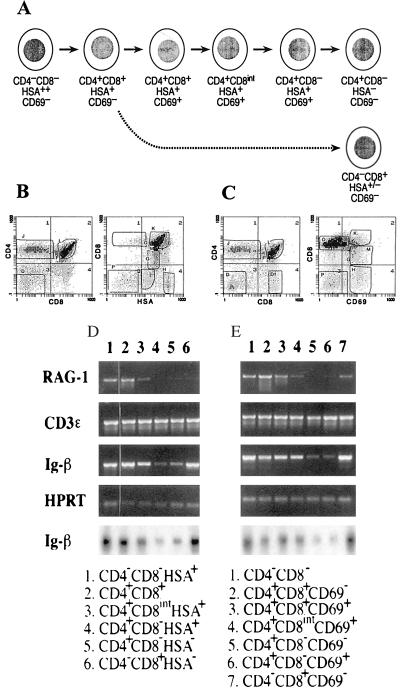
C57BL/6-Tlaa. thymocytes were stained, after Fc receptor blocking, with phycoerythrin-conjugated anti-CD4 (Becton Dickinson Labware), biotin conjugated anti-CD8 (Becton Dickinson Labware), and Streptavidin-RED670 (Life Technologies, Gaithersburg, MD), as well as fluorescein isothiocyanate antibodies against either HSA (sort 1) (CD24, PharMingen) or CD69 (sort 2) (PharMingen). Sorting gates are shown in Fig. 4 (sort 1, B; sort 2, C). Cells were sorted by using single or double two-color gates depending on the markers used. Only the CD4+ CD8+ CD69+ population showed significant (>7%) contamination with other populations upon reanalysis.
Figure 4.
Shutoff of Ig-β expression at a late stage in the positive selection of CD4+ CD8− thymocytes. (A) Summary of phenotypic changes accompanying positive selection. (B and C) Three-color flow cytometry of normal C57BL/6 thymocytes stained for CD4, CD8, and HSA expression (B) or for CD4, CD8, and CD69 expression (C). The sorting gates used in the two sorting experiments to dissect positive selection stages are indicated, as described in Materials and Methods. (D and E) RT-PCR analyses of RNA levels in the populations fractionated from the stained samples in B (D) and in C (E). Also shown (bottom row, D and E) are autoradiograms of the hybridization of an internal probe to the Ig-β PCR products from these samples. Note that in E, the samples in lanes 5 (CD4+ CD8− CD69−) and 6 (CD4+ CD8− CD69+) are reversed with respect to their order in development.
RESULTS
Ordered Induction of T Lineage Genes Before Lineage Commitment.
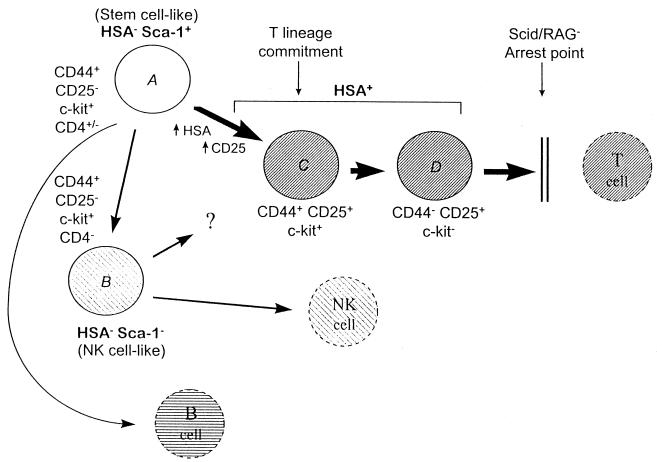
Expression of both lineage-appropriate and lineage-inappropriate genes was monitored by RT-PCR analysis of RNA from defined subsets of immature thymocytes. To obtain the cells in high enrichment, in most cases the starting thymocyte populations were taken from immunodeficient scid/scid and RAG2−/− mice and sorted by multiparameter flow cytometry, as described in detail (13). The developmental potentials of the equivalent populations of normal thymocytes have been studied extensively by cell transfer and in vitro culture approaches (reviewed in refs. 11 and 12), and all interpretations in this report of their relative progress toward T lineage commitment are based on that body of work. Three main populations were examined, as shown in Fig. 1: HSA+ Sca-1+ CD25+ CD44+/− c-kit+/−cells (“HSA+”), which include cells that would normally be undergoing TCR gene rearrangement (populations C and D); HSAlow Sca-1+ CD25− CD44high c-kit+ cells (“HSA− Sca-1+”), which are partially CD4low and resemble stem cells (population A); and HSAlow Sca-1− CD25− CD44high c-kit+ cells (“HSA− Sca-1− ”), which resemble immature NK cells (population B) (13, 19) (H.W., R.A.D., J. A. Yang-Snyder, and E.V.R., unpublished results). To control for biological variation, all conclusions are based on parallel analyses of corresponding subpopulations from different strains of scid/scid mice and from RAG2−/− mice.
Figure 1.
Summary of immature cell populations in the immunodeficient mouse thymus and their developmental relationships. Bold type indicates the names of subsets of cells analyzed in this paper. The extended profiles of phenotypic markers are from refs. 13 and 28 and H.W., R.A.D., and E.V.R., unpublished results. Italic designations A–D key these populations to the samples studied in Fig. 2. Cell fates and developmental relationships among populations A through D are based on work by others using the corresponding populations from normal adult thymus, in which populations A and B are not separated (11, 12). The CD44+ CD25+ subset of HSA+ cells (population C) has lost the potential to give rise to B or NK cells, although it retains the ability to give rise to dendritic cells (not shown). Cells beyond this stage (e.g., population D) are fully T lineage committed. The possibility that the NK-like cells (population B) can give rise to T cells as well as NK cells is based on the properties of a similar subset in fetal thymus (51–53). The CD4+ subset, which has been studied extensively in normal thymus, includes a subset of population A plus some of the transitional cells between A and C that have not yet acquired CD25 (H.W., R.A.D., and E.V.R., unpublished results).
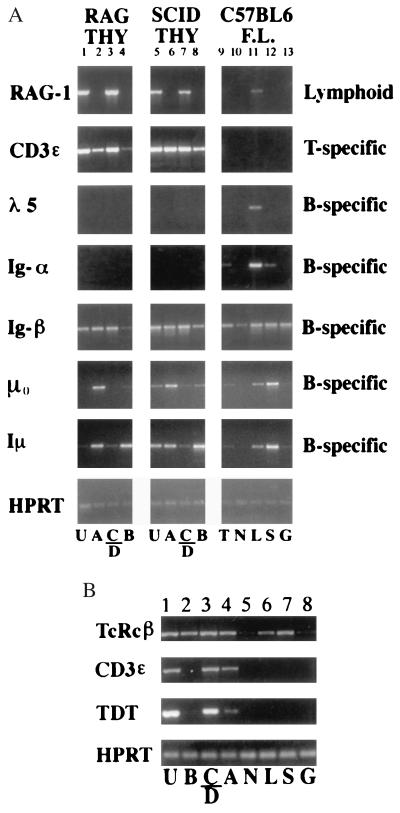
Fig. 2 shows that T lineage genes begin to be expressed noncoordinately in these cell types. RAG1 RNA was expressed only in the HSA+ subpopulations (Fig. 2A), as expected because only at this stage does TCR gene rearrangement normally begin (20). Expression of CD3ɛ and TdT begins earlier, because these were observed in HSA+ cells and, at lower levels, in the HSA− Sca-1+ cells (Fig. 2 A and B). Another T lineage-specific gene, pTα, showed an onset of expression intermediate between those of CD3ɛ and TdT and that of RAG1 (data not shown). However, germ-line transcripts from the TCRβ constant region were observed at similar levels in all thymocyte populations (in these mutant cells even the HSA+ stages must use the germ-line configuration for expression, so only TCRβ enhancer activity is relevant). TCR-Cβ transcripts also were the only T cell-specific genes detectable in fractions from normal E14.5 fetal liver, as shown in Fig. 2B. TCR-Cβ transcripts were expressed both in a fetal liver fraction enriched for B, T, and NK progenitors (fraction “L,” Sca-1+ B220+ Gr-1−; Fig. 2B, lane 6) (21–24) and in a stem cell-enriched fraction (fraction “S,” Sca-1+ B220− Gr-1−) (Fig. 2B, lane 7). Some RAG1 expression also was seen in the fetal liver “L” fraction (Fig. 2A, lane 11), but this appears to be because of B lineage precursors therein. The lack of any CD3ɛ expression in these populations shows that T lineage differentiation in E14.5 fetal liver does not proceed even to the level of the earliest thymocytes.
Figure 2.
Expression of early T cell genes in immature thymocytes and normal fetal liver cells. RT-PCR was performed with the indicated primers to compare subsets of cells from immunodeficient mouse thymocytes with subsets from wild-type C57BL/6 fetal liver. All samples presented in one lane were derived from equal aliquots of the same cDNA sample. Subpopulations analyzed in each lane are indicated by the letters under each sample. For thymocyte subsets: U, unfractionated; A, HSA–Sca-1+; B, HSA– Sca-1–; C/D, HSA+ (Sca-1+). For fetal liver subsets: T, total; N, B220− Sca-1− Gr-1− (triple negative); L, B220+ Sca-1+ Gr-1− (lymphoid-enriched); S, B220− Sca-1+ Gr-1− (stem cell enriched); G, Sca-1− Gr-1+ cells. (A) Lanes: 1–4, RAG2−/− thymus; 5–8, C.B 17-scid/scid thymus; 9–13, C57BL/6 fetal liver subsets. (B) Lanes: 1–4, B6-scid/scid thymus; 5–8, C57BL/6 fetal liver. RAG1 levels are higher in RAG-2−/− thymocytes than in SCID thymocytes because the former survive longer after the induction of recombinase expression (13). TdT is expressed only during postnatal lymphocyte development.
These results suggest that TCRβ germ-line transcription is activated first in lymphoid precursors, followed by CD3ɛ and TdT, then pTα, and last by induction of RAG1. The expression of pTα and TdT seems to be excluded or down-regulated in the NK-like HSA− Sca-1− population (“B”), and RAG1 expression is induced only in those cells that have reached the HSA+ stage (“C/D”). Some expression is precocious: at least two T lineage-specific genes, TCRβ and CD3ɛ, clearly begin to be expressed before the stages (CD25+ CD44+/−; here, population “C” in the HSA+ fraction) when the population is committed to give rise to T cells (25–28).
Selective Ectopic Expression of a B Lineage Gene.
B cell developmental potential is reported to persist in the “CD4low” class of immature thymocytes (25, 26, 28), which consists of cells in a continuum from the HSA− Sca-1+ fraction to the HSA+ fraction (29) (H.W., unpublished results; see Fig. 1). However, the B cell genes λ5 and Ig-α (mB-1, CD79a), which were detected readily in the Sca-1+ B220+ fetal liver fraction, were undetectable in most of the thymocyte subsets (Fig. 2A). A slight signal for Ig-α expression was observed only in the minor, poorly characterized HSA+ Sca-1− fraction (data not shown), possibly because of the presence of a minor population of thymic B cell precursors (30). The absence of Ig-α and λ5 expression initially suggested that the highly immature HSA− Sca-1+ thymocytes (population “A”) do not express any B lineage genes, even though they may be competent to undergo B lineage differentiation. However, as representatives of even earlier B lineage genes (1, 7, 18), expression of the germ-line IgH transcripts μ0 and Iμ and of Ig-β (B29, CD79b) was also monitored (Fig. 2A). The primer pairs used for Ig-β detect the full-length mRNA, not the deleted form (31, 32).
Unlike Ig-α and λ5, these three transcripts were detected in thymocytes, describing distinct patterns of expression. The μ0 amplification product was generated only from the most immature, HSA− Sca-1+ cells (“A”), whereas the Iμ product (generally considered B lineage-specific) was found in both subsets of HSA− cells (“A” and “B”). Both of these transcripts, however, were shut off during the transition from HSA− to HSA+ (Fig. 2A and data not shown). Thus, the germ-line transcription of the IgH locus does initiate in thymocytes, but is repressed during or before the induction of RAG1. Like these transcripts, Ig-β was also found to be expressed in the HSA− Sca-1+ thymocytes (Fig. 2A). Unexpectedly, however, Ig-β was also found in the more advanced HSA+ thymocytes (populations “C” and “D”), which correspond in phenotype (CD44+/− CD25+) to normal immature thymocytes that have completely lost any B cell developmental potential (25, 26, 28). Only the HSA− Sca-1− thymocytes (“B”) expressed this gene at lower levels.
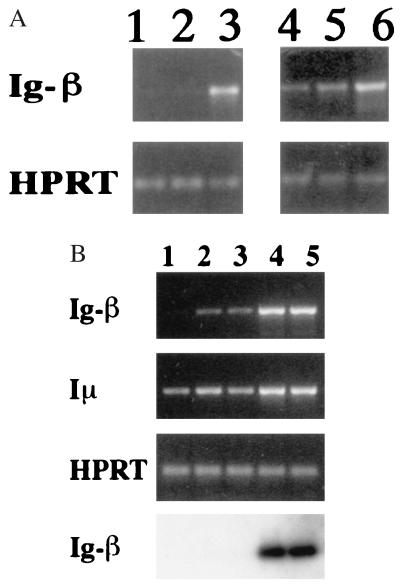
Ig-β initially was reported to be expressed only in B lineage cells, and absent from thymocytes, by the criterion of slot-blot hybridization (33). To confirm that genuine Ig-β was being detected with our primers, normal adult tissues were tested to verify specificity of expression (Fig. 3), and the PCR products were hybridized with a third, labeled oligonucleotide representing an internal Ig-β sequence (Fig. 3B). Although expression was abundant in normal spleen (Fig. 3A, lanes 3 and 6), no product could be amplified from skeletal muscle cDNA (lane 1; ≈102 × less, by hybridization), and much less Ig-β could be detected in cDNA from T enriched normal spleen cells (Fig. 3A, lane 2; Fig. 3B, lane 3). In pure populations of T cells isolated by fluorescence-activated cell sorting (Fig. 3B, lanes 1 and 2), only very low levels of Ig-β RNA were detected in the CD8+ population (lane 2) and even less was detected in the CD4+ population (lane 1). When monitored by hybridization at low numbers of PCR cycles, Ig-β RNA indeed was undetectable in these T cells (Fig. 3B Bottom). By contrast, the μ0 and Iμ transcripts from the germ-line IgH locus were readily detected in mature T cells (Fig. 3B and data not shown). Thus, our results support the identification of Ig-β as highly “B lineage-specific” at the level of mature cells.
Figure 3.
Preferential expression of Ig-β (B29) in populations containing B cells and thymocytes. (A) Analysis of Ig-β RNA expression in different tissues. Lanes: 1, skeletal muscle; 2, T enriched spleen (>93% T cells); 3 and 6, normal mouse splenocytes (≥60% B cells; 4, total C57BL/6-scid/scid thymocytes; 5, normal C57BL/6 thymocytes. (B) Comparison of Ig-β and Iμ expression in subsets of normal mouse splenic T cells and B cells. T cell subsets were purified as described in Materials and Methods. Samples without reverse transcriptase gave no signal (data not shown). (Top) Products of 33 PCR cycles detected by ethidium bromide staining. (Bottom) Products of 24 PCR cycles detected by hybridization. Lanes: 1, CD4+ T cells; 2, CD8+ T cells; 3, T enriched spleen; 4, total spleen; 5, B220+ B enriched cells.
By contrast, unfractionated normal thymus populations with no significant B lineage contamination showed at least as much Ig-β expression as immunodeficient thymus populations (Fig. 3A, lanes 4 and 5). Although there are non-T lineage and primitive cells in scid or RAG− thymus that might express Ig-β, these are diluted ≈100× in normal thymus. Thus, this result rules out the possibility that thymic Ig-β expression is restricted to a rare early or non-T lineage thymocyte type. The level of this expression in unfractionated normal thymus remains ≥9× lower than in spleen (data not shown), explaining the lack of detection using other methods. Even so, Ig-β RNA continues to be expressed in cells before and after TCR surface expression, and in thymocytes of both TCR-αβ and TCR-γδ lineages, as shown by the similar levels of Ig-β RNA per cell in thymocytes from normal, TCRβ−/−, TCRδ−/−, and TCRβ−/− δ−/− mice (data not shown). Thus, although mature T cells express little if any Ig-β, the ongoing transcription of this B cell gene appears to be a common and persistent feature of diverse classes of thymocytes.
Delayed Repression: Shutoff of Ig-β Expression During Positive Selection.
To ascertain whether Ig-β expression might be turned off at any stage of thymic development, normal C57BL/6 thymocytes were fractionated to separate cells through different stages of positive selection and then analyzed for RNA expression (Fig. 4). Two different sorting strategies were used to separate postselection intermediates between the CD4+ CD8+ stage and the CD4+ CD8− stage: one on the basis of CD8 and HSA expression, and another on the basis of CD8 and CD69 expression. As shown in Fig. 4, CD3ɛ expression levels were similar in all populations, as expected, and RAG1 expression declined sharply at the CD4+ CD8int HSA+ CD69+ stage, in agreement with previous reports that this gene is turned off rapidly in response to TCR-mediated selection signals (34–36). Ig-β RNA was not confined to the earliest CD4− CD8− cells: it was expressed comparably in all stages through postselection RAG1 down-regulation, and only reduced in the CD4+ CD8− single positive stages (Fig. 4). This expression pattern was confirmed by hybridization with an internal oligonucleotide probe (Fig. 4 D and E, lowermost panel). Ig-β expression appeared to persist even longer in CD4− CD8+ thymocytes, in agreement with its continued detectable expression in CD8+ peripheral T cells. Thus, the expression of Ig-β RNA persists in thymocyte differentiation at least until after the cells begin to undergo positive selection to the CD4+ lineage. Only at this late stage is Ig-β transcription shut off.
DISCUSSION
We have sought to define the progressive narrowing of developmental potential and the lineage commitment that occur during thymocyte differentiation (11, 12, 25–28) in terms of discrete changes in gene expression. Our results show that T lineage specification and commitment actually involve a cascade of staggered gene regulatory events, not a single clean transition.
The activation of different T lineage “specific” genes begins asynchronously, with several of these genes turned on at stages that apparently precede T lineage commitment. Our data generally agree with previous results on the expression of particular genes in early thymocytes (37–41), but the different onsets of expression of these genes are more sharply defined here than in previous studies. This is largely because of the sensitivity of RT-PCR, the simplified isolation of early subsets from immunodeficient thymocyte populations (13), our separation of the NK-like Sca-1− HSA− (CD4−) cells from the precursor-like Sca-1+ HSA− (CD4+/−) cells within the CD44+ CD25− thymocyte population, and our emphasis on direct, semiquantitative comparison among different T lineage genes. The results imply that some “T cell-specific” genes are initially activated in cells that may never give rise to T cells. Such genes therefore cannot require the action of any “master regulator” factor powerful enough to impose T lineage commitment. Although such a dominant master regulator, if it existed, could participate in later aspects of T lineage gene expression, it could not be required for the initial transcription of RNAs such as CD3ɛ or TCR-Cβ (unrearranged) transcripts.
Control of gene expression through this process is initially lineage-nonspecific, in that two transcripts controlled by the Ig-Eμ enhancer also begin to be expressed in immature thymocytes. These transcripts are down-regulated just before the induction of recombinase activity, indicating a programmed, stage-specific repression mechanism. The role of this repression is underlined by the fact that mature T cells, which no longer express recombinase, appear to resume free expression of these two transcripts, in agreement with an extensive body of evidence for the activity of the Eμ enhancer in later-stage thymocytes and mature T cells. The implication is that negative regulators, not positive regulators dependent on prior lineage commitment, may restrict IgH transcription specifically during one critical period when recombinase must choose between Ig and TCR gene targets.
This developmental gene expression program is still leaky enough to allow the sustained transcription of Ig-β until late stages of thymocyte maturation. There are other cases of apparent interlineage sharing of lymphocyte molecules, such as the expression of CD3 in fetal NK cells (42), but this case adds emphasis to the phenomenon in several ways. The T and B lineage programs are more distinct than the NK and T lineages, yet thymocytes do not repress Ig-β until they have progressed far down this well marked developmental pathway, with any chance to differentiate as B cells left far behind. Also, unlike other, poorly characterized markers shared by B cells and thymocytes, e.g., ThB and HSA itself, Ig-β protein is a critical, cell-type-specific component of the B cell antigen receptor, which acts in early B cell precursors to trigger the initiation of V-DJh Ig gene rearrangements (43). There is still a question of whether Ig-β is really B lineage-specific (note some expression of Ig-β in a heterogeneous, Gr-1+ subset of fetal liver cells, Fig. 2A). Nevertheless, it achieves a highly B-specific pattern of expression in mature cells. Also, although Ig-β is one of the few B lineage genes that continues to be expressed in fetal liver cells of E2A knockout mice (44), it appears to depend on B lineage-specific transcription factors, because it is not expressed in the developmentally arrested B220+ marrow cells of EBF knockout mice (3). Thus, its expression has been viewed as an early, positive marker of B lineage specification or commitment. However, our data show that late T lineage-specific repression, not a requirement for B lineage-specific activating factors, restricts expression of this gene in T cells.
The permissiveness of thymocytes for expression of the “early” B cell gene, Ig-β, contrasts with their lack of expression of the slightly “later” B cell genes, Ig-α and λ5. This suggests that some positive transcription factors needed to drive expression of these “late” genes are excluded by T lineage differentiation, even though the factors that drive expression of Ig-β are not. Whatever the mechanism, this exclusion is temporally and mechanistically separate from the event in which the “inappropriate” Ig-β gene expression is repressed, later in differentiation. Thus, diverse negative as well as positive regulatory mechanisms are required.
Even if the Ig-β transcription does not result in any protein product, it is a valuable indicator of “upstream” regulatory events in thymocytes. The separate issue of whether low-level expression of Ig-β RNA confers any positive function on thymocytes remains to be determined. At a crude steady-state level, it is clear that major thymocyte populations are unchanged in Ig-β knockout mice (43). On the other hand, if the Ig-β were expressed at the protein level, there is evidence that it could interact strongly with the CD3 complexes that are also present in thymocytes at early stages (45). It is conceivable that such hybrid complexes could play a role in pre-T cell receptor function or the establishment of signaling thresholds for viability (46, 47).
To prove a strict correlation between the regulation of various genes in the earliest stages of T cell differentiation and the establishment of developmental commitment, we will need to use single-cell methods. However, our present findings already show that the induction of genes associated with one lineage need not be rigidly linked with repression of genes associated with other potential lineages. Moreover, cells can make substantial progress activating T lineage-specific genes before they lose developmental plasticity and undergo commitment. At least some of this precocious expression appears to be multilineage. There are precedents for early multilineage gene expression, followed by lineage-specific repression, in erythromyeloid precursors (48, 49). Other hematopoietic precursors provide evidence for transdifferentiation (50). Now in the lymphoid case as well, both positive and negative gene regulatory mechanisms can be seen to contribute to lineage specificity, acting at different times. The stage is set now for identification of the lineage-specific transcription factors that are targets and executors of these mechanisms.
Acknowledgments
We are very grateful to Patrick Koen for excellent assistance with the flow cytometry, to Ray Hotz for essential care and management of the mutant mice, and to members of the Rothenberg lab and the Stowers Consortium at the California Institute of Technology for helpful and stimulating discussions. This work was supported by a grant from the U.S. Public Health Service (RO1 AI34041), by a grant from the State of California Tobacco-Related Disease Research Program (4RT 0264), and by funding from the Stowers Institute for Medical Research.
ABBREVIATIONS
- RT-PCR
reverse transcriptase–PCR
- TCR
T cell antigen receptor
- TdT
terminal deoxynucleotidyltransferase
References
- 1.Li Y, Hayakawa K, Hardy R R. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kee B L, Paige C J. Intl Rev Cytol. 1995;157:129–179. doi: 10.1016/s0074-7696(08)62158-0. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Grosschedl R. Nature (London) 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 4.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Nature (London) 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 5.Olson M C, Scott E W, Hack A A, Su G H, Tenen D G, Singh H, Simon M C. Immunity. 1995;3:703–714. doi: 10.1016/1074-7613(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 6.Weiss M J, Orkin S H. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y-S, Wasserman R, Hayakawa K, Hardy R R. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 8.Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F. J Exp Med. 1996;184:2217–2229. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon M C, Olson M, Scott E, Hack A, Su G, Singh H. Curr Top Microbiol Immunol. 1996;211:113–119. doi: 10.1007/978-3-642-85232-9_11. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D E, Hohaus S, Voso M T, Chen H M, Smith L T, Hetherington C J, Tenen D G. Curr Top Microbiol Immunol. 1996;211:137–147. doi: 10.1007/978-3-642-85232-9_14. [DOI] [PubMed] [Google Scholar]
- 11.Shortman K, Wu L. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 12.Zúñiga-Pflücker J C, Lenardo M J. Curr Opin Immunol. 1996;8:215–224. doi: 10.1016/s0952-7915(96)80060-4. [DOI] [PubMed] [Google Scholar]
- 13.Diamond R A, Ward S B, Owada-Makabe K, Wang H, Rothenberg E V. J Immunol. 1997;158:4052–4064. [PubMed] [Google Scholar]
- 14.Murphy E, Hieny S, Sher A, O’Garra A. J Immunol Methods. 1993;162:211–223. doi: 10.1016/0022-1759(93)90386-l. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Levelt C, Salio M, Zheng D X, Sancho J, Liu C-P, She J A, Huang M, Higgins K, Sunshine M-J, et al. Int Immunol. 1995;7:435–448. doi: 10.1093/intimm/7.3.435. [DOI] [PubMed] [Google Scholar]
- 16.Soloff R S K, Wang T-G, Lybarger L, Dempsey D, Chervenak R. J Immunol. 1995;154:3888–3901. [PubMed] [Google Scholar]
- 17.Bruno L, Rocha B, Rolink A, von Boehmer H, Rodewald H-R. Eur J Immunol. 1995;25:1877–1882. doi: 10.1002/eji.1830250713. [DOI] [PubMed] [Google Scholar]
- 18.Schlissel M, Voronova A, Baltimore D. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzaki Y, Gyotoku J-i, Ogawa M, Nishikawa S-i, Katsura Y, Gachelin G, Nakauchi H. J Exp Med. 1993;178:1283–1292. doi: 10.1084/jem.178.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godfrey D I, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 21.Sagara S, Sugaya K, Tokoro Y, Tanaka S, Takano H, Kodama H, Nakauchi H, Takahama Y. J Immunol. 1997;158:666–676. [PubMed] [Google Scholar]
- 22.Rolink A, ten Boekel E, Melchers F, Fearon D T, Krop I, Andersson J. J Exp Med. 1996;183:187–194. doi: 10.1084/jem.183.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cumano A, Paige C J, Iscove N N, Brady G. Nature (London) 1992;356:612–615. doi: 10.1038/356612a0. [DOI] [PubMed] [Google Scholar]
- 24.Spangrude G J, Klein J, Heimfeld S, Aihara Y, Weissman I L. J Immunol. 1989;142:425–430. [PubMed] [Google Scholar]
- 25.Zúñiga-Pflücker J C, Jiang D, Lenardo M J. Science. 1995;268:1906–1909. doi: 10.1126/science.7541554. [DOI] [PubMed] [Google Scholar]
- 26.Moore T A, Zlotnik A. Blood. 1995;86:1850–1860. [PubMed] [Google Scholar]
- 27.Wu L, Vremec D, Ardavin C, Winkel K, Süss G, Georgiou H, Maraskovsky E, Cook W, Shortman K. Eur J Immunol. 1995;25:418–425. doi: 10.1002/eji.1830250217. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Scollay R, Egerton M, Pearse M, Spangrude G J, Shortman K. Nature (London) 1991;349:71–74. doi: 10.1038/349071a0. [DOI] [PubMed] [Google Scholar]
- 29.Rothenberg E V, Chen D, Diamond R A. J Immunol. 1993;151:3530–3546. [PubMed] [Google Scholar]
- 30.Mori S-i, Inaba M, Sugihara A, Taketani S, Doi H, Fukuba Y, Yamamoto Y, Adachi Y, Inaba K, Fukuhara S, Ikehara S. J Immunol. 1997;158:4193–4199. [PubMed] [Google Scholar]
- 31.Hashimoto S, Chiorazzi N, Gregerson P K. Mol Immunol. 1995;32:651–659. doi: 10.1016/0161-5890(95)00023-8. [DOI] [PubMed] [Google Scholar]
- 32.Koyama M, Nakamura T, Higashihara M, Herren B, Kuwata S, Shibata Y, Okumura K, Kurokawa K. Immunol Lett. 1995;47:151–156. doi: 10.1016/0165-2478(95)00071-x. [DOI] [PubMed] [Google Scholar]
- 33.Hermanson G G, Eisenberg D, Kincade P W, Wall R. Proc Natl Acad Sci USA. 1988;85:6890–6894. doi: 10.1073/pnas.85.18.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turka L A, Schatz D G, Oettinger M A, Chun J J M, Gorka C, Lee K, McCormack W T, Thompson C B. Science. 1991;253:778–781. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 35.Brandle D, Muller S, Muller C, Hengartner H, Pircher H. Eur J Immunol. 1994;24:145–151. doi: 10.1002/eji.1830240122. [DOI] [PubMed] [Google Scholar]
- 36.Chan S H, Cosgrove D, Waltzinger C, Benoist C, Mathis D. Cell. 1993;73:225–236. doi: 10.1016/0092-8674(93)90225-f. [DOI] [PubMed] [Google Scholar]
- 37.Wilson A, Held W, MacDonald H R. J Exp Med. 1994;179:1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saint-Ruf C, Ungewiss K, Groettrup L B, Fehling H J, von Boehmer H. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 39.Wilson A, MacDonald H R. Int Immunol. 1995;7:1659–1664. doi: 10.1093/intimm/7.10.1659. [DOI] [PubMed] [Google Scholar]
- 40.Ismaili J, Antica M, Wu L. Eur J Immunol. 1996;26:731–737. doi: 10.1002/eji.1830260402. [DOI] [PubMed] [Google Scholar]
- 41.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. J Exp Med. 1996;184:1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spits H, Lanier L L, Phillips J H. Blood. 1995;85:2654–2670. [PubMed] [Google Scholar]
- 43.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 44.Bain G, Robanus Maandag E C, Izon D J, Amsen D, Kruisbeek A M, Weintraub B C, Krop I, Schissel M S, Feeney A J, van Roon M, et al. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 45.Müller B, Cooper L, Terhorst C. Immunol Lett. 1995;44:97–103. doi: 10.1016/0165-2478(94)00199-2. [DOI] [PubMed] [Google Scholar]
- 46.Groettrup M, von Boehmer H. Immunol Today. 1993;14:610–614. doi: 10.1016/0167-5699(93)90201-U. [DOI] [PubMed] [Google Scholar]
- 47.Wang B, Biron C, She J, Higgins K, Sunshine M-J, Lacy E, Lonberg N, Terhorst C. Proc Natl Acad Sci USA. 1994;91:9402–9406. doi: 10.1073/pnas.91.20.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cross M A, Heyworth C M, Murrell A M, Bockamp E O, Dexter T M, Green A R. Oncogene. 1994;9:3013–3016. [PubMed] [Google Scholar]
- 49.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 50.Kulessa H, Frampton J, Graf T. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 51.Rodewald H-R, Moingeon P, Lucich J L, Dosiou C, Lopez P, Reinherz E L. Cell. 1992;69:139–150. doi: 10.1016/0092-8674(92)90125-v. [DOI] [PubMed] [Google Scholar]
- 52.Leclercq G, Debacker V, De Smedt M, Plum J. J Exp Med. 1996;184:325–336. doi: 10.1084/jem.184.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carlyle J R, Michie A M, Furlonger C, Nakano T, Lenardo M J, Paige C J, Zúñiga-Pflücker J C. J Exp Med. 1997;186:173–182. doi: 10.1084/jem.186.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]