Abstract
Ribonucleoprotein particles (RNPs) are important components of all living systems, and the assembly of these particles is an intricate often multisteped process. The 30S ribosomal subunit is composed of one large RNA (16S rRNA) and 21 ribosomal proteins (r-proteins). In vitro studies have revealed that assembly of 30S subunit is a temperature-dependent process involving sequential binding of r-proteins and conformational changes of 16S rRNA. Additionally, a temperature-dependent conformational rearrangement was reported for a complex of primary r-protein S4 and 16S rRNA. Given these observations, a systematic study of the temperature dependence of 16S rRNA architecture in individual complexes with the other five primary binding proteins (S7, S8, S15, S17 and S20) was performed. While all primary binding r-proteins bind 16S rRNA at low temperature, not all r-proteins/16S rRNA complexes undergo temperature-dependent conformational rearrangements. Some RNPs achieve the same conformation regardless of temperature, others show minor adjustments in 16S rRNA conformation upon heating, and finally others undergo significant temperature-dependent changes. Some of the architectures achieved in these rearrangements are consistent with subsequent downstream assembly events such as assembly of the secondary and tertiary binding r-proteins. The differential interaction of 16S rRNA with r-proteins illustrates a means for controlling the sequential assembly pathway for complex RNPs and may offer insights into aspects of RNP assembly in general.
Keywords: RNA-protein interactions, 30S subunit assembly, conformational rearrangements, temperature dependence, chemical probing
INTRODUCTION
The Escherichia coli (E. coli) 30S ribosomal subunit is composed from 16S ribosomal RNA (rRNA) and 21 ribosomal proteins (r-proteins), and it can be a rich source of information for the student of RNA-protein interactions. The crystal structure of the ribosome from E. coli was determined recently,1 and detailed structures of the individual 30S ribosomal subunit are also available2; 3. As these structures represent end points for the assembly process, they are very useful in analyzing assembly events. Additionally, the structures of some of the unliganded r-proteins have also been determined4; 5; 6; 7; 8; 9. Comparison of free and 30S bound r-proteins allows inferences about changes in r-protein structure, as a result of ribonucleoprotein particle (RNP) assembly to be proposed. However, very few detailed structures of segments of 16S rRNA are available and thus similar inferences about RNA conformational changes during ribosome assembly have not been put forth. While the understanding of RNA-protein interactions has been greatly enhanced by advances in RNP crystallography, a detailed view of conformational changes during RNP assembly is still lacking. This is particularly important for RNPs containing large RNA molecules. Systematic studies, using a well characterized model system, such as the 30S ribosomal subunit, will advance our understanding of events central to RNP assembly.
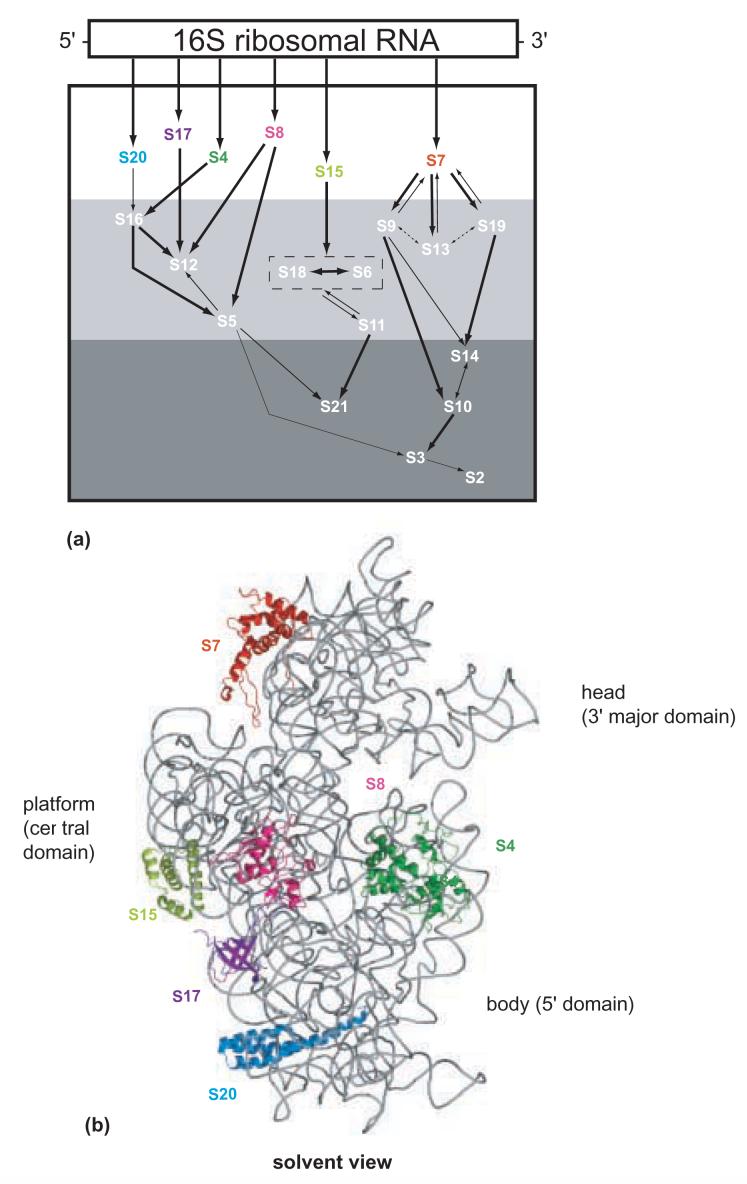
The E. coli 30S subunit is a good model for RNP assembly as it can be reconstituted into a functional conformation in vitro from its isolated components10. This system has allowed analysis of RNPs of varying composition and use of transcribed 16S rRNA or fragments thereof to be studied11; 12. This system has also revealed that the sequential binding of r-proteins to 16S rRNA is a critical step in orchestrating formation of functional 30S subunits. Traditionally, the r-proteins have been categorized into three assembly classes, as indicated in the in vitro assembly map13; 14(Figure 1a). The r-proteins that bind directly and independently to 16S rRNA are classified as primary, and they are S4, S7, S8, S15, S17 and S20. The secondary binding proteins, S5, S6, S9, S11- S13, S16, S18 and S19 bind 16S rRNA after the assembly of at least one primary protein, while the tertiary binding proteins, S2, S3, S10, S14 and S21, require association of at least one primary and one secondary r-protein for their binding. These data suggest that r-proteins associate in an ordered cascade and that primary binding r-proteins play critical roles in domain organization.
Figure 1.
- Modified in vitro 30S subunit assembly map. The 16S rRNA is represented by a rectangle in a 5' to 3' direction. The arrows indicate the co-dependencies for the assembly of the r-proteins. The size of the arrow indicates the relative strength of the assembly dependency between components. The r-proteins shown in the white region are primary binding r-proteins. The r-proteins shown in white in the light gray and dark gray box indicate secondary, and tertiary binding r-proteins, respectively. S6 and S18 are enclosed in a box to indicate that they bind as a heterodimer.
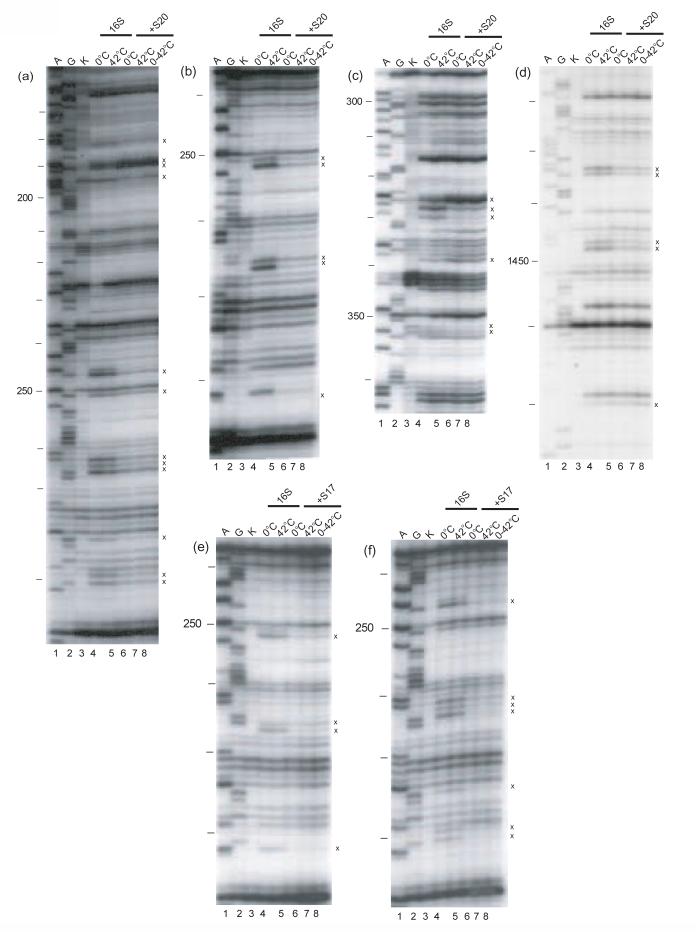
- Crystal structure of the 16S rRNA from the E. coli 30S subunit with all the primary proteins. The 16S rRNA is shown in gray, and the r-proteins are S4 green, S7 red, S8 magenta, S15 bright yellow, S17 dark purple and S20 light blue, as in Figure 1a. The 3-D parts of the 30S subunit are indicated while the corresponding domains from the 16S rRNA secondary structure are specified in paranthesis. All the Figures containing 3-D structures were prepared using Pymol44, and the pdb file 2AW7.
Analysis of 30S subunit assembly in the presence of all or many of the r-proteins, has revealed global trends, without dissecting the role of the individual r-proteins. The presence of all r-proteins allows the concerted changes in the conformation of 16S rRNA to be assessed, but by necessity obscures the contribution of individual proteins. For example, in the study of the temperature-dependent dynamics of 30S subunit assembly, the r-proteins were classified in different kinetic classes based on their footprints observed in previous studies of less complex RNPs15. For some primary binding r-proteins only a subset of their footprints were observed; for example, only a third of the S15-specific16 and about half of the S7-specific footprints17 could be assigned in this ensemble study15. Thus, while these bulk approaches are illuminating, data can be masked or invisible, and further analysis of more minimal systems may be necessary to fully dissect the scope of changes during assembly processes.
Conformational changes play an important role in the assembly of the 30S ribosomal subunit. In vitro 30S subunit assembly involves a large conformational change from one intermediate to another18; 19 en route to 30S subunit formation, and this change can be facilitated by increased temperature20. Changes in 16S rRNA architecture associated with this assembly pathway have been detailed21; 22 using a defined subset or all of the small subunit r-proteins. Our understanding of the roles of r-proteins in orchestrating the architectural changes would be advanced by determining more exactly which r-proteins contribute to these specific conformational changes. Indeed, this approach has proven useful in analyzing the interaction of 16S rRNA with a single r-protein, S4, as function of temperature23. A temperature-dependent conformational change of 16S rRNA in the presence of S4 was observed when the complexes were formed at different temperatures (0, 30 and 42°C)23. These changes were revealed by differences in the chemical modification pattern of 16S rRNA in the minimal complexes. This approach may be particularly fruitful now, as structures of 30S subunits1; 2; 3 allow a better understanding of the implications of temperature influence on the RNP architecture, and subsequently on assembly.
To the best of our knowledge, other than the paper of Powers et. al (1995) there are few studies in which protein-RNA interactions at different temperatures are dissected by structural methods. Some studies have shown a dependence of the kinetics of RNA-protein interactions on temperature24; 25. Additionally, there are a few studies in which small differences in the RNA-protein binding constants at different temperatures were observed,26; 27 but no detailed structural analysis of the complexes was performed. In an attempt to dissect temperature-dependent changes in RNP structure, a footprinting study of the complexes of 16S rRNA and individual primary r-proteins formed at different temperatures was undertaken. This approach allows temperature-dependent changes to be revealed. This systematic analysis of the independent interactions of all of the primary binding r-proteins with 16S rRNA will allow a better understanding of the rRNA/r-protein interactions.
RESULTS
Complex formation and chemical probing of binary RNPs
Individual complexes of natural 16S rRNA with the recombinant primary binding r-proteins S7, S8, S15, S17 and S20 (S4 was generally omitted as it had been studied previously23) were formed by incubating the reaction mixture at either 0°C or 42°C and a third complex was formed by incubating the reaction mixture first at 0°C and then at 42°C, and herein will be referred to as “shifted” complex. Once complexes had been formed, all particles were placed on ice and probing was performed at low temperature. This approach will allow the detection of r-protein facilitated temperature-dependent differences in 16S rRNA architecture and for the “shifted” complex will reveal if either the low or high temperature interactions predominate. The reactivity of 16S rRNA in these complexes and of naked 16S rRNA incubated at the appropriate temperature, toward the base specific probes dimethyl sulfate (DMS) and kethoxal was investigated by primer extension analysis. In the previous work on the S4/16S rRNA RNP, no temperature-dependent differences were observed when the RNA backbone was probed with hydroxyl radicals23 and our findings for other primary r-protein containing RNPs are consistent with this earlier work (data not shown). Given that a large number of direct contacts between the 16S rRNA backbone and these r-proteins were observed in the structure of 30S subunits1; 28, this confirms association of the primary binding proteins under the experimental conditions. Additionally, our results are also consistent with the observation that when temperature-dependent conformational rearrangements are observed, both components must be heated together for the changes to occur (data not shown). The reactivities observed for naked 16S rRNA at 42°C and complexes formed at 42°C were very similar to the ones previously published for similar conditions16; 17; 29. Throughout this manuscript, reference to previous footprints implies those observed for complexes of r-proteins with 16S rRNA that were formed at high temperature, since until this work a comprehensive low temperature analysis was lacking. Our data reveal a similar probing pattern for the shifted complexes and those formed at 42°C, and thus these two sets of RNPs will be discussed as one. The results indicate that the primary binding protein/16S rRNA RNPs can be classified into three distinct groups as regards temperature-dependent conformational rearrangements. One class reveals a large temperature-dependence, as previously reported for S423. A second class reveals slight temperature-dependence, where most of the footprints are observed at low temperature, but the intensity is not fully reached until after heating. Somewhat surprisingly, the proteins from the third class reveal virtually no temperature-dependence. Thus it appears that not all primary r-protein/16S rRNA complexes undergo obvious temperature-dependent conformational rearrangements. Data for each of the r-protein/16S rRNA complexes will be discussed below.
S20/16S rRNA complexes
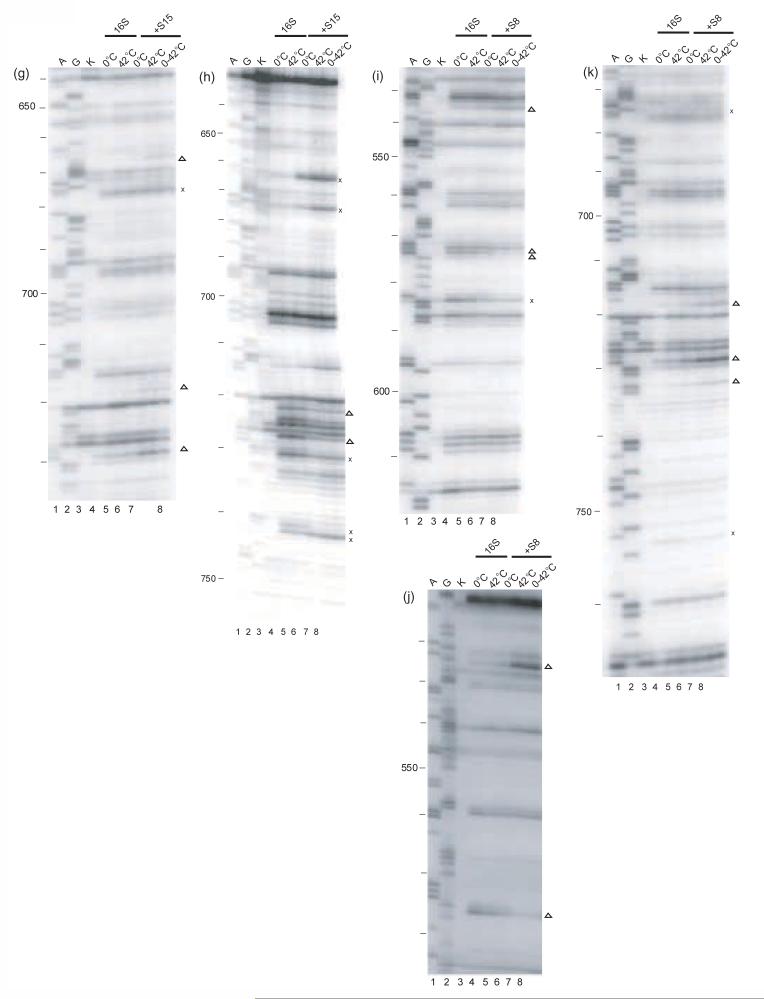
X-ray crystallographic studies of 30S subunits reveal that S20 is one of the few r-proteins that interacts with two different domains of 16S rRNA, the 5' domain (body) and H44 of the 3' minor domain1; 30(Figure 1b). This is consistent with all of S20 footprints being localized in the 5' domain and in H4429. No differences in the footprints were observed between the S20/16S rRNA complexes regardless of the temperature at which they are formed (see Table 1, Figure 2a-d, Figure 3). Thus, it appears that in the minimal binary particle S20 and 16S rRNA interact in a temperature-independent manner. These results are in marked contrast to those previously reported for the S4/16S rRNA RNP23 and suggest that the primary r-proteins/16S rRNA RNPs can behave differently.
Table 1.
16S rRNA nucleotides with protein specific temperature-dependent reactivity differences.
| Nucleotide | S8 | S15 | S7 |
|---|---|---|---|
| 5′ domain | |||
| G524 | E | e | |
| A535 | E | e | |
| Central domain | |||
| A573 | p | ||
| A574 | p | ||
| G575 | p | ||
| A665 | e | ||
| G674 | e | ||
| A718 | e | e | |
| G724 | P | ||
| G727 | P | ||
| A728 | e | e | |
| C732 | E | ||
| G858 | P | ||
| G859 | P | ||
| A860 | p | ||
| G861 | p | ||
| A865 | p | ||
| 3′major domain | |||
| A935 | p | ||
| A937 | p | ||
| A938 | p | ||
| G939 | p | ||
| G944 | p | ||
| G945 | p | ||
| G951 | p | ||
| G953 | e | ||
| G954 | e | ||
| A977 | e | ||
| A978 | e | ||
| C979 | e | ||
| C980 | e | ||
| A1236 | P | ||
| C1237 | p | ||
| A1238 | p | ||
| A1239 | p | ||
| A1248 | p | ||
| C1249 | p | ||
| A1250 | PP | ||
| A1251 | P | ||
| A1252 | P | ||
| A1287 | p | ||
| A1288 | p | ||
| A1289 | P | ||
| G1300 | p | ||
| C1302 | p | ||
| G1304 | p | ||
| G1305 | p | ||
| C1314 | p | ||
| G1316 | p | ||
| C1317 | E | ||
| A1318 | e | ||
| A1319 | e | ||
| C1320 | e | ||
| C1322 | p | ||
| G1331 | p | ||
| A1332 | p | ||
| A1333 | p | ||
| G1334 | p | ||
| G1337 | E | ||
| G1338 | E | ||
| A1349 | p | ||
| A1360 | p | ||
| G1361 | P | ||
| A1362 | p | ||
| A1363 | p | ||
| G1365 | p | ||
| A1374 | p |
E – indicates an enhancement, relative strength is indicated by e or E, from weakest to more intense
P – indicates a protection, relative strength is indicated by p, P or PP, from weakest to more intense
Nucleotides reactivate in RNPs with the same composition formed at 0°C and 42°C, respectively were compared and those where a difference was evident are noted.
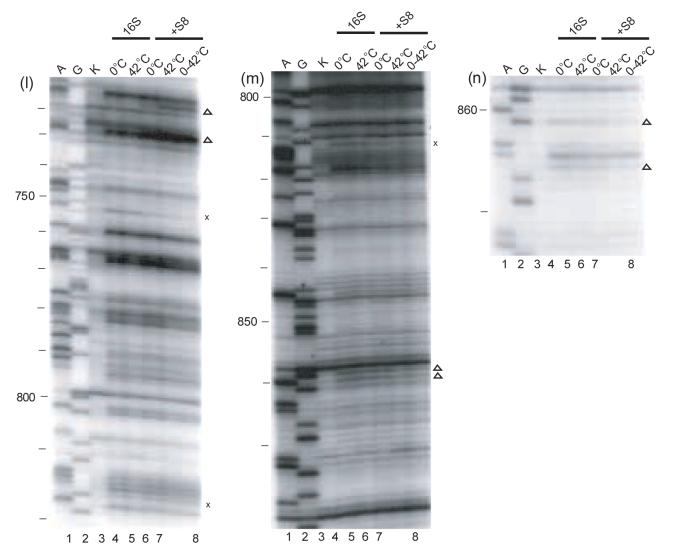
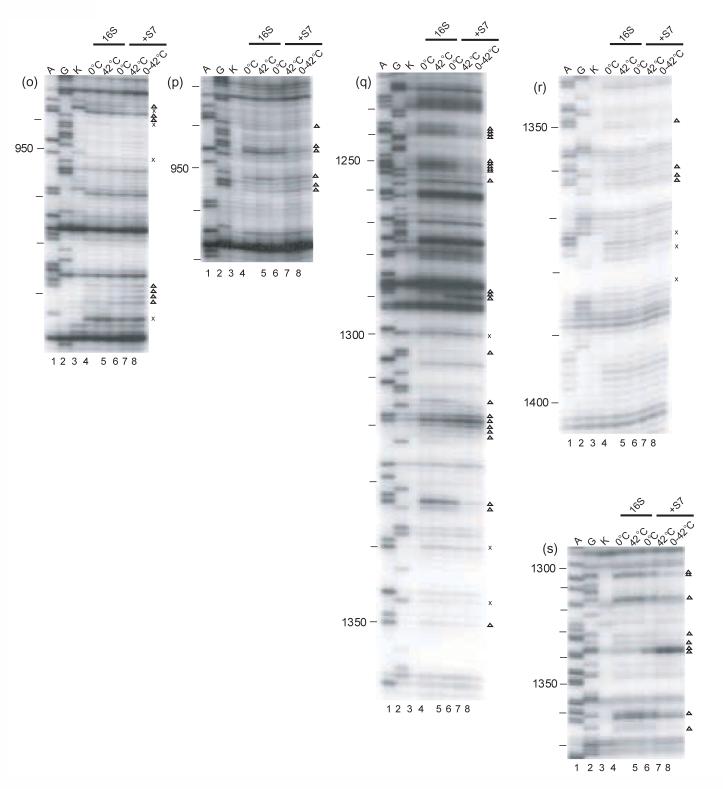
Figure 2.
Primer extension analysis of the r-protein/16S rRNA complexes. Individual gels of the minimal complexes modified by DMS or kethoxal are shown. A and G (lanes 1 and 2) are dideoxy sequencing lanes, K (lane 3): unmodified 16 S rRNA. All the other lanes are treated with the probe indicated below. The other lanes 4-8 are: modified 16S rRNA kept at 0°C (lane 4), or at 42°C (lane 5), Sx/16S rRNA formed at 0°C (lane 6), or at 42°C (lane 7) and the shifted complex (lane 8). Compare lanes 4 and 6 for the complexes formed at 0°C, lanes 5 and 7 for the complexes formed at 42°C, or lanes 6 and 7 for the differences between the two complexes. The probes and primers used for the experiments are indicated below. S20/16S rRNA: a) DMS-323, b) kethoxal-323, c) DMS-480, d) DMS-1508; S17/16S rRNA: e) DMS-323; f) kethoxal, 323; S15/16S rRNA: g) DMS-795, h) kethoxal-795 S8/16S rRNA: i) DMS-683, j) kethoxal-683, k) DMS-795, l) DMS-939, m) kethoxal-939, n) DMS–939; S7/16S rRNA: o) DMS-1046, p) kethoxal-1046, q) DMS-1391, r) DMS -1491, s) kethoxal -1491. The symbols x and Δ indicate temperature-independent and temperature-dependent footprints, respectively.
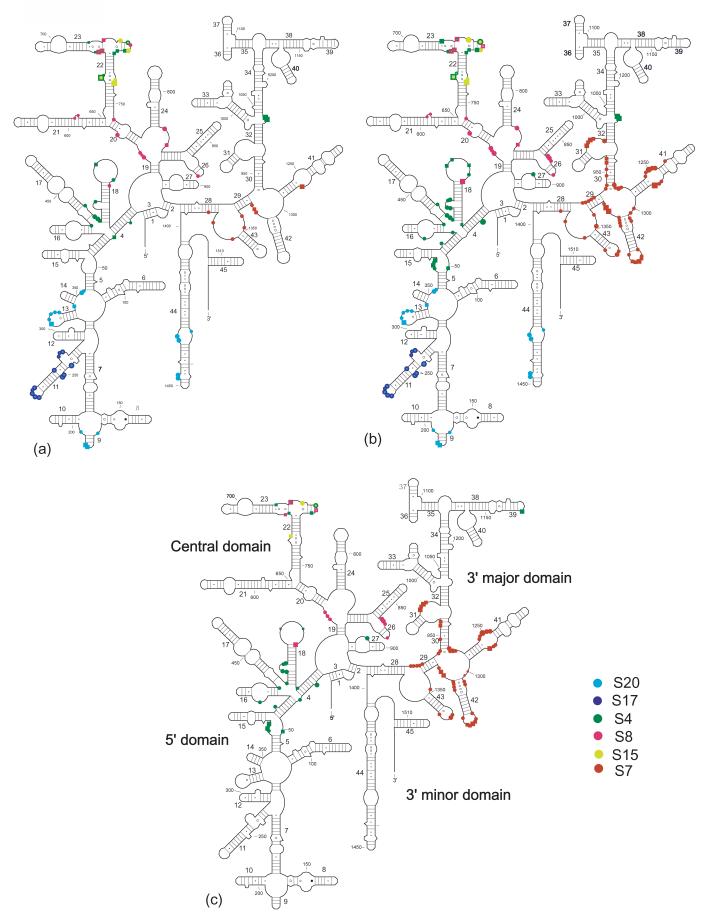
Figure 3.
Nucleotides with altered reactivity as a result of binding of an r-protein to 16S rRNA, for both kethoxal and DMS probing represented on the secondary structure of 16S rRNA45. Circles denote the sites of protections, while squares denote enhancement sites, and the size represents the intensity of the change. The 16S rRNA is shown in dark gray, changes attributed to: S4, green; S7, red; S8, magenta; S15 bright yellow; S17, dark purple, and S20, light blue. Nucleotides enhanced or protected by more than one protein are shown as concentric rings or squares. (a) Changes in modification patterns shown on the secondary structure of 16 S rRNA for complexes formed at 0°C. b) Changes in modification patterns shown on the secondary structure of 16 S rRNA for the interaction at 42°C. c) Difference in the nucleotides with altered reactivity between the complexes formed at 42°C and 0°C.
S17/16S rRNA complexes
Structural studies revealed that in the context of 30S subunits, r-protein S17 makes contacts with helix 11 of the 5' domain and helices 20, 21, 22 of the central domain (Figure 1b) 1; 30. However the footprints in the minimal S17/16S rRNA particle are present almost exclusively (12 out of 15) in helix 1129. Our results reveal that the footprints, which are all protections, are the same regardless of the temperature of S17/16S rRNA complex formation (Table 1, Figure 2e, f, Figure 3). Thus, similar to what was observed with S20 (see above), temperature seems to have no effect on the footprints of r-protein S17 on 16S rRNA. These results suggest that r-protein dependent organization of portions of the 5' domain can occur in a single step and that the temperature of complex formation has no obvious effect on this interaction.
S15/16S rRNA complexes
The footprints specific for S15 in the minimal RNP are localized in helices 22 and 2316 and these are very consistent with S15 binding the three way junction formed by H20, H21 and H22 in the full 30S subunits (Figure 1b)1; 30. Thus, in both minimal and more elaborate particles,31 S15 interactions are restricted to the central domain of 16S rRNA. The slight temperature-dependent conformational adjustments observed for the S15/16S RNP (Table 1, Figure 2g, h, Figure 3) are distinct from those observed for S20 and S17 (see above) and those reported for S423. All of the S15 temperature-dependent footprints follow a similar pattern (Figure 3). S15-dependent protections appear at 0°C, but four of these develop in intensity at higher temperature (Figure 2g, h). Only one temperature-dependent footprint is observed in helix 22, where the majority of the crystal contacts between S15 and 16S rRNA are observed1 (Figure 3). The majority of these changes in reactivity of nucleotides due to altered temperature are present in helix 23, which is not involved in direct RNA-protein contacts in the crystal structure of 30S subunits1. These results suggest that binding occurs at low temperature but the complex of S15 and 16S rRNA is further accommodated at higher temperature.
S8/16S rRNA complexes
The footprints specific for S8 in the minimal particle are present in the 530 loop, 570 region, helices 20, 21 and 23, and also the 820 and 860 regions16. In general, many S8 footprints are near junctions for the 5' and central domains, and for the central and the 3' domains (Table 1, Figure 2i-n, Figure 3). No temperature-dependent footprinting changes are observed in the 3-way helical junction, H20/H21/H22, in the S8/16S rRNA complex (Figure 3). In the RNP containing S8 and 16S rRNA formed at 0°C, many of the enhancements and protections specific to the binding of S8 at 42°C are observed (Figure 2i-n, Figure 3). However, most nucleotides in helix 23 (Figure 2k, l), 530 and 570 loops (Figure 2i, j), which are footprinted by S8, reveal a temperature-dependent requirement for attaining the full extent of footprinting (Figure 3). Hence, the majority of the S8 specific footprints are not as intense at 0°C as at higher temperature. The largest temperature-dependent differences in the reactivities are observed for the nucleotides from helix 26 and the 860 region (Figure 2m, n; Figure 3c). These results suggest that a conformational rearrangement of the S8/16S rRNA complex may be involved in organizing the more 3' elements of the S8 binding site.
S7/16S rRNA complexes
S7 nucleates the assembly of the head of the 30S subunit, by binding to two multiple-stem junctions of the 3' domain of 16S rRNA, H28/H29/H43 and H29/H30/H41/H42 (Figure 1b)1; 30. Consistent with its RNA interactions in the 30S subunit, binding of S7 has been shown to affect the reactivity of many 16S rRNA nucleotides in footprinting experiments17. Our data indicate that the S7/16S rRNA RNP undergoes extensive temperature-dependent conformational rearrangements (Table 1, Figure 2o-s, Figure 3). For all the regions footprinted by S7 temperature-dependent alterations in reactivity are observed, suggesting that conformational rearrangements are prevalent for this complex (Figure 3c). When the S7/16S rRNA containing RNP is formed at low temperature, approximately 16% of the high temperature footprints are detected. Interestingly, there is a correlation between footprints that are observed at 0°C and direct contacts between S7 and 16S rRNA that are apparent in the 30S subunit1. Direct contacts between S7 and the 1350/1370 loop of 16S rRNA are present in the structure of the 30S subunit and protections in this region are observed at 0°C (Figure 2q-s, Figure 3a). Almost all of the other S7-specific footprints are absent in complexes formed at low temperature. Strong temperature dependence is observed at the three way junction H28/H29/H43 and the multiple-stem junction H29/H30/H41/H42, suggesting that these elements are altered as a consequence of temperature-dependent rearrangements (Figure 3c). Thus, the extent of temperature-dependent changes observed with the S7/16S rRNA RNP are the most similar to those reported for S4/16S rRNA complexes23.
DISCUSSION
The results presented in this manuscript and those published earlier by Powers and Noller23 clearly illustrate that the interaction of primary binding r-proteins with 16S rRNA can be significantly influenced by temperature. However, not all the primary binding r-protein/16S rRNA particles appear to undergo temperature-dependent conformational rearrangements. The study of these relatively simple RNPs, in isolation from the remaining small subunit components, has allowed a detailed analysis of their specific interactions. Our data suggest that the assembly of 16S rRNA containing RNPs can occur at distinct stages and that some of these RNPs can progress from one conformation to another in a temperature-dependent manner, while others appear less dynamic. These studies allow insight into multiple mechanisms of primary binding protein interaction with 16S rRNA and underscore the complexity of 30S subunit assembly and RNP formation in general.
For the RNPs that display strong temperature-dependent conformational rearrangement, a concern might be whether binding occurs at low temperatures or if binding constants are similar at various temperatures. While there are only a few studies in which binding of r-proteins to 16S rRNA is analyzed as a function of temperature, those that have been done are supportive of association of r-proteins with 16S rRNA under such conditions24; 25; 32; 33. Binding constants were determined at 0°C and 42°C for S432 and S824; 33. The binding constants are of the same order of magnitude (107) but the values are slightly less at low temperature than observed at high temperature. The binding rates during assembly for the majority of the 30S subunit r-proteins were determined by pulse chase quantitative mass spectrometry at 15°C and 40°C25. Binding is observed at 15°C for all the primary r-proteins25, although there are some temperature-dependent rate differences. In addition, in vitro 30S subunit assembly studies indicate that primary and secondary binding r-proteins can bind at low temperature, as an intermediate composed of 16S rRNA and these r-proteins is readily detectable20; 34. Thus, the temperature-dependent footprinting changes reported herein are not likely due merely to association events, but most probably reflect RNP rearrangements.
There are many protections that do not overlap with direct contacts between r-proteins and 16S rRNA in the 30S subunit1; 34. These likely represent regions of 16S rRNA which change environment upon r-protein binding or represent contacts that form transiently, but are not observed in the fully assembled 30S subunit. Nonetheless the comparison of our protection data (the enhancements can be ignored in this context) and base-specific direct contacts reveals some interesting trends. Many of the nucleotides which are protected at low temperature correspond with sites that are in direct contact with r-proteins in the 30S structure1; 35. While the correlation is far from absolute, it raises an interesting possibility for the effects on 30S subunit assembly. At low temperature the r-proteins may make initial contact with a major binding site in 16S rRNA and these interactions may be largely maintained throughout RNP assembly. At high temperature, the structure of the r-proteins/16S rRNA RNPs can be altered, thus resulting in changes in reactivity of specific nucleotides. These changes are often more remote from the actual contact sites of r-proteins with 16S rRNA and may have profound effects on subsequent assembly events.
Another interesting correlation exists between temperature-dependent footprinting changes and the size and structure of the r-protein (Figure 6). RNPs containing the two smallest primary binding proteins S17 and S20 show no temperature dependence, while only a slight temperature dependence is observed for the RNPs containing the next smallest primary r-protein, S15. Additionally, S8/16S rRNA shows more temperature dependence than the three smaller r-proteins mentioned above while RNPs containing S4 and S7, the largest primary binding r-proteins, show the highest degree of temperature-dependent conformational rearrangement. One possible explanation of these observations relates to the domain organization of these r-proteins. The smaller r-proteins tend to contain only one domain1; 28. Structural studies have revealed that the core structures of S151; 28 and S177;1; 28are very similar when unliganded or part of the 30S subunit, thus correlating well with the minimal temperature-dependence observed in these RNPs. The larger r-proteins tend to be composed from multiple “domains” or a more globular domain and some extended less canonical protein structures28; 1. Thus differential interactions of r-proteins revealed in this and previous23 work can be correlated with structural characteristics of these primary binding r-proteins.
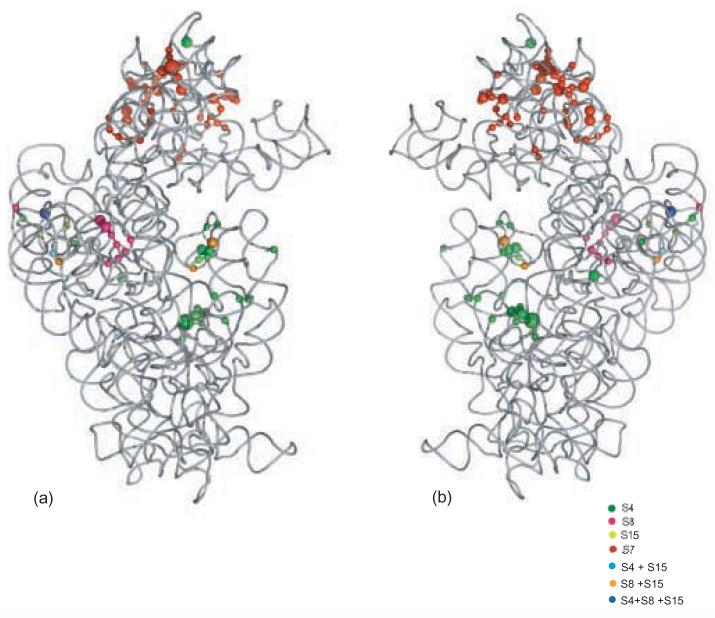
Figure 6.
The temperature-dependent footprints for each primary r-protein represented on the crystal structure of 16S rRNA from the E. coli 30S subunit. Nucleotides with altered reactivities are represented as spheres. The size of the spheres is indicative of the intensity of the change. The r-proteins are omitted for clarity.
The primary r-proteins S17 and S20 show little or no temperature dependence for the interaction with 16S rRNA and bind mainly to the 5' domain of 16S rRNA. Interestingly, it was shown that the 5' domain of naked 16S rRNA has structural features similar to those observed in the 30S subunit36, consistent with fewer r-protein dependent structural accommodations. Our findings for S17 and S20 are consistent with other studies looking at assembly dynamics in the context of all the small subunit r-proteins15; 21; 22. S17 and S20 are the only r-proteins that have all of the corresponding footprints in one kinetic class: the early binders15. Thus it appears that S17 and S20 have similar properties when studied in isolation or in ensemble studies.
The r-protein S15 is one of the proteins where the advantages of studying minimal complexes are obvious. Only five footprints specific for S15 were identifiable in the assembly dynamics study15, while in our experiments we were able to assess the majority of S15 dependent footprints. Additionally, while we observe many footprints at 0°C, only one footprint was attributable to S15 at that temperature, when all the r-proteins are present15. Also, some of its footprints coincide with direct RNA-protein contacts in the 30S subunit1; 16. Thus our experiments give a more detailed understanding of the role of S15 in facilitating changes in 16S rRNA architecture which are consistent with the contacts observed in the fully assembled 30S subunit.
Additionally, our studies using the minimal S8/16S rRNA binary complexes reveal more details, than assembly studies using a full complement of r-proteins. When S8 footprints are followed in the presence of all the r-proteins, no footprints specific for S8 were observed at 0°C15. Based on these data one cannot conclude if S8 binds to 16S rRNA at low temperature. However, when the minimal S8/16S rRNA complex is formed at the same temperature, the majority of S8 specific footprints are observed (Figure 3a). In fact, the majority of the footprints specific for S8 are observed at low temperatures, although many of them are only partial (Figure 3a, b). Thus our results are not in complete agreement with the classification of S8 as a mid-binding r-protein15. Overall these data might support a designation of early-mid binding protein for S8 since it can bind at an early phase, as epitomized by low temperature association, but it is further accommodated later in assembly in a temperature-dependent manner.
The S8 footprints which reveal differential temperature-dependent conformational changes are also somewhat clustered in the mature 30S subunit. On the three dimensional structure of 16S rRNA from 30S subunits1 the footprinting classes almost appear to be layered (Figure 4e, f). All the protections that appear at 0°C are clustered, and they are in the lower part of the 30S subunit. The protections that emerge only at 42°C are also clustered and are localized more toward the head of the 30S subunit (Figure 4e). The protections that appear at 0°C and achieve a higher level of footprinting at 42°C are proximal to one another and are somewhat in between the other two sets. Thus it appears that there is a relative spatial context to the conformational rearrangement associated with the S8/16S rRNA particle.
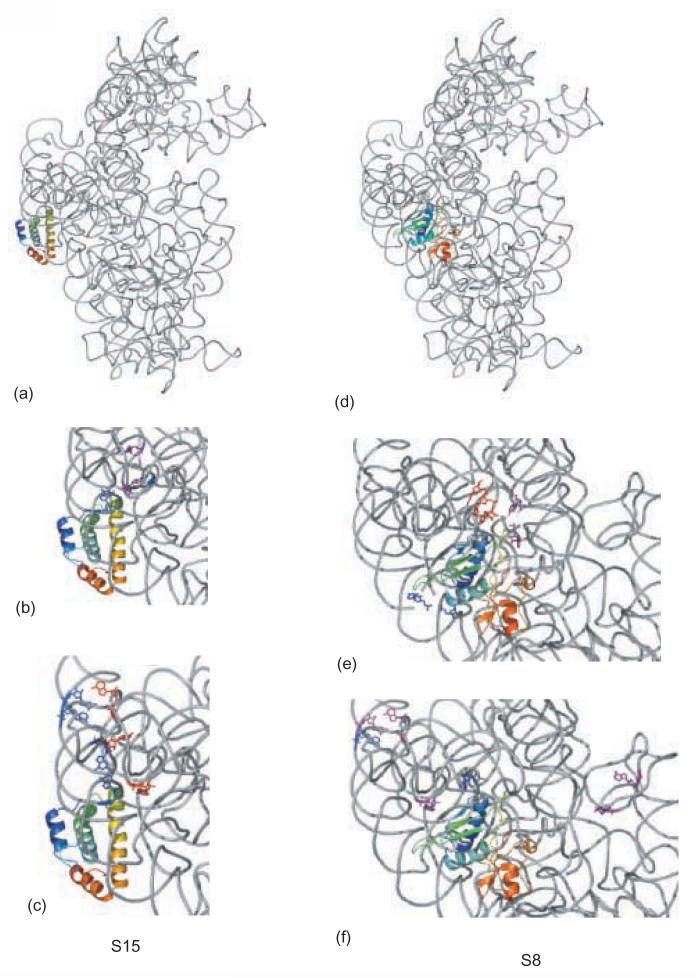
Figure 4.
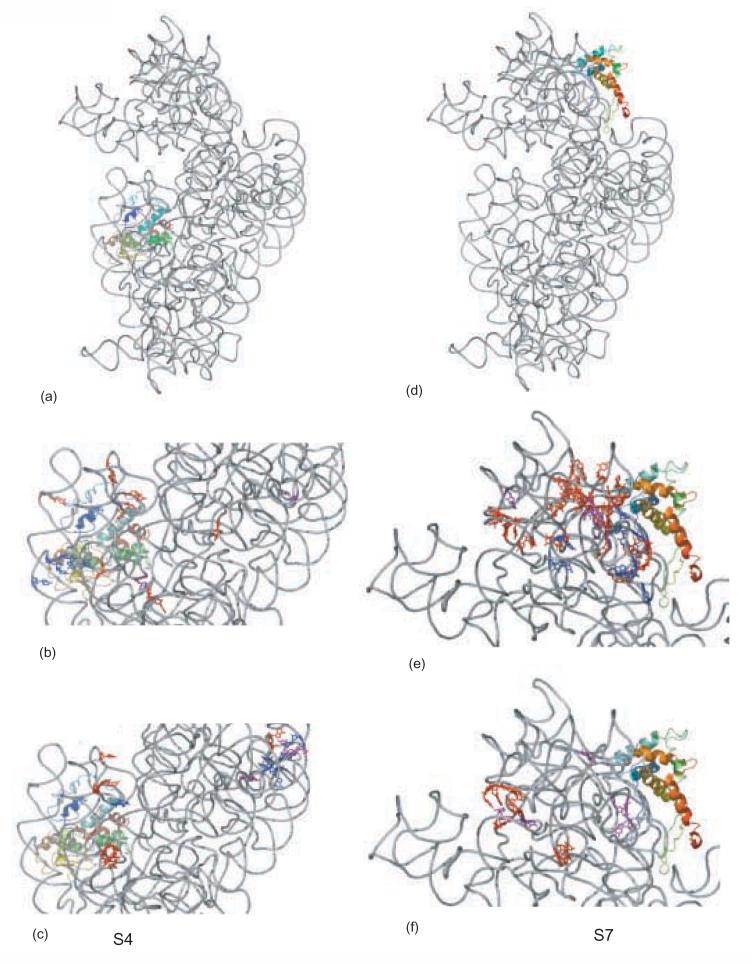
Details of footprints for r-proteins S15 and S8. The 16S rRNA is shown in gray, S15 and S8 are shown in rainbow, from blue (N-terminus) to red (C-terminus). Footprints which are not temperature-dependent are shown in blue, footprints that appear at 0°C and continue to develop in intensity at 42°C are shown in purple, and footprints that appear only at 42°C are shown in red. a) 16S rRNA r-protein and S15 from the crystal structure of E. coli 30S subunit, b) S15-dependent protections; c) S15-dependent enhancements; d) 16S rRNA r-protein and S8 from the crystal structure of E. coli 30S subunit, e)S8-dependent protections; f) S8-dependent enhancements.
The majority of the S7 footprints that appear at 0°C are grouped together on the three dimensional structure of 16S rRNA from 30S subunits1 (Figure 5e, f). They are localized in the region of 16S rRNA that is near the N-terminus of S7. The protections that appear at 42°C are more dispersed; nonetheless many of them are clustered along one region of the head (Figure 5e). There is also a trend that can be related to the proximity of the sites to S7 and the extent of temperature dependence observed: the enhancements which are more proximal to S7 are initially observed at 0°C and become more intense at 42°C, while the ones that are more distal from S7 mostly appear only in the complex formed at 42°C (Figure 5f). This may reflect that many direct contacts are formed at low temperature and long range changes are augmented at higher temperature. Thus it appears that S7/16S rRNA undergoes an extensive temperature-dependent conformational rearrangement and that this rearrangement is consistent with the architecture of the 30S subunit and allows long range conformational rearrangements to be revealed.
Figure 5.
Details of footprints for r-proteins S4 and S7. The 16 S rRNA is shown in gray, S15 and S8 are shown in rainbow, from blue (N-terminus) to red (C-terminus). Coloring of footprints as described in Figure 4. a) 16S rRNA r-protein and S4 from the crystal structure of E. coli 30S subunit, b) S4-dependent protections; c) S4-dependent enhancements; d) 16S rRNA r-protein and S7 from the crystal structure of E. coli 30S subunit, e) S7-dependent protections; f) S7-dependent enhancements.
Our data reveal that binding of S7 can be dissected into two separable phases, allowing a model for the sequential interaction of S7 with 16S rRNA to be proposed. In this model, the highly charged N-terminus and the first helical elements of S7 would associate with 16S rRNA in the initial phase, while the second binding event involving the C-terminal portion of S7 would occur in the second phase, as revealed by temperature-dependent changes (Figure 5e, f). This proposal is consistent with studies in which the binding constants for the complexes formed between 16S rRNA and fragments of S7 were determined37. If the N-terminus of S7 is deleted binding to 16S rRNA is destroyed. However, when the N-terminus of S7 is intact but other parts of S7 are deleted, the binding constant decreases but the protein-RNA interaction still takes place37. These findings are also consistent with in vivo studies which indicate that when the N-terminus of S7 r-protein is deleted, the assembly efficiency is reduced to about 3% of that observed for full length S738. Thus our data reveal a model for a two stage association of S7 with 16S rRNA that is supported by other in vitro and in vivo studies, and likely reveal details of bipartite association of r-proteins with 16S rRNA.
As suggested earlier for S7, the binding of S4 could occur in two steps which are revealed by the temperature-dependence of the footprints23. First, the more globular central domain of S4 would bind, and then the N- and C-termini would bind latter, as revealed by the temperature-dependent rearrangement of this RNP23. As the temperature-dependent S4 specific footprints are more dispersed throughout the 16S rRNA, and many of them are quite remote from the area of S4/16S rRNA direct contact (Figure 5c), it could be suggested that S4 facilitates long range conformational rearrangements during the course of 30S subunit assembly. For S4, it seems that the core protein structure is similar before and after binding to 16S rRNA1; 6; 9; 28 although information is only available for a subfragment of S4 in the free form9.
The temperature-dependent changes observed for some of the 16S rRNA-primary r-protein containing RNPs have significant implications for subsequent assembly events. The majority of the experiments performed to analyze the assembly dependence of the small subunit r-proteins were carried out at elevated temperature. Thus the work presented here aids in our understanding of the requisite order that has been observed. One interesting example of this is found in the functionally important 530 loop. Enhancements of specific nucleotides in this region requires both S816 and elevated temperature. Previous work has revealed that in subsequent stages of assembly these enhanced nucleotides become less reactive to chemical probes in an S5-dependent manner. Thus r-protein mediated temperature-dependent conformational changes could be critical for perpetuation of the 30S subunit assembly cascade.
The complexity of the spectrum of interactions between RNA and proteins is well illustrated in our model system. Even in the same RNP, differential folding of segments of the RNA molecule with a single protein can be observed. Simple single phase interactions are observed, in general, with RNA binding proteins that are very small, and well structured. In other instances, a more regulated process appears to be utilized. Alteration of temperature can be used to reveal modulation of folding of RNA within these RNP. Changes in RNA structure within the RNP can be subtle, such as fine adjustments, or quite substantial. Our simple study suggests that the r-proteins can interact with 16S rRNA differentially and that at least two types of induced fit are observed: when only the RNA is changing its conformation after binding and when both the RNA and the protein are changing conformation at binding.
Materials and Methods
16S rRNA/r-protein complex formation
The complexes were prepared from 16S rRNA and the 30S subunit recombinant r-proteins isolated as described previously39; 40; 41. The buffers used at the formation of the RNPs are: reconstitution A minus buffer (RA−) which is 20 mM K+-Hepes (pH 7.6), 20 mM MgCl2, 6 mM β-mercaptoethanol; reconstitution A plus buffer (RA+) which has the same composition as RA−, plus 330 mM KCl. The complexes were formed as follows: natural 16S rRNA in RA− was incubated at 42°C for 15 minutes, followed by 10 minutes on ice prior to complex formation. 40 pmoles of 16S rRNA were mixed with 200 or 240 pmoles of the appropriate r-protein, and the final KCl concentration was adjusted to 330 mM, by using the appropriate ratios of RA+ and RA−, and taking into account that the protein solutions are 1 M in KCl. The reaction mixture was incubated at the desired temperature, 0°C or 42°C for 1 h, or for the shifted complex, 30 min at 0°C and 30 minutes at 42°C. Two samples containing only 16S rRNA were also incubated at 0°C or 42°C, for comparison. All samples were then incubated on ice for ten minutes, before probing.
Chemical probing and primer extension analysis of the 16S rRNA/r-protein complexes
Chemical probing of 16S rRNA, and the RNPs with kethoxal and DMS was performed as previously described39; 42 except that the reactions were performed on ice. The probing times were: kethoxal, 60 min and DMS, 120 min. Each complex was independently probed 2-5 times using 16SrRNA from two different preparations.
Primer extension was performed essentially as described39; 42. Each primer extension was performed multiple times with primers designed to redundantly cover appropriate regions of 16S rRNA. The footprints were evaluated visually, and some were confirmed with the ImageJ software43.
Acknowledgements
We would like to thank Kristi Holmes for providing some of the 16S rRNA used in these experiments and Kirthi Narayanaswami for critical reading of the manuscript. This work was supported by the National Institute of Health grant GM62432 to GMC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 2.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–8. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 3.Wimberly BT, Brodersen DE, Clemons WM, Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 4.Hosaka H, Nakagawa A, Tanaka I, Harada N, Sano K, Kimura M, Yao M, Wakatsuki S. Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor. Structure. 1997;5:1199–1208. doi: 10.1016/s0969-2126(97)00270-0. [DOI] [PubMed] [Google Scholar]
- 5.Wimberly BT, White SW, Ramakrishnan V. The structure of ribosomal protein S7 at 1.9 A resolution reveals a beta-hairpin motif that binds double-stranded nucleic acids. Structure. 1997;5:1187–98. doi: 10.1016/s0969-2126(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 6.Sayers EW, Gerstner RB, Draper DE, Torchia DA. Structural Preordering in the N-Terminal Region of Ribosomal Protein S4 Revealed by Heteronuclear NMR Spectroscopy. Biochemistry. 2000;39:13602–13613. doi: 10.1021/bi0013391. [DOI] [PubMed] [Google Scholar]
- 7.Golden BL, Hoffman DW, Ramakrishnan V, White SW. Ribosomal protein S17: characterization of the three-dimensional structure by 1H and 15N NMR. Biochemistry. 1993;32:12812–20. doi: 10.1021/bi00210a033. [DOI] [PubMed] [Google Scholar]
- 8.Berglund H, Rak A, Serganov A, Garber M, Hard T. Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus. Nat. Struct. Biol. 1997;4:20–3. doi: 10.1038/nsb0197-20. [DOI] [PubMed] [Google Scholar]
- 9.Davies C, Gerstner RB, Draper DE, Ramakrishnan V, White SW. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998;17:4545–58. doi: 10.1093/emboj/17.16.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Nat. Acad. Sci. U.S.A. 1968;59:777–84. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–80. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 12.Samaha RR, O'Brien B, O'Brien TW, Noller HF. Independent in vitro assembly of a ribonucleoprotein particle containing the 3' domain of 16S rRNA. Proc. Nat. Acad. Sci. U.S.A. 1994;91:7884–8. doi: 10.1073/pnas.91.17.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from Escherichia coli. Nature. 1970;226:1214–18. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 14.Grondek JF, Culver GM. Assembly of the 30S ribosomal subunit: Positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–1866. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 1993;232:362–74. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 16.Svensson P, Changchien LM, Craven GR, Noller HF. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16S ribosomal RNA. J. Mol. Biol. 1988;200:301–8. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- 17.Powers T, Changchien LM, Craven GR, Noller HF. Probing the assembly of the 3′ major domain of 16 S ribosomal RNA. Quaternary interactions involving ribosomal proteins S7, S9 and S19. J. Mol. Biol. 1988;200:309–19. doi: 10.1016/0022-2836(88)90243-4. [DOI] [PubMed] [Google Scholar]
- 18.Dunn JM, Wong K-P. Molecular mechanism of in vitro 30S ribosome assembly. I. Conformational changes of the 16S RNA. J. Biol. Chem. 1979;254:7705–11. [PubMed] [Google Scholar]
- 19.Dunn JM, Wong K-P. Structure and stability of the reconstituted intermediate particles involved in the in vitro self-assembly of the 30S ribosomal subunit. Biochemistry. 1979;18:4380–5. doi: 10.1021/bi00587a018. [DOI] [PubMed] [Google Scholar]
- 20.Held WA, Nomura M. Rate determining step in the reconstitution of Escherichia coli 30S ribosomal subunits. Biochemistry. 1973;12:3273–81. doi: 10.1021/bi00741a020. [DOI] [PubMed] [Google Scholar]
- 21.Holmes KL, Culver GM. Analysis of Conformational Changes in 16S rRNA During the Course of 30S Subunit Assembly. J. Mol. Biol. 2005;354:340–357. doi: 10.1016/j.jmb.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 22.Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- 23.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J. Biol. Chem. 1995;270:1238–42. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 24.Mougel M, Ehresmann B, Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16S rRNA: kinetic and thermodynamic characterization. Biochemistry. 1986;25:2756–65. doi: 10.1021/bi00358a003. [DOI] [PubMed] [Google Scholar]
- 25.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumann C, Otridge J, Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 1996;271:12269–74. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- 27.Baumann C, Xirasagar S, Gollnick P. The trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis binds to unstacked trp leader RNA. J. Biol. Chem. 1997;272:19863–9. doi: 10.1074/jbc.272.32.19863. [DOI] [PubMed] [Google Scholar]
- 28.Brodersen DE, Clemons WM, Jr., Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–68. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 29.Stern S, Changchien LM, Craven GR, Noller HF. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988;200:291–9. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- 30.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 31.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science. 2000;288:107–13. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 32.Gerstner RB, Pak Y, Draper DE. Recognition of 16S rRNA by ribosomal protein S4 from Bacillus stearothermophilus. Biochemistry. 2001;40:7165–73. doi: 10.1021/bi010026i. [DOI] [PubMed] [Google Scholar]
- 33.Mougel M, Eyermann F, Westhof E, Romby P, Expert-Bezancon A, Ebel JP, Ehresmann B, Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J. Mol. Biol. 1987;198:91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- 34.Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 s ribosomes studied in vitro. J. Mol. Biol. 1969;40:391–413. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- 35.Allers J, Shamoo Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 36.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5' domain. J. Mol. Biol. 2005;351:508–19. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Robert F, Gagnon M, Sans D, Michnick S, Brakier-Gingras L. Mapping of the RNA recognition site of Escherichia coli ribosomal protein S7. RNA. 2000;6:1649–59. doi: 10.1017/s1355838200001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fredrick K, Dunny GM, Noller HF. Tagging Ribosomal Protein S7 Allows Rapid Identification of Mutants Defective in Assembly and Function of 30 S Subunits. J. Mol. Biol. 2000;298:379–394. doi: 10.1006/jmbi.2000.3563. [DOI] [PubMed] [Google Scholar]
- 39.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 40.Culver GM, Noller HF. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 2000;318:446–460. doi: 10.1016/s0076-6879(00)18069-3. [DOI] [PubMed] [Google Scholar]
- 41.Culver GM, Noller HF. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–43. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merryman C, Noller HF. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith CWJ, editor. RNA:Protein Interactions. A Practical Approach. Oxford University Press; New York: 1998. [Google Scholar]
- 43.Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda Maryland, USA: 19972006. [Google Scholar]
- 44.DeLano WL. DeLano Scientific. San Carlos; CA, USA: 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 45.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]