Abstract
The role of the proximal promoter GC-box in regulating basal and cAMP-dependent GTP Cyclohydrolase I gene transcription was investigated using a variety of cell lines and techniques. These studies show that the GC-box is composed of a triad of cis-elements that in vitro bind specificity proteins Sp1 and Sp3. Sp1 and Sp3 were found associated with the native proximal promoter in PC12 cells but were not recruited to the promoter during cAMP-dependent transcription. Studies using Drosophila SL2 cells showed that Sp3 occupies two sites within the GC-box and enhances transcription when acting alone and synergistically when combined with nuclear factor-Y (NF-Y) and CCAAT/Enhancer-Binding Protein (C/EBP)β, cognate binding proteins for the adjacent cAMP response element (CRE) and CCAAT-box cAMP response elements. In contrast, Sp1 bound only one site within the GC-box and did not enhance transcription unless combined with NF-Y and C/EBPβ. Studies in SL2 cells also showed that Sp1 and Sp3 do not co-occupy the GC-box, and accordingly Sp1 competes for Sp3 binding to repress Sp3-dependent transcription. In PC12 cells, complete mutation of the GC-box reduced basal but not cAMP-dependent transcription, resulting in an overall increase in the cAMP response and demonstrating that formation of this enhanceosome does not require Sp1 or Sp3. Experiments in which the GC-box was replaced with a Gal4 element and the promoter challenged with Gal4 fusion proteins support this conclusion and a role for Sp3 in maintaining high levels of basal transcription in PC12 cells. Equivalent amounts of Sp1 and Sp3 were found associated with the native proximal promoter in PC12 and Rat2 cells, which differ 10-fold in basal transcription. Similar levels of methylation of CpG dinucleotides located within the GC-box were also observed in these two cells lines. These results suggest that Sp1 and Sp3 bound to the GC-box might help to preserve an open chromatin configuration at the proximal promoter in cells which constitutively express low levels of GTP Cyclohydrolase I.
Keywords: GTP Cyclohydrolase I, PC12 cell, Sp1, Sp3 transcription
GTP cyclohydrolase I (GCH1; EC 3.5.4.16) is the first and rate-limiting enzyme in tetrahydrobiopterin (BH4) biosynthesis, the essential cofactor for monoamine and nitric oxide production and the detoxification of L-phenylalanine (Thony et al. 2000). GCH1 transcription is dynamic and can be enhanced by the second messenger cAMP in only a handful of cell types, including adrenal chromaffin cells (Abou-Donia et al. 1986), midbrain dopamine neurons (Zhu et al. 1994; Bauer et al. 2002), mesangial cells (Pluss et al. 1996), and PC12 cells (Anastasiadis et al. 1998; Kapatos et al. 2000). While this specificity implies novel signaling mechanisms, the effect of cAMP on GCH1 gene transcription is mediated entirely through the ubiquitous protein kinase A (Kapatos et al. 2007) which suggests that cAMP responsiveness is determined by the cellular complement of transcription factors made available to the GCH1 gene promoter.
Studies of the rat and human GCH1 promoters have identified the first 140 bp upstream from the transcription start sites as the minimal sequence necessary for cell type-specific cAMP-dependent transcription (Kapatos et al. 2000; Hirayama et al. 2001). Within this sequence lie a GC-box, a CRE and a CCAAT-box that are evolutionarily conserved. Both the CRE and the CCAAT-box are required for maximum basal and cAMP-dependent transcription (Kapatos et al. 2000; Kapatos et al. 2007). While the CRE binds members of the basic leucine zipper family of transcription factors, including cAMP-response element binding protein (CREB), ATF-2, c-Jun and C/EBPβ, the CCAAT-box binds the obligate heterotrimeric protein NF-Y (Kapatos et al. 2000; Hirayama et al. 2001; Sarraj et al. 2005; Wu et al. 2004; Kapatos et al. 2007). A recent examination of the endogenous GCH1 gene functioning within intact PC12 cells has confirmed these observations and also showed that cAMP treatment causes the recruitment of C/EBPβ and NF-Y along with Pol II to the proximal promoter (Kapatos et al. 2007).
Previous research using in vitro footprinting and PC12 cell nuclear extracts concluded that the GCH1 proximal promoter GC-box binds members of the stimulatory protein-1 (Sp1) family of transcription factors (Kapatos et al. 2000). This same study showed that the GC-box reduces cAMP-dependent transcription conferred by the CRE and CCAAT-box cAMP-response elements on a heterologous promoter, suggesting an inhibitory role for Sp-proteins in GCH1 transcription. Sp1, Sp3, and Sp4 proteins each recognize the identical GC-rich cis-element with the same affinity through a highly conserved C-terminal zinc-finger DNA binding domain (Suske 1999; Bouwman and Philipsen 2002), yet differentially regulate gene expression through subtle differences in N-terminal activation domains (Hagen et al. 1995; Ahlgren et al. 1999). Sp1 and Sp3 are both substrates for protein kinase A and phosphorylation is reported to enhance DNA binding and trans-activation (Rohlff et al. 1997; Ge et al. 2001). Sp-proteins typically affect transcription through interactions with components of the general transcriptional machinery (Smale et al. 1990; Hoey et al. 1993; Gill et al. 1994; Saluja et al. 1998) as well as through interactions with co-activators (Ryu et al. 1999). Sp-proteins also interact with proteins known to be associated with the GCH1 promoter, including C/EBPβ (Lee et al. 1997), NF-Y (Roder et al. 1999; Borestrom et al. 2003) and ring finger protein 4 (Poukka et al. 2000).
We now present data in support of a triad model of the rat GCH1 proximal promoter GC-box in which three distinct cis-elements have the potential to bind Sp1 and Sp3 proteins. We also show that while endogenous Sp1 and Sp3 are associated with the native GCH1 proximal promoter and are important for maintaining basal transcription neither protein is recruited to the native promoter in response to cAMP or absolutely required for the cAMP response. Finally, we find no relationship between the basal rate of GCH1 transcription, the amounts of Sp1 and Sp3 protein associated with the proximal promoter and levels of CpG dinucleotide methylation within the GC-box, suggesting that Sp1 and Sp3 might also have a role in preserving an open chromatin configuration at the proximal promoter.
Materials and methods
Plasmids
The construction and characterization of wild-type and mutated forms of the pGL3-based luciferase reporter construct p0.27GCH-GL3 has been reported previously (Kapatos et al. 2000). The Renilla reporter plasmid pRL-Null and the 5XGal4-luciferase construct pFR-luc were purchased (Promega, Madison WI, USA). Plasmids pPac-0, pPac-Sp1 and pPac-Sp3 were a gift from Dr Robert Tjian, UCLA, CA, USA. Plasmids pPac-NF-YA, pPac-NF-YB and pPac-NF-YC were gifts from Dr. Timothy F. Osborne, University of California, Los Angeles, USA. Gal4-Sp1(amino acids 1–603) and Gal4-Sp3(amino acids 1–527) were a gift from Dr Guntram Suske, Philipps-University, Germany. Gal4-C/EBPβ (amino acids 1–83) was a gift from Dr Peter Johnson, NCI, MD, USA. All plasmid DNA was purified by ion-exchange chromatography (Qiagen, Valencia, CA, USA).
Site-directed mutagenesis
Site-directed mutagenesis of the GC-box was performed using the technique described by Wang and Malcolm 1999. Briefly, p0.27GCH1-GL3 or p0.27GCH1-GL3-CREmtCATmt was used as template to generate either luciferase constructs or electrophoretic mobility shift assay (EMSA) probes, respectively. The forward primers for mutagenesis of the GC-box were as follows (reverse primers are the inverse compliment): mutation M1 (M1, mutation underlined) TCCAGGATTTCGGGGCGGAGGGGAGGGGCGAGCCCTT; mutation M2 (M2) CCCCTCCAGGAGGGCTTTGC GGAGGGGAGGGGC; mutation M3 (M3) CTCCAGGAGGGCGGGGCGGATTTGAGGGGCGAG; mutation M4 CTCCAGGAGGGCGGGGCGGAGGGGATTTGCGAG; mutation 1234 CCCCTCCAGGATTTCTTTGCGGATTTGATTTGCGAGC. The Gal4-GCH1-GL3 construct was generated by replacing the GC-box in the p0.27GCH1-GL3 wild-type plasmid with a single Gal4 element after first introducing restriction sites flanking the GC-box, digesting to remove the GC-box and religation of the plasmid. This construct was then used as the template for mutagenesis with forward primer (Gal4 element underlined) GCT CTT ACG CGT GCT AGC CCA ACG GAG TAC TGT CCT CCG AGT TCC TTG ACG CAA GAG GCT CGG. All mutations were confirmed by sequencing.
Cell culture
PC12 cells were maintained on collagen in media composed of Dulbecco’s modified Eagle medium containing penicillin–streptomycin, supplemented with 5% fetal calf serum and 10% horse serum at 37°C and 10% CO2. Rat2 cells were grown in Dulbecco’s modified Eagle medium containing penicillin–streptomycin and 10% fetal calf serum at 37°C and 10% CO2. SL2 cells were maintained at 24°C in air in Schneider’s Drosophila SL2 media (Invitrogen, Carlsbad, CA, USA) supplemented with penicillin–streptomycin and 10% fetal bovine serum.
Transient transfections and luciferase assays
PC12 and SL2 cells were plated at a density of 250–300,000 cells per well in poly-D-lysine-coated 24-well plates. The next day, each well was transfected with 800 ng of DNA using Lipofectamine 2000 (Invitrogen). Cells were harvested 24–48 h later and assayed using the dual luciferase assay (Promega). In some experiments, PC12 cells were treated with 5 mmol/L 8Br-cAMP for 4 h prior to harvesting. Luciferase assays were performed in replicates of four and experiments repeated at least three times. Data were normalized across experiments relative to control values and analyzed by one-way ANOVA with post hoc Bonferroni tests to correct for multiple comparisons (PRISM, Graphpad, San Diego, CA, USA). Differences were accepted as significant at p < 0.05.
Nuclear extracts
PC12 and SL2 cell nuclear extracts were prepared by plating 20 million cells on 100 mm poly-D-lysine-coated dishes. In some experiments, PC12 cells were incubated with 5 mmol/L 8Br-cAMP for 4 h prior to harvest. In others, SL2 cells were transfected with 24 μg of pPac-0, pPac-Sp1, or pPac-Sp3 and harvested 48 h later. After washing, cells were scraped into 1.5 mL of Dulbecco’s phosphate-buffered saline containing protease and phosphatase inhibitors (Sigma, St Louis, MO, USA) and centrifuged at 400 g at 4°C for 5 min. Cell pellets were suspended in ice-cold Buffer A (Dignam et al. 1983; 10 mmol/L HEPES-KOH pH 7.9 at 4°C, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 1 mmol/L NaF, 0.5 mmol/L dithiothreitol (DTT), 0.2 mmol/L phenyl methyl sulfonyl fluoride (PMSF), 10 μg/mL leupeptin, 1 μg/mL aprotinin and 10 μg/mL pepstatin A and phosphatase inhibitors), allowed to swell on ice for 10 min and centrifuged at 400 g for 5 min at 4°C. The pellet was suspended in 100 μL of Buffer C (20 mmol/L HEPES-KOH, pH 7.9 at 4°C, 25% glycerol, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 1 mmol/L NaF, 0.5 mmol/L DTT, 0.2 mmol/L PMSF, 10 μg/mL leupeptin, 1 μg/mL aprotinin and 10 μg/mL pepstatin A and phosphatase inhibitors), incubated on ice for 20 min and centrifuged at 4°C at 21 000 g for 10 min. The supernantant was collected and assayed for protein content (Bradford 1976).
Electrophoretic mobility shift assay
EMSA probes for wild-type GCH1 and mutations M2, M3, and M4 were generated by PCR using as template p0.27GCH1-GL3-CREmtCATmt and primers forward −148 TAGCCCCTCCAG GAGGGC −130 and reverse −68 ACTCTGCCGGCACCG −54. For generation of probes containing mutations M1 and M1234 the forward primer was −148 TAGCCCTCCAGGAT −134. PCR products were isolated by agarose gel electrophoresis and purified using the Qiaquik Gel extraction kit (Qiagen). Probes were end-labeled with T4 polynucleotide kinase and γ32P ATP and purified using G25 spin columns. The EMSA reaction buffer contained 12.5 mmol/L HEPES-KOH, pH 7.9, 10% glycerol, 100 mmol/L KCl, 1 mmol/L EDTA, 1 mmol/L DTT, 1 μg poly dI-dC, 5 μg acetylated BSA and 0.2 mmol/L PMSF (Kapatos et al. 2000). The order of addition on ice was reaction buffer, unlabelled competitor (250–1000×), or antibody (1 μg), nuclear extract (1 μg), and labeled probe (~10 fmoles) followed by incubation at 25°C for 20 min. Samples were loaded and run on 6% polyacrylamide gels using 0.5× tris borate EDTA running buffer. Gels were dried and either exposed to film or phosphorimager screens (Typhoon, GE Healthcare, Piscataway, NJ, USA). Phosphorimage screens were scanned and the relative optical density of bands determined using ImageQuant software (Molecular Dynamics).
Western blotting
About 5–10 μg of protein was loaded onto 4–12% acrylamide gels and electrophoresed using morpholino propane sulfonic acid running buffer (NuPAGE, Invitrogen). Proteins were transferred to nitrocellulose using a semi-dry apparatus. Membranes were blocked using dry milk and incubated overnight with a 1: 500 dilution of rabbit antibodies directed against Sp1 (Upstate, Lake Placid, NY, USA), Sp3 or Sp4 (both from Santa Cruz, Santa Cruz, CA, USA). Membranes were washed, incubated with a 1: 25,000 dilution of horseradish peroxidase-coupled goat anti-rabbit, developed using a chemiluminescent horseradish peroxidase substrate (Pierce, Rockford, IL, USA) and simultaneously exposed to film. To insure equal loading of nuclear proteins in experiments using PC12 cells membranes were stripped and re-probed using an antibody to TFIID (Santa Cruz).
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay was performed as described (West et al. 2004) with minor modifications (Kapatos et al. 2007). Briefly, 20 million PC12 cells or 10 million Rat2 cells were plated on poly-D-lysine-coated 100 mm dishes. The next day cells were treated with 5 mmol/L 8Br-cAMP for 1 h or directly cross-linked by addition of serum-free media containing 1% fresh formaldehyde for 10 min at 25°C. Cells were washed with phosphate-buffered saline and cross-linking terminated by the addition of 0.125 M glycine for 10 min. All further procedures were performed at 4°C in the presence of protease and phosphatase inhibitors. Cells were scraped into phosphate-buffered saline and collected by centrifugation. Cell pellets were suspended in 1 mL of lysis buffer and incubated for 10 min at 4°C. DNA was fragmented by sonicating to an average size of 400 bp. Cell debris was pelleted, supernatant harvested and 5% set aside as input DNA. After reversing cross-links the DNA content of input DNA was determined and equal amounts of DNA were then diluted 10-fold with dilution buffer and pre-cleared by the addition of rabbit IgG and salmon sperm DNA/BSA blocked Protein A-agarose. Supernatants were then divided into 4 parts, each receiving 5 μg of antibody directed against either the large subunit of RNA polymerase II, (Santa Cruz) Sp1 or Sp3. Anti-Green Fluorescent Protein (GFP; Santa Cruz) or normal rabbit immunoglobulin G (IgG; Santa Cruz) served as negative controls. Immune complexes were collected and washed sequentially with low-salt, high-salt, LiCl, and finally TE. Immune complexes and input DNA were incubated with RnaseA, overnight at 65°C to reverse cross-links and then digested with Proteinase K. DNA was purified using the Qiaquick PCR Purification system (Qiagen). Samples were analyzed by real-time quantitative PCR (Roche LightCycler, Mannheim, Germany) using QuantiTect SYBR-Green PCR reagent (Qiagen). PCR primers amplified either the proximal (forward −46 GCCGCGCCTCTCTTTTTATG −27; reverse −146 TGTGCAACTGCGGGGTTTAG −167) or distal (forward −5396 TCACTGGCTCATGTACTGAATG −5374; reverse −5439 GAC-CTGCTTCCTCTCAATACAG −5417) sequence of the rat GCH1 promoter. All samples were run in triplicate and experiments repeated at least three times. Standard curves for each PCR product were generated by plotting Ct versus serial dilutions of pooled input DNA. The enrichment for Pol II, Sp1, and Sp3 was calculated by first subtracting values for non-specific immunoprecipitation and then dividing by input DNA. Data were normalized relative to control and analyzed by one-way ANOVA with post hoc t-tests. Differences were accepted as significant at p < 0.05. In experiments comparing PC12 and Rat2 cells sample data were expressed relative to the PC12 standard curve of input DNA.
Quantitative real-time RT-PCR analysis
Total RNA was isolated from control or 4 h 8Br-cAMP-treated cultures of PC12 or Rat2 cells. RNA was treated with DNase I as recommended by the supplier (Qiagen, RNeasy) and 25–150 ng of RNA was reverse transcribed (Qiagen, Omniscript) in a reaction primed with random hexamers. 10–20% of this reaction served as template for quantitative real-time RT-PCR analysis using Quanti-Tect SYBR-Green PCR reagents (Qiagen), to amplify either rat GCH1 (forward, caagggataccaggagacca; reverse, tctcgtcatggtcctcatca) or β-actin (forward, gtcgtaccactggcattgtg; reverse, ctctcagctgtggtggtgaa) cDNA. Control reactions minus reverse transcriptase were included for each primer pair and quantity of RNA. Standard curves for each transcript were generated using serial dilutions of pooled RNA from control samples and distinct reverse transcription reactions. GCH1 mRNA abundance were expressed relative to that of β-actin.
DNA methylation analysis
Genomic DNA was isolated from 5 × 106 PC12 and Rat2 cells (Zymo Research, Orange, CA, USA). 500 ng of PC12 and Rat2 cell DNA was then treated with bisulfite to convert unmethylated cytosines to uracils (Zymo Research). Bisulfite-treated GCH1 promoter DNA was amplified by methylation-specific PCR using primers designed by MethylPrimer Express software v1.0 (Applied Biosystems, CA, USA) to amplify a 288 bp product encompassing the GC-box, CRE and CCAAT-box (forward, −182 ATTTGAGGGTTGTTTTGTGTAA −161; reverse + 86 ACACCCRAA AATACTACCAAA + 107). PCR products were directly cloned into the pGEM-T vector (Promega), transformed into Top10 bacteria and DNA mini-preps prepared from 10 colonies for each cell type. DNA was sequenced using the M13 forward primer and analyzed by comparison to the sequence of untreated DNA.
Results
Bioinformatics analysis of the proximal promoter GC-box
A bioinformatics analysis of the entire 142 bp of the GCH1 proximal promoter (transcription element search system; Schug and Overton, 1997) determined that the three Sp-protein binding sites with the highest probability scores are all clustered within the GC-box footprint (Fig. 1a). These cis-elements will be referred to as Site I (−134 to −125), Site II (−130 to −121) and Site III (−124 to −114) and form the basis of the triad model of the GCH1 GC-box (Fig. 1a). Sites I and III are located at either ends of the GC-box and do not overlap whereas Site II is located in the center of the GC-box and overlaps Sites I and III (Fig. 1a and b). Because Sites I and III do not overlap, Sites I and III can potentially be occupied simultaneously by Sp-proteins (Fig. 1a and b). Since Sites I and III overlap with Site II, however, steric hindrance between binding proteins would prevent occupation of Site II if either Site I or III are bound (Fig. 1a and b). Similarly, because Site II overlaps Sites I and III, occupation of Site II would block protein binding to Sites I and III (Fig. 1a and b).
Fig. 1.
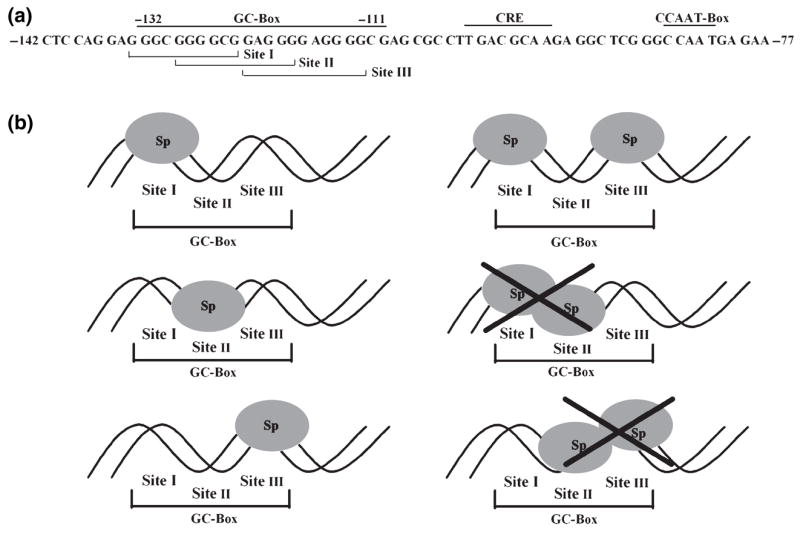
The triad model of the rat GCH1 proximal promoter GC-box. (a) The rat GCH1 proximal promoter sequence spanning from −142 to −77 is shown with the GC-box, the CRE and the CCAAT-box labeled. TESS (Transcription Element Search System) analysis of the entire proximal promoter detected three high probability Sp1 binding sites clustered within the GC-box footprint spanning from −132 to −111. Site I (−134 GGGCGGGGCG −125), Site II (−130 GGGGCGGAGGG −121) and Site III ((−124 GAGGGGAGGGG (−114) are bracketed and labeled. These three sites are the basis for the triad model of the GC-box. (b) In the triad model of the GC-box each of the three sites within the GC-box is capable of binding Sp-proteins. Sites I and III can be occupied simultaneously by Sp-proteins because these two sites do not overlap. Because Sites I and III overlap with Site II occupation of Sites I or III would prevent binding to Site II. Similarly, because Site II overlaps Sites I and III occupation of Site II would block binding to Sites I and III.
PC12 cells express Sp1 and Sp3 proteins
Before experiments were begun on the role of the GC-box and cognate binding proteins in GCH1 transcription it was imperative to identify which members of the Sp-protein immediate family are expressed by PC12 cells. Moreover, it was critical to know whether incubation with 8Br-cAMP alters Sp-protein levels, as is known to occur in this cell line following treatment with phorbol esters (Papanikolaou and Sabban 2000) or nerve growth factor (Sobue et al. 2005). Western blotting of nuclear extracts from control PC12 cells and PC12 cells treated for 4 h with 5 mmol/L 8Br-cAMP detected both Sp1 and Sp3 proteins but not Sp4 (Fig. 2a). Although a single Sp1 protein of approximately 100 kDa was observed, three different Sp3 proteins were found; one large abundant protein of approximately 100 kDa and two smaller less abundant proteins of approximately 77 and 73 kDa. Incubation with 8Br-cAMP did not alter the abundance of Sp1 or Sp3 proteins or result in the appearance of Sp4. We have therefore focused our attention on Sp1 and Sp3 proteins.
Fig. 2.
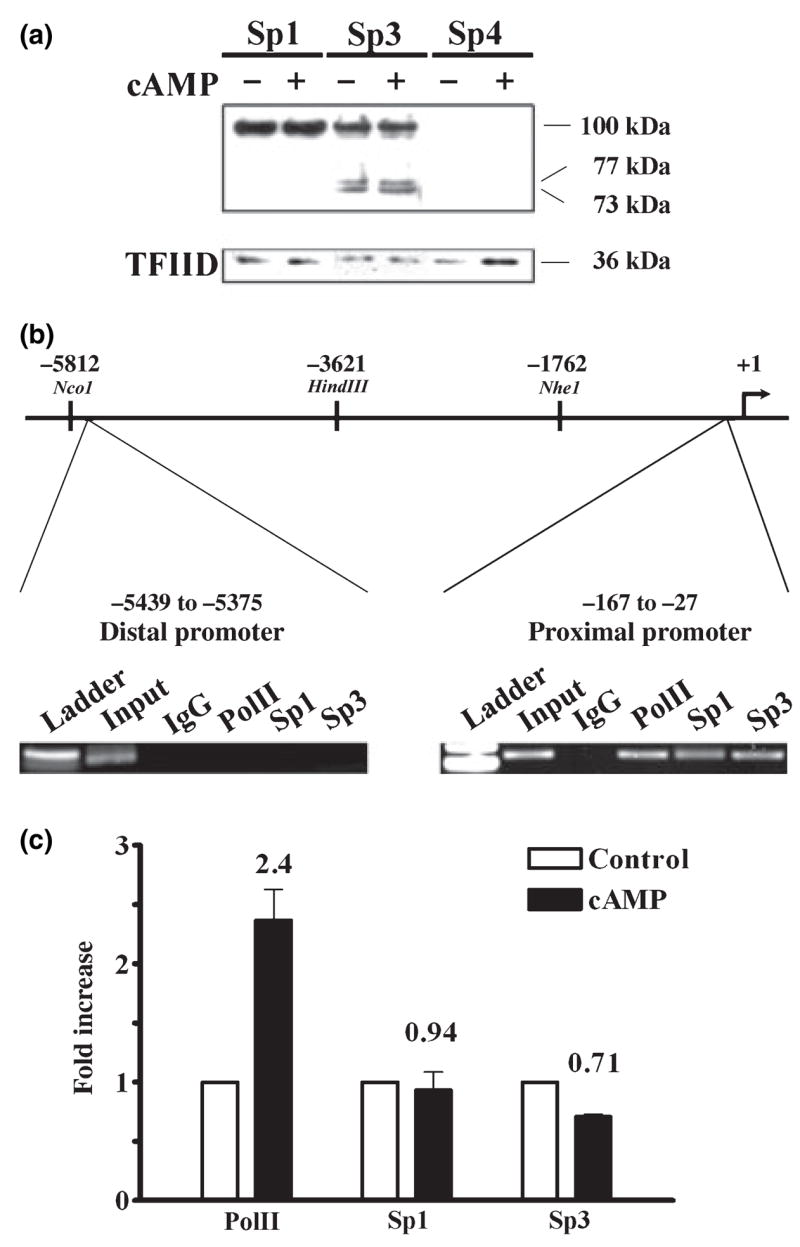
Sp1 and Sp3 protein expression and association with the native promoter in PC12 cells. (a) Three western blots of 5 μg of nuclear protein from control PC12 cells (−) and PC12 cells treated with 5 mmol/L 8-Br-cAMP for 4 h (cAMP, +). Proteins were separated on 4–12% gradient acrylamide gels and probed with antibodies directed against Sp1, Sp3 or Sp4. A single Sp1 protein of approximately 100 kDa was observed, as were three isoforms of Sp3 protein of approximately 100, 77, and 73 kDa. No Sp4 protein was detected. As a loading control the three blots were stripped and probed again with an antibody directed against the nuclear protein TFIID. (b) Schematic representation of 5812 bp of the rat GCH1 5′ flanking sequence showing the proximal (−167 to −27) and distal (−5439 to −5375) promoter regions amplified by PCR during ChIP analysis of the native promoter. Cross-linked and sheared DNA from control PC12 cells was precipitated using antibodies directed against Pol II, Sp1, Sp3 or control IgG, amplified by PCR and analyzed by agarose gel electrophoresis. Amplicons of the appropriate size were found in the proximal (141 bp) but not distal (65 bp) promoter reactions, indicating that under basal conditions Pol II, Sp1 and Sp3 are each associated with the GCH1 proximal promoter. (c) Control and 8Br-cAMP-treated (5 mmol/L for 1 h, cAMP) PC12 cells were analyzed by ChIP using antibodies directed against Pol II, Sp1 and Sp3 and precipitated proximal promoter DNA quantified by real-time PCR. Data were normalized relative to control immunoprecipitations and analyzed by ANOVA with post hoc Bonferroni tests. Only Pol II was significantly different between control and cAMP.
Sp1 and Sp3 are associated with the native GCH1 proximal promoter in PC12 cells
Another essential early step towards understanding the role played by the GC-box in GCH1 transcription was to use the ChIP technique to determine whether endogenous Sp1 and Sp3 proteins are localized to the native GCH1 proximal promoter under basal conditions and, like C/EBPβ and NF-Y (Kapatos et al. 2007), are recruited in response to cAMP. Sheared DNA from untreated PC12 cells was immunoprecipitated using antibodies specific to the large subunit of Pol II, Sp1, Sp3, and control IgG. GCH1 proximal promoter DNA was detected by PCR using primers that amplify a 141 bp product (−167 to −27) which includes the GC-box, CRE and CCAAT-box (Fig. 2b). A 65 bp product from the distal promoter (−5439 to −5375) located 5.4 kb upstream from the cap site was also amplified and served as a gene-specific control (Fig. 2b). Gel analysis showed that amplifications of proximal but not distal promoter DNA resulted in PCR products of the correct size for Pol II, Sp1, and Sp3 immunoprecipitations whereas no product was observed for the IgG control (Fig. 2b). Both Sp1 and Sp3 are therefore specifically associated with the proximal GCH1 promoter active within PC12 cells. Quantitative real-time PCR analysis of ChIP samples subsequently revealed that Sp1 and Sp3 are not recruited to the proximal promoter in response to cAMP treatment, although the amount of Pol II was increased by more than twofold indicating enhanced gene transcription (Fig. 2c).
EMSA identifies Sp1 and Sp3 binding to the GC-box in PC12 cells
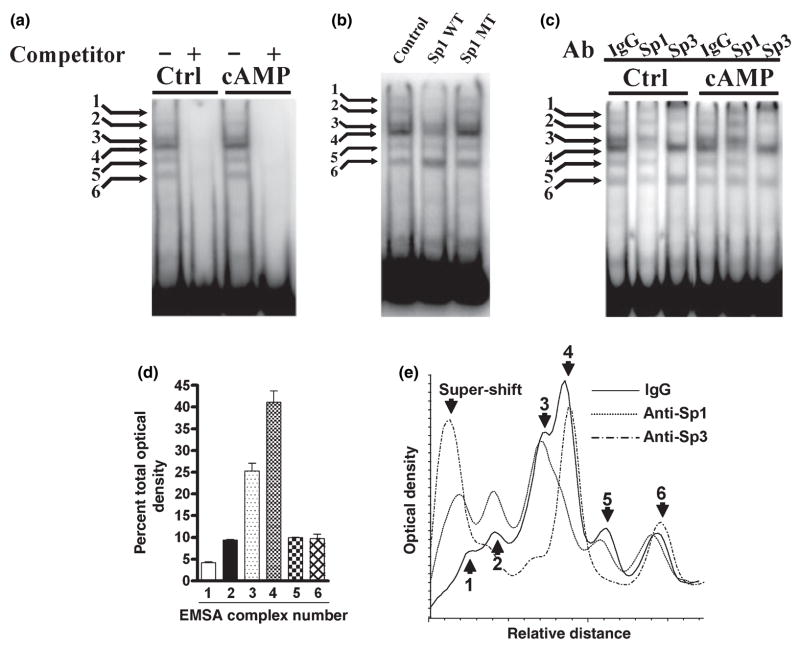
Chromatin immunoprecipitation is not necessarily informative about the binding of a protein to its cis-element and cannot confirm that Sp1 and Sp3 are actually bound to the GCH1 GC-box DNA in vivo. EMSA was therefore used to determine whether the GC-box binds the Sp1 and Sp3 proteins found in PC12 nuclear extracts and ChIP assays. Control PC12 cells and PC12 cells treated with 5 mmol/L 8Br-cAMP for 4 h were also compared in these experiments to determine whether a direct assay of protein-DNA interactions can detect phosphorylation-induced changes in Sp-protein binding. The probe used for EMSA spanned from position −148 to −54 and contained the wild-type GC-box but mutated forms of the CRE and CCAAT-box.
Initial experiments with short film exposure times at −80°C revealed two protein-DNA complexes, a major bottom band and minor top band (data not shown). Application of the more sensitive and linear phosphorimaging technique to EMSA revealed six protein-DNA complexes (complexes 1–6) all of which were competed away with excess probe and did not appear to be altered by 4 h treatment with 8Br-cAMP (Fig. 3a). Densitometric analysis of these six protein-DNA complexes indicated that complex 1 contains 5%, complex 2 contains 10%, complex 3 contains 25%, complex 4 contains 40%, complex 5 contains 10% and complex 6 contains 10% of total binding activity (Fig. 3d). Additional studies showed that complexes 1–5 but not complex 6 were competed by a consensus but not mutated Sp1 element (Fig. 3b), demonstrating that Sp-proteins are components of at least five of the six complexes. EMSA super-shift assays are notoriously difficult to interpret when there are multiple complexes present. Nonetheless, EMSA using antibodies directed against Sp1 and Sp3 (Fig. 3c) combined with densitometric analysis (Fig. 3e) allowed definitive identification of Sp1 in complex 4, Sp3 in complexes 3 and 5 and tentative identification of Sp3 in complexes 1 and 2. Based upon these results and the fraction of total binding made up by each complex we estimate that Sp1 comprises approximately 40% and Sp3 upwards of 50% of the GC-box binding proteins found in PC12 nuclear extracts. Treatment of cells with 8Br-cAMP did not modify the pattern of super-shifted complexes (Fig. 3c). Complex 6 was not shifted by antibodies to either Sp1 or Sp3 and was not studied further.
Fig. 3.
Binding of Sp1 and Sp3 to the GC-box in PC12 cells. (a) EMSA using 1 μg of nuclear protein from control PC12 cells (Ctrl) and PC12 cells treated with 5 mmol/L 8-Br-cAMP for 4 h (cAMP) identified six protein-DNA complexes (labeled 1–6) that could be competed away by excess unlabeled probe (+) and appeared unchanged by cAMP treatment. (b) EMSA using 1 μg of PC12 nuclear protein showed competition for binding of complexes 1–5 by a double-stranded oligonucleotide containing a wild-type Sp1 binding site (WT; ATTCGATCGGGGCGGGGCGAGC) but not a mutated Sp1 site (MT; ATTCGATCGGTTCGGGGCGAGC). (c) EMSA using 1 μg of nuclear protein from control (Ctrl) and 8-Br-cAMP treated (cAMP) PC12 cells combined with 1 μg of control IgG, anti-Sp1 or anti-Sp3 detected Sp1 in complex 4 and Sp3 in complexes 3 and 5 and probably in complexes 1 and 2. Complex 6 was not shifted by either antibody. (d) Analysis of EMSA band densities of the control lanes shown in Fig. 3(a)–(c) indicates that complex 1 contains 5%, complex 2 contains 10%, complex 3 contains 25%, complex 4 contains 40%, complex 5 contains 10% and complex 6 contains 10% of the total binding. (e) Densitometry traces of the three Ctrl lanes shown in Fig. 3(c). The solid line represents the IgG control and complexes 1–6 are identified with arrows. The dotted line represents the sample treated with anti-Sp1. Note the almost complete loss of complex 4. The dashed line represents the sample treated with anti-Sp3. Note the super-shift of complexes 3 and 5 and the probable shifts of complexes 1 and 2.
Sp3 but not Sp1 forms multiple complexes with the GC-box in SL2 cells
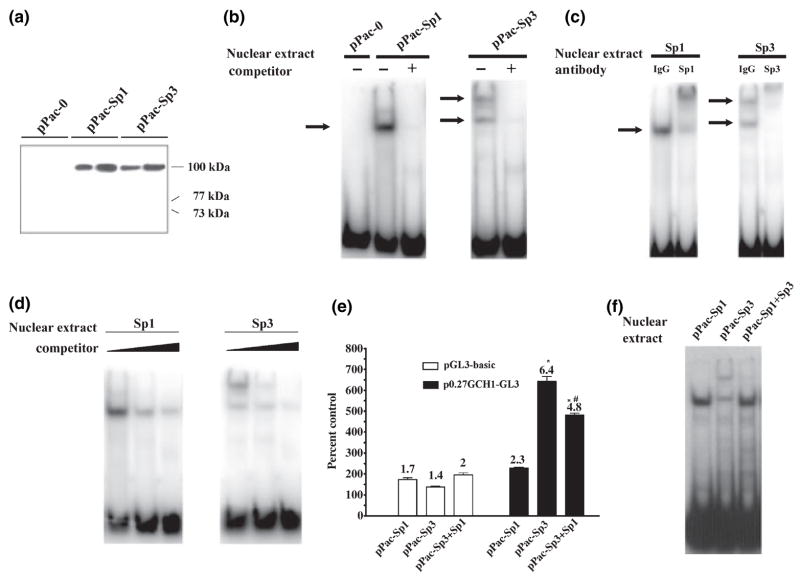
The complexity of the GC-box, expression of Sp1 and multiple forms of Sp3 and the EMSA binding pattern make it very difficult to study the role of Sp1 and Sp3 in GCH1 transcription in PC12 cells. Drosophila SL2 cells do not express the Sp family of transcription factors (Courey and Tjian 1988) and thus offer a cellular milieu to establish a basic understanding of Sp1 and Sp3 function at the GCH1 promoter. Western blots of nuclear extracts were prepared from SL2 cells transfected with vector driving Sp1 or Sp3 expression under the control of the Drosophila β-actin promoter (Fig. 4a). SL2 cells transfected with the empty pPac-0 vector contained no endogenous proteins that react with the antibodies to Sp1 and Sp3 used here. Moreover, Sp1 and Sp3 expressed by SL2 cells were of similar size (~100 kDa) and abundance, with no truncated forms of Sp3 present (Fig 4a). EMSA using nuclear extracts prepared from transfected SL2 cells also clearly showed that there are no endogenous proteins capable of binding to the GC-box (Fig. 4b, see pPac-0) and that the GC-box specifically recruits Sp1 and Sp3 (Fig. 4b). However, the patterns of Sp1 and Sp3 binding to the GC-box are different. Sp1 overwhelmingly formed a single complex whereas Sp3 formed two complexes in approximately equal abundance, one similar in size to that produced by Sp1 and the other significantly larger. Super-shift assay using antibodies to Sp1 and Sp3 confirmed that Sp1 is responsible for the single complex while Sp3 forms both the slow migrating large and fast migrating small complexes (Fig. 4b). Competition experiments subsequently determined that Sp1 and Sp3 binding is displaced by increasing concentrations of unlabeled probe and that the slow migrating Sp3 complex is displaced at lower probe concentrations than is the fast migrating complex, suggesting that Sp3 binds with lower affinity to a second site within the GC-box (Fig. 4d).
Fig. 4.
Sp1 and Sp3 binding to the GC-box and activation of transcription in SL2 cells. (a) Three simultaneously developed western blots of 5 and 10 μg of nuclear protein isolated from Drosophila SL2 cells transiently transfected with the pPac-0 empty vector, pPac-Sp1 or pPac-Sp3, harvested 48 h later and probed with antibodies to Sp1 and Sp3. The blot from cells transfected with the pPac-0 empty vector was probed with a combination of antibodies to Sp1 and Sp3. Note that no proteins were detected in the pPac-0 sample and that Sp1 and Sp3 proteins are produced at comparable levels with no small isoforms of Sp3 present. (b) EMSA using 1 μg of nuclear protein prepared from SL2 cells transiently transfected to express Sp1 or Sp3 shows that there are no proteins in SL2 cells which bind the GC-box (see pPac-0), that Sp1 forms a single complex (arrow) while Sp3 forms two complexes (arrows), and that Sp1 and Sp3 binding is competed away by excess unlabeled probe (+). (c) EMSA using 1 μg of nuclear protein prepared from SL2 cells transiently transfected to express Sp1 or Sp3 shows that anti-Sp1 super-shifts the Sp1 complex while anti-Sp3 super-shifts both the large and small Sp3 complexes. (d) EMSA using 1 μg of nuclear protein prepared from SL2 cells transiently transfected to express Sp1 or Sp3 shows that increasing amounts of unlabeled probe displaces Sp1 and Sp3 binding and that the large Sp3 complex is displaced at lower probe concentrations than is the small complex. (e) Luciferase assays of SL2 cells transiently co-transfected with 600 ng of p0.27-GCH1-GL3 or pGL3-basic along with 20 ng of pPac-Sp1, pPac-Sp3 or 20 ng pPac-Sp1 + 20 ng pPac-Sp3 and pPac-0 carrier DNA. Data were normalized relative to pPac-0 and analyzed by ANOVA with post hoc Bonferroni tests (*p ≤ 0.05 vs. pGL3-basic; #p ≤ 0.05 vs. pPac-Sp3). (f) EMSA using 1 μg of nuclear protein from SL2 cells expressing Sp1 or Sp3 or the combination of 0.5 μg of Sp1 and 0.5 μg of Sp3 nuclear protein shows that Sp1 competes to prevent formation of the large Sp3 complex.
In SL2 cells Sp3 activates transcription while Sp1 competes to repress Sp3 activation and formation of the large Sp3 complex
The next series of experiments investigated the effects of Sp1 and Sp3 on GCH1 promoter function. Luciferase assays were carried out in SL2 cells co-transfected with either the wild-type GCH1 proximal promoter reporter construct p0.27GCH1-GL3 or the empty pGL3 parental vector along with equal amounts of pPac-Sp1 or pPac-Sp3 expression vector DNA. Equal amounts of Sp1 or Sp3 plasmid DNA generate equal amounts of functional Sp1 and Sp3 proteins, as shown by analysis of band densities from EMSA experiments using nuclear extracts derived from different preparations of transfected SL2 cells (see Figs 4b–d, 5b and c). Preliminary experiments noted that both Sp1 and Sp3 enhanced activity from cryptic Sp-protein binding sites within the pGL3 vector itself (Fig. 4e). Indeed, Sp1 was found to have no statistically significant effect on transcription from the GCH1 promoter when luciferase activity induced by transfection with pPac-Sp1was corrected for this background activity (Fig. 4e). In contrast, the effect of Sp3 was robust, enhancing transcription by more than fivefold over the empty pGL3 vector (Fig. 4e). Co-transfection of equal amounts of Sp1 and Sp3 plasmid DNA showed that Sp1 actually inhibits Sp3-dependent activation by 50% (Fig. 4e). In agreement with the functional competition of Sp3 by Sp1, EMSA combining Sp1- and Sp3-containing nuclear extracts demonstrated that Sp1 is capable of competing away the large Sp3 complex (Fig. 4f). No doubt, Sp1 also competes with Sp3 and vice versa to form the smaller complex that is common to both proteins. Importantly, the combination of Sp1 and Sp3 proteins did not produce a large complex like that observed for Sp3.
Fig. 5.
Mutation of the GC-box supports a triad model in which Sp1 and Sp3 interact with C/EBPβ and NF-Y to activate transcription. (a) Mutagenesis was performed based upon the triad model of the GC-box. In each case a GGG within a site was replaced with a TTT and is labeled above. Mutation M1 within Site I forces Sp-protein binding to Sites II or III while mutation M2 within Sites I and II forces binding to Site III. Similarly, mutation M3 within Sites II and III forces binding to Site I while mutation M4 within Site III forces binding to Sites I or II. Mutation M1234 in Sites I, II, and III eliminates Sp-protein binding completely. (b) EMSA using 1 μg of nuclear protein from SL2 cells transiently transfected to express Sp1 demonstrates that mutations M1, M2, M3 and M4 do not affect binding to the GC-box whereas mutation M1234 completely eliminates Sp1 binding. (c) EMSA using 1 μg of nuclear protein from SL2 cells transiently transfected to express Sp3 shows that mutations M1, M2, M3, and M4 prevent formation of the large Sp3 complex without affecting formation of the smaller complex whereas mutation M1234 completely eliminates Sp3 binding. (d) Luciferase assays of SL2 cells transiently co-transfected with 780 ng of wild-type (WT) p0.27GCH1-GL3 or the reporter constructs containing GC-box mutations shown in Fig. 5a, M1, M2, M3, M4 or M1234 along with 20 ng of pPac-0 or 20 ng of pPac-Sp3. Data were analyzed by ANOVA and post hoc Bonferroni tests (*p < 0.05 vs. WT pPac-Sp3). (e) Luciferase assays of SL2 cells transiently transfected with the 600 ng of wild-type p0.27-GCH1-GL3 reporter along with various combinations of 50 ng pPac-NF-YA, B and C, 50 ng pPac-C/EBPβ, 20 ng pPac-Sp3, 20 ng of pPac-Sp1 and varying amounts of pPac-0. Data were analyzed by ANOVA and post-hoc Bonferroni tests (*p < 0.05 vs. pPac-0; #p < 0.05 vs. pPac-C/EBPβ; @p < 0.05 vs. pPac-Sp3; !p < 0.05 vs. pPac-Sp3 + pPac-C/EBPβ; + p < 0.05 vs. pPac-Sp3 + pPac-NFY or pPac-C/EBPβ; &p < 0.05 vs. pPac-Sp3 + pPac-NF-Y + pPac-C/EBPβ).
In SL2 cells mutation of any single site within the GC-box has no effect on Sp1 binding but eliminates formation of the large Sp3 complex
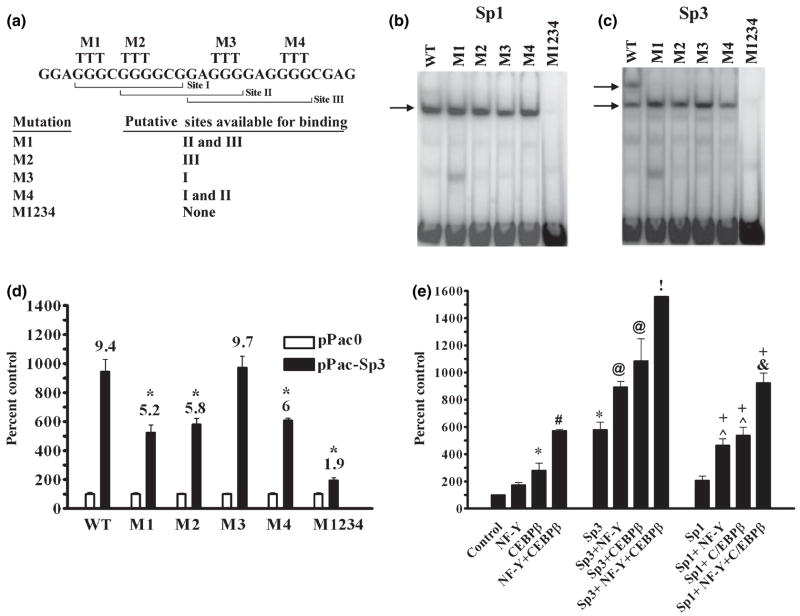
To further characterize the GC-box and Sp1 and Sp3 binding mutagenesis was performed under the guidance of the triad model. Accordingly, GGG sequences within the GC-box were strategically replaced by TTT mutations so as to inactivate each or all of the three putative Sp-protein binding sites. Figure 5a shows that mutation M1 within Site I forces Sp-protein binding to Sites II or III while mutation M2 within Sites I and II forces binding to Site III. Similarly, mutation M3 within Sites II and III forces binding to Site I while mutation M4 within Site III forces binding to Sites I or II. Mutation M1234 in Sites I, II, and III eliminates Sp-protein binding completely.
Repeated EMSA analysis of the effect of these mutations on binding of Sp1 and Sp3 using nuclear extracts from transfected SL2 cells showed that individual mutations M1, M2, M3, or M4 had no reproducible effect on Sp1 binding whereas binding was eliminated by the complete mutation of the GC-box in M1234 (Fig. 5b). We interpret this to mean that each individual mutation leaves at least one site within the triad available for Sp1 binding. In contrast, each individual mutation prevented formation of the large Sp3 complex without affecting the small Sp3 complex (Fig. 5c). Mutation M1234 eliminated Sp3 binding (Fig. 5c). Because Sp3 does not homodimerize (Yu et al. 2003) these findings indicate that the slow migrating large Sp3 complex is composed of Sp3 bound to non-overlapping Sites I and III. Similarly, the fast migrating small complexes formed by Sp1 and Sp3 must result from occupation of any one of the three sites within the GC-box. These results serve to validate the triad model and demonstrate that the GC-box is the only cis-element located between positions −142 and −54 of the proximal promoter that is capable of binding Sp-proteins.
Sp3 activation in SL2 cells is prevented by mutation of all three sites within the GC-box
The functional impact of GC-box mutations on GCH1 transcription was next examined in SL2 cells transfected with the Sp3 expression vector and proximal promoter luciferase constructs containing M1, M2, M3, M4, or M1234 mutations. In these experiments, Sp3 activated transcription from the wild-type promoter by ninefold. Mutations M1, M2, and M4 each reduced by 40–50% the response of the promoter to Sp3, commensurate with their capacity to prevent formation of the large Sp3 complex. However, mutation M3 did not affect the ability of Sp3 to enhance transcription. One might conclude from this that formation of the large Sp3 complex is not required for maximum activation of transcription. An alternate interpretation is that forced occupation of Site I by Sp3 best supports activation (Fig. 5a). While mutation M4 also forces Sp3 to Site I, Site II in this mutation is left open for occupation which would reduce binding to Site I and thereby decrease activation (Fig. 5a). As predicted by EMSA, mutation M1234 decreased transcription to levels observed with the empty pGL3 vector, demonstrating that the GC-box offers the only functional binding sites for Sp3 within the entire 142 bp proximal promoter.
Sp1 and Sp3 act synergistically with C/EBPβ and NF-Y to enhance transcription in SL2 cells
Although Sp1 and Sp3 in PC12 cells are not recruited to the GCH1 promoter in response to cAMP, Sp1, or Sp3 protein–protein interactions might play a role in stabilizing already bound or recently recruited C/EBPβ and NF-Y. Fortunately, SL2 cells are not only null for Sp-proteins but also for CREB, C/EBPβ, and NF-Y (Lee et al. 1997; Dooley et al. 1999). Experiments were therefore performed in SL2 cells to investigate and compare interactions between Sp1, Sp3, NF-Y, and C/EBPβ at the GCH1 promoter. Luciferase assays in SL2 cells co-transfected with the reporter p0.27GCH1-GL3 in combination with plasmid DNA encoding for Sp1 or Sp3, the three NF-Y subunits A, B, and C, or C/EBPβ showed that when expressed alone Sp3 and C/EBPβ each significantly enhanced transcription while Sp1 and NF-Y did not (Fig. 5e). NF-Y and C/EBPβ interacted at the promoter to stimulate transcription to a greater degree than did C/EBPβ acting alone. The promoter response to NF-Y or C/EBPβ was further enhanced when either of these proteins was combined with Sp3 or Sp1. Finally, the combination of Sp3 or Sp1 with NF-Y and C/EBPβ synergistically enhanced transcription. These results indicate that both Sp1 and Sp3 are capable of nucleating an enhanceosome complex at the GCH1 proximal promoter that includes C/EBPβ and NF-Y, although Sp3 is significantly more effective than is Sp1.
Mutations of the GC-box differentially modify Sp3 binding in PC12 cells
Based upon knowledge gained using SL2 cells we began a limited series of experiments using GC-box mutations to characterize Sp-protein binding in PC12 cells. We first questioned whether the large EMSA complexes 1 and 2 in PC12 nuclear extracts represent binding of Sp3 to two sites within the GC-box. EMSA with super-shift was used to determine the effect of the M1 mutation on Sp1 and Sp3 binding patterns. Similar to what was observed using SL2 nuclear extracts, mutation M1 had no obvious affect on formation of the major Sp1 complex 4 (Fig. 6a). M1 also did not modify formation of the major Sp3 complex 3 or the minor Sp3 complex 5. Complexes 3, 4, and 5 therefore presumably represent binding of Sp1 and Sp3 to single sites within the GC-box. However, mutation M1 eliminated large complexes 1 and 2. By analogy with data obtained using SL2 cells, these results support the conclusion that complexes 1 and 2 are produced by binding of Sp3 to two sites within the GC-box. Mutation M1234 abolished binding to the GC-box (including complex 6), demonstrating that all binding in EMSA produced by PC12 nuclear extracts is dependent upon sequences located within the GC-box (Fig. 6a).
Fig. 6.
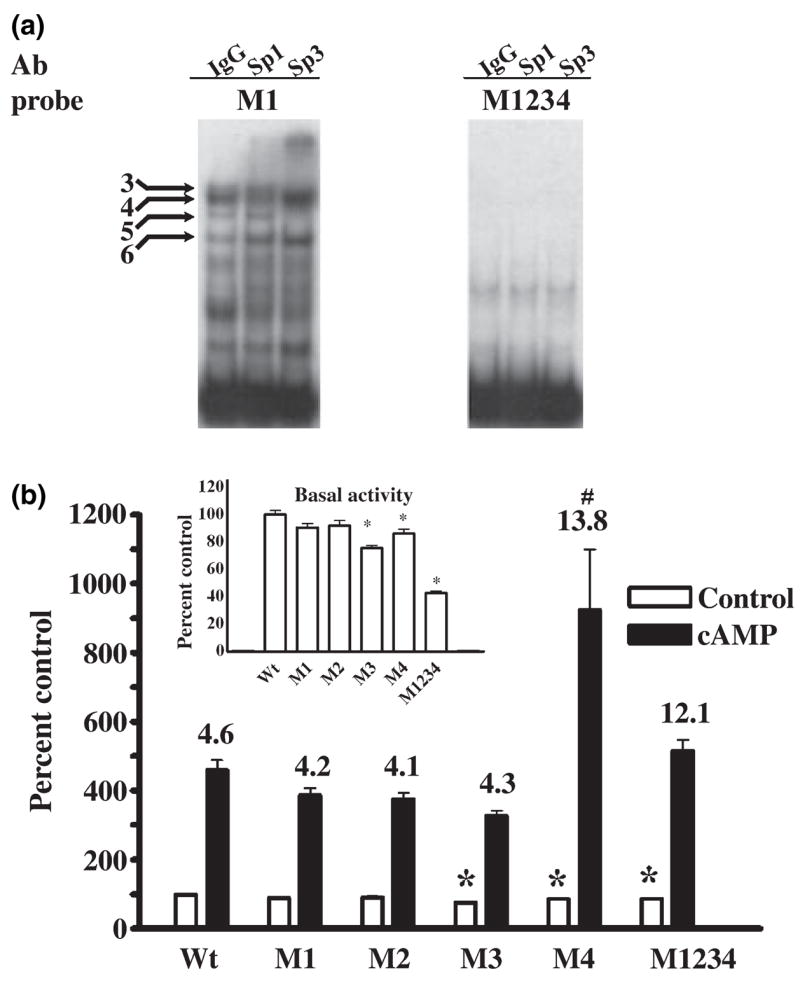
Mutation of the GC-box decreases Sp3 binding yet can enhance cAMP-dependent transcription in PC12 cells. (a) EMSA using 1 μg of PC12 nuclear protein and 1 μg of control IgG, anti-Sp1 or anti-Sp3 shows that mutation M1 of the GC-box does not affect formation of the Sp1- and Sp3-containing complexes 3, 4 and 5 or complex 6 but does eliminate complexes 1 and 2, indicating that these complexes contain Sp3. Mutation M1234 completely eliminates PC12 nuclear protein binding to the GC-box. (b) Luciferase assays of PC12 cells transiently transfected with the wild-type (WT) 400 ng of p0.27GCH1-GL3 or reporter constructs containing the GC-box mutations M1, M2, M3, M4, or M1234, 40 ng of pRL-null and 360 ng of carrier plasmid DNA and then challenged with 5 mmol/L 8Br-cAMP (cAMP) for 4 h. The insert shows control activity plotted on a smaller scale. Data were analyzed by ANOVA with post hoc Bonferroni tests (*p < 0.05 vs. WT control or cAMP).
Mutations of the GC-box differentially modify basal and cAMP-dependent transcription in PC12 cells
The next series of experiments were designed to determine the effect of GC-box mutations on basal and cAMP-dependent transcription in PC12 cells using GCH1 proximal promoter constructs in which the CRE and CCAAT-box cAMP-response elements are left intact. Incubation with 8Br-cAMP for 4 h increased activity from the wild-type promoter by 4- to 5-fold (Fig. 6b). Mutations M1 and M2 which force binding of Sp-proteins to Sites II and III close to the CRE and CCAAT-box cassette had no significant effect on basal or cAMP-dependent transcription. In contrast, mutations M3 and M4 which force binding to Sites I and II furthest away from the cAMP-response elements significantly reduced basal activity (see Fig. 6b insert). Furthermore, while mutation M3 did not alter cAMP-dependent transcription, mutation M4 clearly doubled the absolute response of the promoter to cAMP. Complete mutation of the GC-box in M1234 decreased basal transcription by 60% (see Fig. 6b insert) without affecting the absolute promoter response to cAMP, resulting in a fold stimulation that was three times that of the wild-type promoter.
Sp3 increases basal but not cAMP-dependent transcription in PC12 cells
Experiments in PC12 cells using over-expression of Sp1 and Sp3 or dominant negative forms of Sp1 and Sp3 failed to further define distinct roles for these proteins in GCH1 transcription (data not shown). We therefore chose a strategy in which the 22 bp GC-box in the wild-type GCH1 luciferase promoter construct was replaced with a single copy of the 19 bp yeast Gal4 cis-element and then challenged by co-expression of fusion proteins composed of amino acids 1–147 of the yeast Gal4 DNA binding domain and the N-terminal activation domains of Sp1 (Gal4 Sp1; amino acids 1–603), Sp3 (Gal4-Sp3; amino acids 1–527) or C/EBPβ (Gal4- C/EBPβ; amino acids 1–83) (Fig. 7a). This paradigm eliminates differences in Sp1 and Sp3 binding as well as any preferences for binding sites within the GC-box yet the cAMP response can still be studied because the CRE and CCAAT-box cassette is intact and the assay is performed in PC12 cells.
Fig. 7.
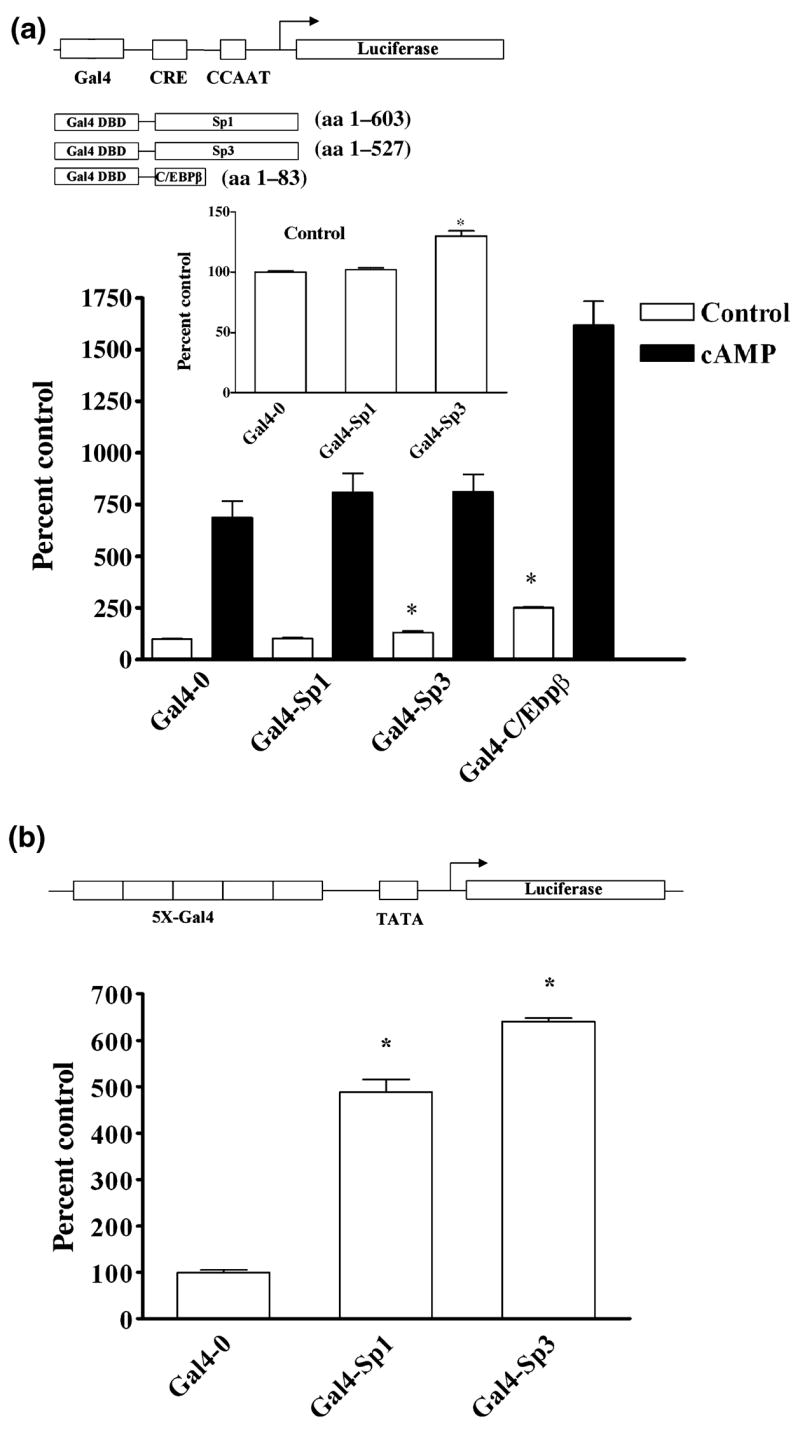
Replacement of the GC-box with a single Gal4 binding element and challenge with Gal4 fusion proteins supports a role for Sp3 in basal transcription. (a) A schematic diagram of the Gal4-0.27GCH1-GL3 luciferase reporter construct in which the 22 bp GC-box has been replaced with a single 19 bp Gal4 cis-element. Fusion proteins of the Gal4 DNA binding domain and the activation domains of Sp1, Sp3, and C/EBPβ are also shown. Luciferase assays of PC12 cells transiently transfected with 420 ng of Gal4-0.27GCH1-GL3, 40 ng of pRL-null, 150 ng Gal4-0, Gal4-Sp1, Gal4-Sp3 or Gal4-C/EBPβ or carrier plasmid DNA and then incubated with 5 mmol/L 8Br-cAMP (cAMP) for 4 h. The insert shows control activity plotted on a smaller scale. Data were analyzed by ANOVA with post hoc Bonferroni tests (*p < 0.05 vs. Gal4-0 control). (b) A schematic diagram of the 5XGal4-heterologous minimal luciferase reporter pFR-luc. Luciferase assays of PC12 cells transiently transfected with 420 ng 5XGal4-heterologous minimal luciferase reporter, 40 ng pRL-null, 150 ng Gal4-0, Gal4-Sp1 or Gal4-Sp3 and carrier plasmid DNA. Data were analyzed by ANOVA with post hoc Bonferroni tests (*p < 0.05 vs. Gal4-0).
Substitution of the GC-box with a single Gal4 DNA binding element decreased basal transcription by greater than 60% (data not shown), a value virtually identical to that produced by complete mutation of the GC-box. This decrease in basal promoter activity was significantly reversed by co-transfection with Gal4-Sp3 but not by Gal4-Sp1 or the Gal4-0 empty vector (Fig 7a and insert). In addition, although Gal4-Sp3 increased basal transcription the absolute response of the promoter to cAMP remained unchanged (Fig. 7a). Experiments with Gal4-C/EBPβ demonstrated that the Gal4-GCH1 reporter construct can respond to introduction of an activation domain with large increases in basal as well as cAMP-dependent transcription (Fig. 7a). The functionality of both Gal4-Sp1 and Gal4-Sp3 was also established by experiments in PC12 cells showing five to sixfold enhancement of transcription from a heterologus luciferase reporter containing five tandem Gal4 sites (Fig. 7b). None of the Gal4-fusion proteins affected transcription from the empty pGL3 vector (data not shown). These results support our contention that the GC-box and Sp3 are important for maintaining basal but not cAMP-dependent transcription in PC12 cells.
The presence of Sp1 and Sp3 at the promoter and the methylation of CpG dinucleotides within the GC-box do not correlate with basal or cAMP-dependent transcription in PC12 and Rat2 cells
A comparison of Sp1 and Sp3 association with the GCH1 proximal promoter GC-box in cell types with divergent basal GCH1 mRNA levels was next undertaken to test the hypothesis that the localization of Sp3 at the proximal promoter correlates with basal GCH1 transcription. With this in mind, resting levels of GCH1 mRNA in rat adrenal chromaffin-derived PC12 cells were determined to be 10-fold higher than in rat fibroblast-derived Rat2 cells (Fig. 8a). Further comparison showed that PC12 cells respond to 4 h treatment with 8Br-cAMP with a sevenfold increase in GCH1 mRNA levels whereas the response of Rat2 cells to this stimulus was insignificant (Fig. 8a). Despite these robust differences in transcription ChIP analysis of the two cell lines demonstrated equivalent amounts of Sp1 and Sp3 at the native proximal promoter (Fig. 8b). Moreover, ChIP showed that while the quantity of Pol II at the Rat2 cell promoter was significantly lower than in PC12 cells, as would be expected based upon the lower transcription rate, Pol II was still three times higher than predicted given the 10-fold difference in basal levels of GCH1 transcription (Fig. 8b).
Fig. 8.
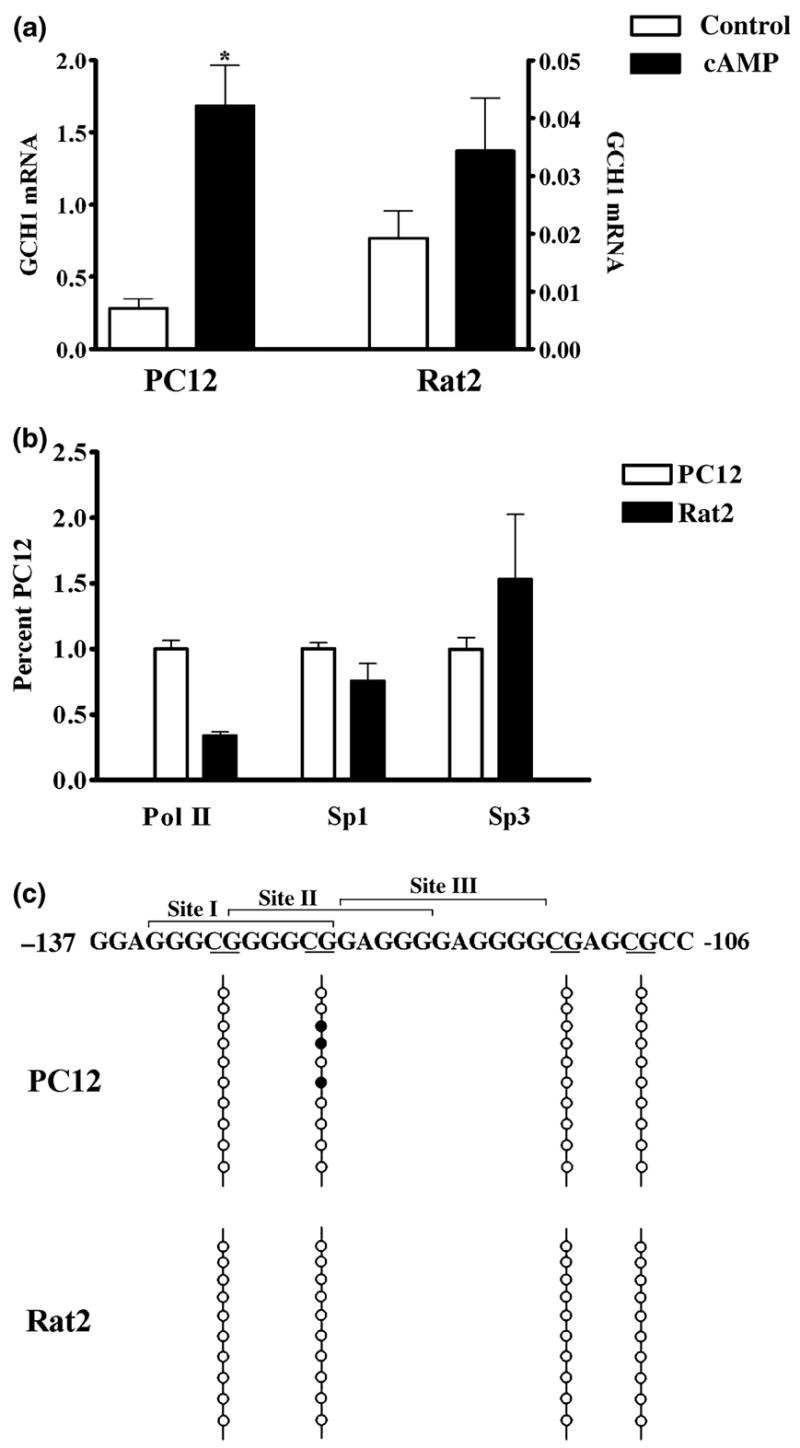
Cells that differ 10-fold in GCH1 gene expression have equal amounts of Sp1 and Sp3 associated with the native promoter and similar patterns of CpG methylation within the GC-box. (a) GCH1 mRNA levels in control cells (control) and cells treated with 5 mmol/L 8Br-cAMP for 4 h (cAMP). PC12 cells (left) and Rat2 cells (right) show 10-fold differences in basal and 47-fold differences in cAMP-stimulated transcription (note the scale change). Data were analyzed by ANOVA with post hoc Bonferroni tests (*p < 0.05 vs. PC12 control). (b) ChIP with quantitative real-time PCR analysis shows that equivalent amounts of Sp1 and Sp3 are associated with the GCH1 proximal promoter in PC12 and Rat2 cells but that significantly less Pol II is associated with the Rat2 promoter. Data are presented as percentages of values obtained for PC12 cells. Data were analyzed by ANOVA with post hoc Bonferroni tests (*p < 0.05 vs. PC12 Pol II). (c) The GCH1 GC-box showing Sites I, II, and III above and CpG dinucleotides underlined below. The four CpG dinucleotides within or adjacent to the GC-box are marked by four vertical lines. Bisulfite-treated GCH1 promoter DNA was amplified by methylation-specific PCR using primers which amplify a 288 bp product encompassing the GC-box, CRE and CCAAT-box. The ten vertical circles represent the ten PCR clones sequenced for each cell line. Filled circles represent CpG dinucleotides in which the cytosine was found to be methylated.
That presence of equivalent amounts of Sp1and Sp3 at the GCH1 proximal promoter in cells with such disparate levels of transcription suggests that these proteins might have multiple functions in GCH1 transcription. Sp-proteins are known to protect genes from repression by blocking methylation of CpG dinucleotides located within and around GC-boxes (Brandeis et al. 1994; Macleod et al. 1994). With this in mind, we performed a bioinformatics analysis of −5000 bp to + 500 bp of the rat GCH1 gene using MethPrimer (Li and Dahiya 2002). This identified a single 238 bp CpG island spanning from −162 to + 77 that encompasses the entire proximal promoter. Located strategically within or immediately adjacent to the GC-box are located four CpG dinucleotides (Fig. 8c). Cloning and sequencing of PC12 and Rat2 GCH1 methylation-specific PCR products revealed that of the four possible methyl acceptor sites only a single CpG within the GC-box is methylated and this occurred in 30% of the PC12 clones and in none of the Rat2 cell clones examined (Fig. 8b).
Discussion
The region −142 through −77 of the rat GCH1 proximal promoter contains the GC-box, the CRE and the CCAAT-box, each originally identified by in vitro footprinting using PC12 cell nuclear extracts or recombinant protein (Kapatos et al. 2000). The boundaries of the GC-box were defined by these experiments as a 22 bp footprint spanning from nucleotides −132 to −111 that was competed away by the sequence GGGGCGGGGGCG, thus implicating Sp1, Sp3, and Sp4 as the likely binding partners. This earlier study also showed using heterologous promoter constructs that the GC-box reduces cAMP-dependent transcription conferred by the CRE and CCAAT-box cassette, suggesting an inhibitory role for Sp-proteins in GCH1 transcription. The present studies clearly demonstrate that the GCH1 GC-box binds Sp1 and Sp3 proteins. Sp1 is generally known to activate transcription while Sp3 is a dual role transcription factor that behaves as an activator or repressor depending on promoter context but in most cases acts as a repressor in the presence of Sp1 (Hagen et al. 1994, 1995; Majello et al. 1994; Yu et al. 2003). Our observation made in SL2 cells that Sp3 activates transcription from the GCH1 promoter while Sp1 acts as a repressor in the presence of Sp3 may therefore appear to be unique but, a similar circumstance has been reported for the mouse growth hormone receptor promoter (Yu et al. 1999). The present studies also show that although Sp1 and Sp3 are not absolutely required for the cAMP response by enhancing basal levels of transcription these proteins decrease the responsiveness of the promoter to cAMP. A number of additional conclusions can be reached from the current studies regarding Sp-proteins, the GC-box and the regulation of GCH1 transcription.
First, incubation with 8Br-cAMP for 4 h did not alter the abundance of Sp1 in PC12 cells. This is in contrast to the decline in Sp1 protein observed following treatment of PC12 cells with phorbol esters (Papanikolaou and Sabban 2000) and the increase in Sp1 protein following nerve growth factor treatment of PC12 cells (Sobue et al. 2005 but see Yan and Ziff 1997). A 95 kDa form of Sp1 is also known to result from phosphorylation of Sp1 (Jackson et al. 1990) and has been observed in some (Billon et al. 1999; Sobue et al. 2005) but not all (Papanikolaou and Sabban 2000; Liu et al. 2001; Persengiev et al. 2001) studies using PC12 cells. While Sp1 is a substrate for protein kinase A (Rohlff et al. 1997) we did not detect the 95 kDa form in nuclear extracts from control or 8Br-cAMP treated PC12 cells prepared in the presence of protein phosphatase inhibitors. The reason for this is unclear but might be related to cell passage number or culture conditions.
Second, in PC12 nuclear extracts we also observed what we interpret to be three isoforms of Sp3, a major protein of 100 kDa and two less abundant proteins of 77 and 73 kDa, the relative amounts of which were not altered by cAMP treatment. It is possible that the 77 and 73 kDa immunore-active proteins represent products of Sp3 proteolysis or non-specific antibody interactions. However, similar studies of Sp3 expression in PC12 cells (Billon et al. 1999) and other cell types (Wang and Bannon 2005; Ishimaru et al. 2006) report three forms of Sp3 of approximately the same molecular weight and relative abundance we show here. The Sp3 proteins we observed are thus likely to be derived from the four alternative Sp3 mRNA ribosomal entry sites, with the 100 kDa band composed of one or both of the two full-length forms and the 77 and 73 kDa bands the two N-terminal truncated forms (Suske 1999; Bouwman and Philipsen 2002; Sapetschnig et al. 2004). Truncated isoforms of Sp3 retain the C-terminal DNA binding domain but are missing the activation domain and thus serve as dominant negative regulators of full-length Sp-proteins (Kennett et al. 1997; Sapetschnig et al. 2004). These small Sp3 isoforms are likely responsible for EMSA complex 5 which comprises 10% of the total GC-box binding activity in PC12 nuclear extracts. By competing with full-length forms of Sp1 and Sp3 for binding to the native GCH1 promoter dominant negative forms of Sp3 are predicted to reduce basal transcription and thereby enhance the overall response to cAMP.
Third, Sp4 is not expressed by PC12 cells. This is not a surprise given that PC12 cells are derived from the adrenal medulla and Sp4 appears to be selectively expressed in the nervous system (Suske 1999; Bouwman and Philipsen 2002). Accordingly, Sp4 is involved in the regulation of neuronal genes, such as neurotrophin-3 (Ishimaru et al. 2006) and the β-subunit of rod cGMP-phosphodiesterase (Lerner et al. 2002) and might play a role in the control of GCH1 transcription within central monoaminergic neurons (Lentz and Kapatos 1996). Our unpublished data indicate that like Sp1, Sp4 binds to a single site within the GCH1 GC-box and is unable to activate transcription but does inhibit Sp3-dependent transcription. Sp4 has properties that are functionally distinct from Sp1 and Sp3 (Hagen et al. 1995), however, and represents another dimension in the regulation of GCH1 transcription that should be explored in the future.
Fourth, Sp1 and Sp3 bind to three sites within the 142 bp proximal promoter and these sites are all located within the GC-box. Sites I (−134 GGGCGGGGCG−125) and III (−124 GAGGGGAGGGG −114) do not overlap while Site II (−130 GGGGCGGAGGG −121) overlaps both Sites I and III. Sites I and III thus have the potential to be occupied simultaneously by Sp-proteins whereas occupation of Sites I and II or Sites II and III would be prohibited by steric hindrance. Repeated EMSA experiments using SL2 nuclear extracts and mutated probes suggest that Sp1 and Sp3 have no obvious preference for binding to any one site within the GC-box. Nonetheless, a common theme that arises from studies of both SL2 and PC12 cells is that forced binding of Sp-proteins to Sites I and II, which are furthest away from the CRE, CCAAT-box and transcription start site, has distinct effects on transcription. In the SL2 nuclear environment the transcriptionally preferred binding site for Sp3 appears to be Site I. In PC12 cells evidence for a preferred Sp-protein binding site comes from the ability of mutation M4 to enhance cAMP-dependent transcription. Inasmuch as this effect of M4 was reversed by complete mutation of the GC-box it would appear that Sp-proteins bound by Sites I or II are somehow involved.
Unlike Sp1, Sp3 binds to the GCH1 GC-box as two protein-DNA complexes, one similar in size to that produced by Sp1 and the other significantly larger. Also unlike Sp1 (Courey et al. 1989) the N-terminal domain of Sp3 does not contain a dimerization domain (Yu et al. 2003). This means that the slowly migrating large Sp3 complexes observed using nuclear extracts from transfected SL2 cells and PC12 cells is not produced by multiple Sp3 proteins tethered to a single cis-element but rather from more than one Sp3 protein simultaneously bound to the GC-box. The slow migrating Sp3 complex must therefore represent occupation of non-overlapping Sites I and III. Because Sp3 shows no obvious preference for binding to any one site it is likely that the first site bound by Sp3 can be either Site I or III. Competition experiments suggest that Sp3 binds to the second site with lower affinity, likely because of steric hindrance produced by Sp3 binding to the first site. Unlike what was observed in SL2 cells, mutations M1 and M2 did not decrease basal transcription in PC12 cells even though mutation M1 was shown by EMSA to eliminate the large Sp3 containing complexes produced by PC12 nuclear extracts. The reason for this disparity is not known but may be related to the fact that PC12 cells express additional activators of GCH1 transcription, such as CREB, C/EBPβ and NF-Y, while SL2 cells do not. The DNA binding domains of Sp1 and Sp3 are essentially identical in size, conformation and DNA recognition (Suske 1999; Bouwman and Philipsen 2002), making the distinction between Sp1 and Sp3 binding at the GCH1 GC-box a topic for further research. Our observations that Sp1 competes with Sp3 to prevent occupation of two sites by Sp3 and that Sp1 and Sp3 cannot occupy the GC-box at the same time strongly suggest, however, that steric hindrance arising from the N-terminal of Sp1 restricts the binding of a second molecule of Sp1 to the promoter.
Fifth, Sp1 and Sp3 are each associated with the native GCH1 proximal promoter but are not recruited or required during cAMP-dependent transcription. Despite the results of these ChIP experiments it is likely that Sp1 and Sp3 proteins are involved in some subtle aspects of the cAMP response. For example, as shown here Sp1 and Sp3 might assist in the recruitment of C/EBPβ and NF-Y to the promoter (Kapatos et al. 2007). As reported for other genes (Krikun et al. 2000; Xu et al. 2000; Wang and Bannon 2005), competition between Sp1 and Sp3 proteins for binding to the GC-box might also determine the basal rate of GCH1 transcription and thereby the relative magnitude of the cAMP response. Our EMSA results using SL2 cells indicate that Sp1 and Sp3 cannot co-occupy the GC-box yet ChIP experiments in PC12 cells show that Sp1 and Sp3 are both associated with the native promoter. One interpretation of this apparent contradiction is that Sp1 and Sp3 are bound to different copies of the native GCH1 promoter. A GCH1 promoter with Sp3 bound has greater basal activity and therefore a smaller fold response to cAMP than does a promoter which has bound Sp1, even when that promoter is occupied by C/EBPβ and NF-Y.
Sixth, association of Sp1 and Sp3 with the native proximal promoter does not predict levels of GCH1 transcription but does correlate with hypo-methylation of four CpG dinucleotides located within or around the GC-box. Cytosine methylation within consensus Sp-binding sites does not affect binding of Sp1 and Sp3 (Harrington et al. 1988; Zhu et al. 2003) whereas methylation of CpG immediately adjacent to a consensus site can reduce Sp1 and Sp3 binding (Zhu et al. 2003). The single cytosine found methylated in 30% of the PC12 cell GCH1 promoters is located immediately adjacent to Site III and in the native promoter would be predicted to drive Sp-protein binding to Sites I or II. Mutation M4 also forces Sp-protein binding to Sites I or II and increases in the response of the promoter to cAMP. The select methylation of a single CpG dinucleotide located with the GCH1 proximal promoter of PC12 cells might therefore be involved in the cell type-specific response to cAMP.
In cells that express very low levels of GCH1 transcription the native proximal promoter is characterized by levels of bound Pol II and acetylated nucleosomes that are normally associated with active chromatin (Kapatos et al. 2007). As shown here, cells such as these typically do not respond to cAMP but they do respond to inflammatory cytokines with a rapid induction of GCH1 transcription that is part of the program required for the BH4-dependent production of nitric oxide (NO) by the inducible form of nitric oxide synthase (Kwon et al. 1989; Werner et al. 1990; D’Sa et al. 1996). In order to retain this cytokine response these cells must maintain the GCH1 promoter in a paused but transcriptionally competent state. Methylation of cytosines in CpG dinucleotides is widely accepted as a gene-silencing signal (Herman and Baylin 2003; Jaenisch and Bird 2003; Klose and Bird, 2003) and the binding of Sp-proteins is known to interfere with promoter DNA methylation both within and surrounding Sp-binding sites (Brandeis et al. 1994; Macleod et al. 1994; Clark et al. 1997; Siegfried et al. 1999; Chan et al. 2004; Pang et al. 2004). We hypothesize that in cell types that express low levels of GCH1 the association of Sp1 and Sp3 proteins with the GCH1 proximal promoter serves to protect promoter bound nucleosomes from repressive histone marks and the subsequent recruitment of DNA methyltransferases. In this way, even naïve cells would be able to respond to cytokine stimulation with an increase in GCH1 gene expression and BH4 biosynthesis.
Acknowledgments
This study was supported by NINDS grant NS26081.
Abbreviations
- ChIP
chromatin immunoprecipitation
- CRE
cAMP response element
- DTT
dithiothreitol
- C/EBP
CCAAT/Enhancer-Binding Protein
- EMSA
electrophoretic mobility shift assay
- GCH1
GTP cyclohydrolase I
- PMSF
phenyl methyl sulfonyl fluoride
- NF-Y
nuclear factor-Y
References
- Abou-Donia MM, Wilson SP, Zimmerman TP, Nichol CA, Viveros OH. Regulation of guanosine triphosphate cyclohydrolase and tetrahydrobiopterin levels and the role of the cofactor in tyrosine hydroxylation in primary cultures of adrenomedullary chromaffin cells. J Neurochem. 1986;46:1190–1199. doi: 10.1111/j.1471-4159.1986.tb00637.x. [DOI] [PubMed] [Google Scholar]
- Ahlgren R, Suske G, Waterman MR, Lund J. Role of Sp1 in cAMP-dependent transcriptional regulation of the bovine CYP11A gene. J Biol Chem. 1999;274:19422–19428. doi: 10.1074/jbc.274.27.19422. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Bezin L, Gordon LJ, Imerman B, Blitz J, Kuhn DM, Levine RA. Vasoactive intestinal peptide induces both tyrosine hydroxylase activity and tetrahydrobiopterin biosynthesis in PC12 cells. Neuroscience. 1998;86:179–189. doi: 10.1016/s0306-4522(97)00611-8. [DOI] [PubMed] [Google Scholar]
- Bauer M, Suppman S, Meyer M, Hesslinger C, Gasser T, Widner HR, Ueffing M. Glial cell line-derived neurotrophic factor up-regulates GTP-cyclohydrolase I activity and tetrahydrobiopterin levels in primary dopaminergic neurons. J Neurochem. 2002;82:1300–1310. doi: 10.1046/j.1471-4159.2002.01074.x. [DOI] [PubMed] [Google Scholar]
- Billon N, Carlisi D, Datto MB, van Grunsven L, Watt A, Wang X-F, Rudkin BB. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene. 1999;18:2872–2882. doi: 10.1038/sj.onc.1202712. [DOI] [PubMed] [Google Scholar]
- Borestrom C, Zetterberg H, Liff K, Rymo L. Functional interaction of nuclear factor y and Sp1 is required for activation of the Epstein-Barr virus C promoter. J Virol. 2003;77:821–829. doi: 10.1128/JVI.77.2.821-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Chan Y, Fish JE, D’Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by (m)cp(m) CpG methylation. Gene. 1999;197:67–71. doi: 10.1016/s0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Courey AJ, Holtzman DA, Jackson SP, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- D’Sa C, Hirayama K, West A, Hahn M, Zhu M, Kapatos G. Tetrahydrobiopterin biosynthesis in C6 glioma cells: induction of GTP cyclohydrolase I gene expression by lipopolysaccharide and cytokine treatment. Brain Res Mol Brain Res. 1996;41:105–110. doi: 10.1016/0169-328x(96)00073-3. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley KA, Millinder S, Osbourne TF. Sterol regulation of 3-hydroxy-3-methlglutaryl-coenzyme A synthase Gene through a direct interaction between sterol regulatory element binding protein and trimeric CCAAT-binding factor NF-Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- Ge Y, Matherly LH, Taub JW. Transcriptional regulation of cell-specific expression of the human cystathionine β-synthase gene by differential binding of Sp1/Sp3 to the -1b promoter. J Biol Chem. 2001;276:43570–43579. doi: 10.1074/jbc.M104930200. [DOI] [PubMed] [Google Scholar]
- Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTA-FII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- Harrington MA, Jones PA, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci USA. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hirayama K, Shimoji M, Swick L, Meyer A, Kapatos G. Characterization of GTP cyclohydrolase I gene expression in the human neuroblastoma SKN-BE(2)M17: enhanced transcription in response to cAMP is conferred by the proximal promoter. J Neurochem. 2001;79:576–587. doi: 10.1046/j.1471-4159.2001.00583.x. [DOI] [PubMed] [Google Scholar]
- Hoey T, Weinzlerl ROJ, Gill G, Chen JL, Dynlacht BD, Tiyan R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Ishimaru N, Tabuchi A, Hara D, Hayashi H, Sugimoto T, Yasuhara M, Shiota J, Tsuda M. Regulation of neurotrophin-3 gene transcription by Sp3 and Sp4 in neurons. J Neurochem. 2006;97:1–13. doi: 10.1111/j.1471-4159.2006.04216.x. [DOI] [PubMed] [Google Scholar]
- Jackson SP, MacDonald JJ, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kapatos G, Vunnava P, Wu Y. Protein Kinase A-dependent recruitment of RNA Polymerase II, NF-Y and C/EBPβ to the GTP cyclohydrolase I Proximal promoter without alterations in histone acetylation. J Neurochem. 2007;101:1119–1133. doi: 10.1111/j.1471-4159.2007.04486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatos G, Stegenga SL, Hirayama K. Identification and characterization of basal and cyclic AMP response elements in the promoter of the rat GTP cyclohydrolase I gene. J Biol Chem. 2000;275:5947–5957. doi: 10.1074/jbc.275.8.5947. [DOI] [PubMed] [Google Scholar]
- Kennett SB, Udvadia AJ, Horowitz JM. Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 1997;25:3110–3117. doi: 10.1093/nar/25.15.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R, Bird A. MeCP2 repression goes non-global. Science. 2003;302:793–795. doi: 10.1126/science.1091762. [DOI] [PubMed] [Google Scholar]
- Krikun G, Schatz F, Mackman N, Guller S, Demopoulos R, Lockwood CJ. Regulation of tissue factor gene expression in human endometrium by transcription factors Sp1 and Sp3. Mol Endocrinol. 2000;14:393–400. doi: 10.1210/mend.14.3.0430. [DOI] [PubMed] [Google Scholar]
- Kwon NS, Nathan CF, Stuehr DJ. Reduced biopterin as a cofactor in the generation of nitrogen oxides by murine macrophages. J Biol Chem. 1989;264:20496–20501. [PubMed] [Google Scholar]
- Lee YH, Williams SC, Baer M, Sterneck E, Gonzalez FJ, Johnson PF. The ability of C/EBP beta but not C/EBP alpha to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz SI, Kapatos G. Tetrahydrobiopterin biosynthesis in the rat brain: heterogeneity of GTP cyclohydrolase I mRNA expression in monoamine-containing neurons. Neurochem Int. 1996;28:569–582. doi: 10.1016/0197-0186(95)00124-7. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Gribanova YE, Whitaker L, Knox BE, Farber DB. The rod cGMP-phosphodiesterase beta-subunit promoter is a specific target for Sp4 and is not activated by other Sp proteins or CRX. J Biol Chem. 2002;277:25877–25883. doi: 10.1074/jbc.M201407200. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. Methprimer: designing primers for methylation PCRS. Bioinfomatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Liu A, Prenger MS, Norton DD, Mei L, Kusiak JW, Bai G. Nerve growth factor uses Ras/ERK and phosphatidylinositol 3-kinase cascades to up-regulate the N-methyl-d-aspartate receptor 1 promoter. J Biol Chem. 2001;276:45372–45379. doi: 10.1074/jbc.M105399200. [DOI] [PubMed] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- Majello B, De Luca P, Hagen G, Suske G, Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22:4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang RT, Lee LT, Ng SS, Yung WH, Chow BK. CpG methylation and transcription factors Sp1 and Sp3 regulate the expression of the human secretin receptor gene. Mol Endocrinol. 2004;18:471–483. doi: 10.1210/me.2003-0245. [DOI] [PubMed] [Google Scholar]
- Papanikolaou NA, Sabban EL. Ability of Egr1 to activate tyrosine hydroxylase transcription in PC12 cells. J Biol Chem. 2000;275:26683–26689. doi: 10.1074/jbc.M000049200. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Li J, Poulin ML, Kilpatrick DL. E2F2 converts reversibly differentiated PC12 cells to an irreversible, neurotrophin-dependent state. Oncogene. 2001;20:5124–5131. doi: 10.1038/sj.onc.1204663. [DOI] [PubMed] [Google Scholar]
- Pluss C, Werner ER, Blau N, Wachter H, Pfeilschifter J. Interleukin 1β and cAMP trigger the expression of GTP cyclohydrolase I in rat mesangial cell. Biochem J. 1996;318:665–671. doi: 10.1042/bj3180665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H, Aarnisalo P, Santti H, Janne OA, Palvimo JJ. Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor mediated transcription by different mechanisms. J Biol Chem. 2000;275:571–579. doi: 10.1074/jbc.275.1.571. [DOI] [PubMed] [Google Scholar]
- Roder K, Wolf SS, Larkin KJ, Schweizer M. Interaction between the two ubiquitously expressed transcription factors NF-Y and Sp1. Gene. 1999;234:61–69. doi: 10.1016/s0378-1119(99)00180-8. [DOI] [PubMed] [Google Scholar]
- Rohlff C, Ahmad S, Borellini F, Lei J, Glazer RI. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J Biol Chem. 1997;272:21137–21141. doi: 10.1074/jbc.272.34.21137. [DOI] [PubMed] [Google Scholar]
- Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- Saluja D, Vassallo MF, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapetschnig A, Koch F, Rischitor G, Mennenga T, Suske G. Complexity of translationally controlled transcription factor Sp3 isoform expression. J Biol Chem. 2004;279:42095–42105. doi: 10.1074/jbc.M404989200. [DOI] [PubMed] [Google Scholar]
- Sarraj JA, Vinson C, Han J, Thiel G. Regulation of GTP cyclohydrolase I gene transcription by basic region leucine zipper transcription factors. J Cell Biochem. 2005;96:1003–1020. doi: 10.1002/jcb.20580. [DOI] [PubMed] [Google Scholar]
- Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- Smale ST, Schmidt MC, Berk AJ, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Aci. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobue S, Hagiwara K, Banno Y, Tamiya-Koizumi K, Suzuki M, Kojima T, Asano H, Nozawa Y, Murate T. Transcription factor specificity protein 1 (Sp1) is the main regulator of nerve growth factor-induced sphingosine kinase 1 gene expression of the rat pheochromocytoma cell line, PC12. J Neurochem. 2005;95:940–949. doi: 10.1111/j.1471-4159.2005.03399.x. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Thony B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J. 2000;347(Pt 1):1–16. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bannon MJ. Sp1 and Sp3 activate transcription of the human dopamine transporter gene. J Neurochem. 2005;93:474–482. doi: 10.1111/j.1471-4159.2005.03051.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Werner ER, Werner-Felmayer G, Fuchs D, Hausen A, Reibnegger G, Yim JJ, Pfleiderer W, Wachter H. Tetrahydrobiopterin biosynthetic activities in human macrophages, fiBroblasts, THP-1, and T 24 cells. GTP-cyclohydrolase I is stimulated by interferon-gamma, and 6-pyruvoyl tetrahydropterin synthase and sepiapterin reductase are constitutively present. J Biol Chem. 1990;265:3189–3192. [PubMed] [Google Scholar]
- West AB, Kapatos G, O’Farrell C, Gonzalez-de-Chavez F, Chiu K, Farrer MJ, Maidment NT. N-myc regulates parkin expression. J Biol Chem. 2004;279:28896–28902. doi: 10.1074/jbc.M400126200. [DOI] [PubMed] [Google Scholar]
- Wu SM, Kuo WC, Hwu WL, Hwa KY, Mantovani R, Lee YM. RNF4 is a coactivator for nuclear factor Y on GTP cyclohydrolase I proximal promoter. Mol Pharmacol. 2004;66:1317–1324. doi: 10.1124/mol.66.5.. [DOI] [PubMed] [Google Scholar]
- Xu Q, Ji YS, Schmedtje JF., Jr Sp1 increases expression of cyclooxygenase-2 in hypoxic vascular endothelium. Implications for the mechanisms of aortic aneurysm and heart failure. J Biol Chem. 2000;275:24583–24589. doi: 10.1074/jbc.M003894200. [DOI] [PubMed] [Google Scholar]
- Yan G-Z, Ziff EB. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J Neurosci. 1997;17:6122–6132. doi: 10.1523/JNEUROSCI.17-16-06122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Schwartzbauer G, Kazlman A, Menon RK. Role of the Sp family of transcription factors in the ontogeny of growth hormone receptor gene expression. J Biol Chem. 1999;274:34327–34336. doi: 10.1074/jbc.274.48.34327. [DOI] [PubMed] [Google Scholar]
- Yu B, Datta PK, Bagchi S. Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 2003;31:5368–5376. doi: 10.1093/nar/gkg706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Hirayama K, Kapatos G. Regulation of tetrahydrobiopterin biosynthesis in cultured dopamine neurons by depolarization and cAMP. J Biol Chem. 1994;269:11825–11829. [PubMed] [Google Scholar]
- Zhu W-G, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, Otterson GA. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21Cip1 promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]