Abstract
Human N-acetyltransferase 1 (NAT1) alleles are characterized by one or more single nucleotide polymorphisms (SNPs) associated with rapid and slow acetylation phenotypes. NAT1 both activates and deactivates arylamine drugs and carcinogens, and NAT1 polymorphisms are associated with increased frequencies of many cancers and birth defects. The recently resolved human NAT1 crystal structure was used to evaluate SNPs resulting in the protein substitutions R64W, V149I, R187Q, M205V, S214A, D251V, E261K, and I263V. The analysis enhances knowledge of NAT1 structure-function relationships, important for understanding associations of NAT1 SNPs with genetic predisposition to cancer, birth defects, and other diseases.
Keywords: NAT1, N-acetyltransferase 1; single nucleotide polymorphisms (SNPs); structure-function relationships; cancer susceptibility; birth defects
INTRODUCTION
The human genome contains two functional arylamine N-acetyltransferase genes, which code for active N-acetyltransferase 1 (NAT1) and 2 (NAT2) enzymes (EC 2.3.1.5). Although NAT1 was originally thought to be monomorphic, the human population contains over 25 NAT1 alleles or haplotypes possessing single nucleotide polymorphisms (SNPs) (Hein, 2002; Boukouvala and Fakis, 2005).
Like all mammalian arylamine N-acetyltransferase proteins, NAT1 contains a functional Cys-His-Asp catalytic triad, which resembles that of cysteine proteases (Sinclair et al., 2000). In a ping-pong bi-bi reaction mechanism, the catalytic Cys68 is acetylated by acetyl-coenzyme A (AcCoA) (Dupret and Grant, 1992), followed by acetyl group transfer to the substrate. NAT1 substrate selectivity is strongly influenced by the three key active site loop residues F125, Y127, and R129 (Goodfellow et al., 2000; Liu et al., 2007; Minchin et al., 2007; Wu et al., 2007).
Molecular epidemiological studies have reported associations between NAT1 genetic polymorphisms and individual risk to urinary bladder (Taylor et al., 1998; Katoh et al., 1999; Gago-Dominguez et al., 2003; Sanderson et al., 2007), colorectal (Bell et al., 1995; Chen et al., 1998; Ishibe et al., 2002; Lilla et al., 2006), breast (Millikan et al., 1998; Zheng et al., 1999; Lee et al., 2003; Ambrosone et al., 2007), lung (Wikman et al., 2001; Gemignan et al., 2007), prostate (Hein, 2002; Rovito, Jr. et al., 2005), and pancreatic cancers (Li et al., 2006; Jiao et al., 2007), and non-Hodgkin lymphoma (Morton et al., 2006; 2007). NAT1 is highly expressed in breast cancer (Sorlie et al., 2001), conveys enhanced growth and resistance to etoposide (Adam et al., 2003), and improved clinical outcome in estrogen receptor alpha positive breast cancer (Bieche et al., 2004). In addition, a reported association of NAT1 polymorphisms with birth defects (Lammer et al., 2004a;b; Jensen et al., 2005; Carmichael et al., 2006) may be related to an important role for NAT1 in folate metabolism (Wakefield et al., 2007).
Previous studies have generated molecular homology models to better understand structural characteristics, substrate specificity, and the effect of polymorphisms in NAT1 (Rodrigues-Lima and Dupret, 2001; Kawamura et al., 2005; Lau et al., 2006; Liu et al., 2006). These models were generated based on homology to N-acetyltransferase enzyme crystal structures from Salmonella typhimurium (Sinclair et al., 2000) or Mycobacterium smegmatis (Sandy et al., 2002). However, the alignment scores between bacterial and mammalian N-acetyltransferase protein sequences are low (~30%), and important differences exist between the bacterial and mammalian N-acetyltransferase protein structures (Walraven et al., 2007).
High resolution crystal structures of human NAT1 and NAT2 were recently reported (Wu et al., 2007). We obtained NAT1 crystal structure coordinates from the Protein Data Bank (PDB) under code 2PQT. Prior to analysis, the residue name HIS was changed to HIE in the PDB file to define histidine residues as neutral species that are protonated in the epsilon position. Hydrogen atoms were added to the structure using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (Pettersen et al., 2004). Figures were generated using a ribbon color scheme that distinguishes the previously defined N-acetyltransferase enzyme domains (Payton et al., 2001). Solvent accessible surface areas (SASA) were measured for residue side-chains using Naccess V2.1.1 and reported as percent of total side-chain surface area (Hubbard and Thornton, 1993). Hydrogen bonding interactions and donor-acceptor hydrogen bonding distances were calculated using HBPLUS and are reported in angstroms (Å) (McDonald and Thornton, 1994). The location and molecular interactions of NAT1 residues R64, V149, R187, M205, S214, D251, E261, and I263 were evaluated. Molecular images depicting the nature and location of these interactions were generated using Chimera.
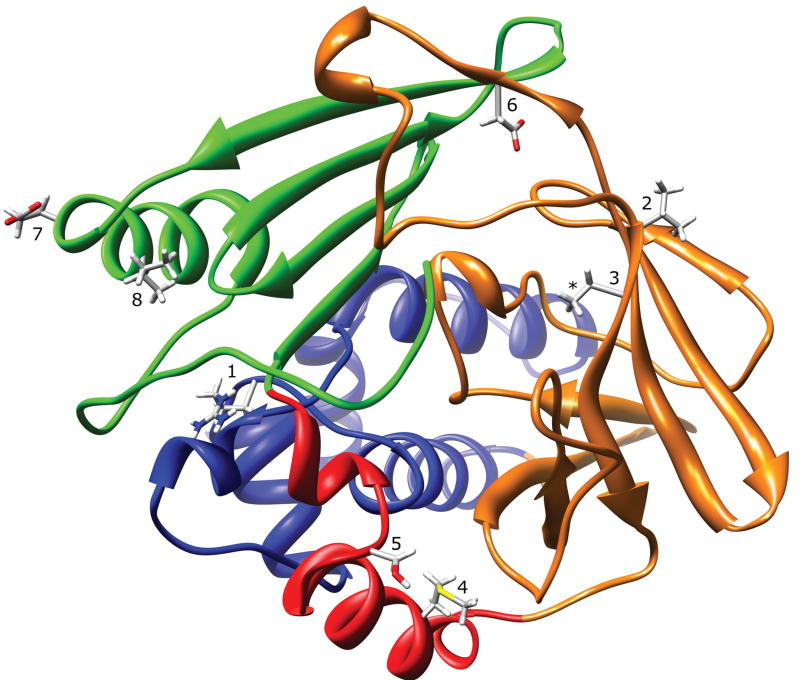
Two major structural features distinguish mammalian N-acetyltransferase structure from the bacterial N-acetyltransferases. First, the domain II loop (165–185) in mammalian N-acetyltransferases is associated with the domain III beta sheet, apparently providing structural stability and limiting active site access (Fig. 1). Second, the mammalian N-acetyltransferase C-terminal tail is not alpha helical, as is the case for the bacterial N-acetyltransferases, but instead is a coil that reaches around and associates with the active site pocket, thereby playing a key role in defining the size and shape of the active site pocket (Fig. 1). Due to the lack of appropriate structural template information for these regions, and the limitations of current homology modeling techniques, these structural features were impossible to model correctly using the bacterial N-acetyltransferase structures as templates. Homology modeling generates protein structure for sequence that is aligned with a structure template, but cannot predict secondary or tertiary structure in the absence of a template. The structural and functional effects of mammalian NAT1 SNP-induced residue changes are summarized in Table 1 and described below.
Figure 1.
NAT1 Crystal Structure (2PQT) Ribbon Diagram. The ribbon is colored to indicate N-acetyltransferase protein domain I (blue), the interdomain region (red), domain II (orange), and domain III (green). The location of residues R64 (1), V149 (2), R187 (3), M205 (4), S214 (5), D251 (6), E261 (7), and I263 (8) are shown. The 2PQT PDB file is missing coordinates for the arginine side-chain guanidine group at residue R187 (*).
Table 1.
Structural effects of single nucleotide polymorphisms in NAT1
| NAT1 SNP | NAT1 Allele | Domain | Secondary Structure | Side-Chain Location | Type of Change | Functional Effect | Structural Effect(s) |
|---|---|---|---|---|---|---|---|
| R64W | NAT1*17 | I | Coil | 13.4% Surface Exposed | ↓Flexibility
↓H-bond Capacity ↑Hydrophobicity ↑Aromaticity |
↓Stability | Loss of Electrostatic Interactions |
| V149I |
NAT1*11A
NAT1*11B |
II | β-Barrel | 39.0% Surface Exposed | Conservative | ↑Activity/Protein Possible | None Expected |
| S214A |
NAT1*11A
NAT1*11B NAT1*11C |
Interdomain | Coil | Active Site Opening | H-bond Capacity Lost
Polarity Lost ↑Hydrophobicity |
↑Activity/Protein Possible | Altered Substrate/AcCoA Binding |
| R187Q |
NAT1*14A
NAT1*14B |
II | β-Barrel | Active Site Pocket | ↑Hydrophobicity
↓H-bond Capacity (+) Charge Lost ↓Size |
↓Stability | Altered Active Site Size/Shape |
| M205V | NAT1*21 | Interdomain | α-Helix | 19.2% Surface Exposed | ↓Flexibility
↓Size |
None Detected | None Expected |
| D251V | NAT1*22 | III | β-Sheet | Core | ↓Flexibility
H-bond Capacity Lost ↑Hydrophobicity Polarity Lost (−) Charge Lost |
↓Stability | Loss of Electrostatic Interactions |
| E261K | NAT1*24 | III | α-Helix | Surface | ↓H-bond Capacity
Different Charge ↓Hydrophobicity |
None Detected | None Expected |
| I263V | NAT1*25 | III | α-Helix | Hydrophobic Core | Conservative | None Detected | None Expected |
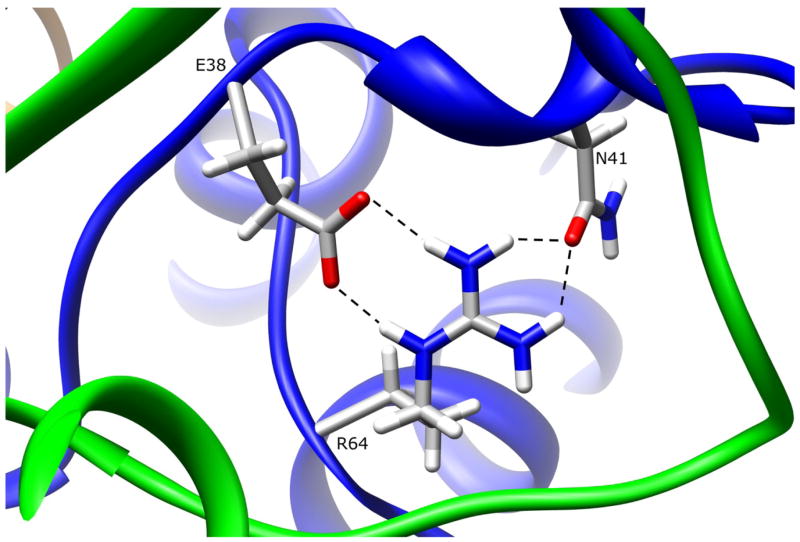
R64W (190C>T)
The side-chain of residue R64 is 13.4% surface exposed in domain I, and hydrogen bonds to E38 (R64:NE-E38:OE1 3.02Å, R64:NH2-E38:OE2 2.89 Å) and N41 (R64:NH2-N41:OD1 2.86 Å, R64:NH1-N41:OD1 3.00 Å), both of which are in domain III (Fig. 2). The hydrogen bonding interactions between positively charged R64 and negatively charged E38 are salt bridges, and are stronger than normal hydrogen bonds. Replacing arginine with either glutamine or tryptophan at residue 64 will result in loss of these interactions, which contribute to the conformation and dynamics of the first domain tertiary structure. Since residues R64 and E38 are completely conserved in all known mammalian N-acetyltransferases these interactions could be important for the function and/or stability of the enzyme. Residue N41 is also highly conserved among N-acetyltransferases. Structural changes in this region could affect the positioning of the catalytic triad C68, and thereby alter its interactions with catalytic H107, and/or affect its acetylation status leading to increased proteasomal degradation (Butcher et al., 2004). However, many other interactions are involved in holding the C68 in place, making it improbable that the R64W substitution results in significant displacement of the catalytic C68. Some hydrogen bonding interactions may be preserved with the W64, but tryptophan is largely hydrophobic and does not have the hydrogen bonding capacity of arginine. The native interactions formed by R64 are likely important to the integrity of the tertiary structure in that region. Functional studies of the R64W variant in yeast demonstrated reduced activity, reduced protein levels, and reduced thermostability (Fretland et al., 2001; 2002). A loss of critical hydrogen bonding interactions in the R64W variant, as observed in the NAT1 crystal structure, is consistent with reduced protein thermostability. Another study suggested that variant NAT1 proteins are more susceptible to proteasomal degradation due to altered acetyl-CoA binding capability, and also reported the reduced thermostability of the R64W variant (Butcher et al., 2004). Based on NAT2 crystal structure 2PFR which has AcCoA bound to the active site, AcCoA appears to be dependent on interactions with Q73, T103, T208, T214, S216, and S287, none of which are influenced by the R64W substitution. It is unclear how the R64W substitution would alter AcCoA binding. Additionally, the reduced AcCoA binding capacity reported for the R64W variant (Butcher et al., 2004) could be explained by a loss of enzyme integrity due to low thermostability. A study of purported NAT1 ortholog Syrian hamster NAT2 reported that the R64W substitution does not alter protein stability, but instead leads to increased protein aggregation and ubiquitylation (Liu et al., 2006). This study also determined that the R64W variant did not have acetylation deficiencies. Using a homology model of NAT1, the authors explained their functional observations in terms of steric clashes and de-shielding of the variant W64 residue. However, the clash with R277 suggested by their model does not exist in the crystal structure since the R277 side chain is directed away from R64. The seemingly contradictory results for protein stability found by the Syrian hamster NAT2 study could be attributed to differences between the NAT1 and hamster NAT2 enzymes, or the differences in how the enzyme preparations were treated and measured since this study used a different method to determine structural stability. A loss of critical hydrogen bonding interactions in the R64W variant, as observed in the NAT1 crystal structure, is consistent with reduced protein thermostability. Under certain conditions this loss of structural integrity could be manifest as protein aggregation or could become the target of protein degradation machinery.
Figure 2.
NAT1 SNP Residue Arginine 64. Residue R64 is partially surface exposed in domain I. The arginine side-chain makes multiple hydrogen bonds to E38 and N41, both of which are also in domain I. These interactions are lost when R64 is mutated to tryptophan.
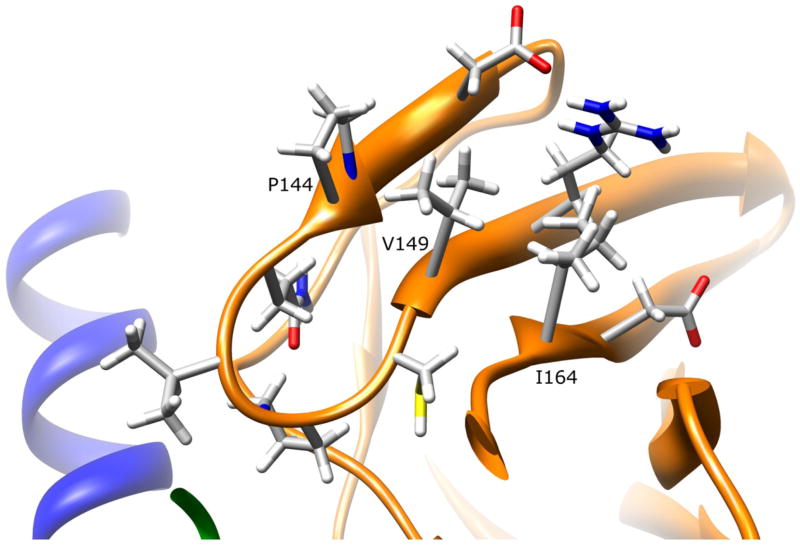
V149I (445G>A) and S214A (640G>T)
The side-chain of V149 is 39.0% solvent exposed on the surface of the domain II beta-barrel (Fig. 3). Hydrophobic interactions are possible with P144 and I164, but because these residues are on adjacent beta strands, they do not interact closely with V149, and do not contribute to secondary structure stability. Since conservative substitution of isoleucine for valine at 149 would not alter those interactions, structural changes are not expected for this variant.
Figure 3.
NAT1 SNP Residue Valine 149. Residue V149 is partially surface exposed on the domain II beta barrel. The valine side-chain has no apparent interactions with other residues. Mutating this residue to isoleucine is not expected to influence enzyme stability or function.
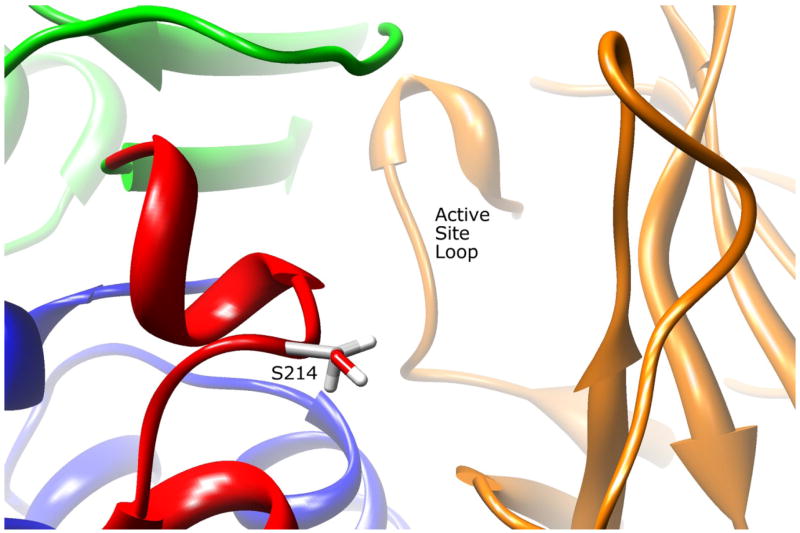
The side-chain of residue S214 is 72.2% solvent exposed in the inter-domain region near the active site pocket opening (Fig. 4). The S214 side-chain makes no apparent molecular interactions with the surrounding residues. Replacing serine with a smaller alanine residue would have no apparent consequences for active site access, and would not affect the structure since the side-chain is near the surface and does not interact with other residues. Analysis of the NAT2 structure complexed with CoA (PDB ID# 2PFR) demonstrates that residue threonine 214 is involved in hydrogen bonding to CoA. Since the NAT1 S214 may also interact with CoA, substituting alanine for serine could affect this interaction.
Figure 4.
NAT2 SNP Residue Serine 214. Residue S214 is located adjacent to the active site pocket opening in the interdomain region. The serine side-chain has no apparent interactions with other residues. Mutating this residue to an alanine could influence AcCoA binding efficiency.
Residue changes V149I and S214A exist together as a result of SNPs in the NAT1*11 allele. The V149I variation is not known to exist alone, while the S214A substitution is found alone in NAT1 enzyme encoded by the NAT1*11C allele. Based on this analysis of NAT1 structure, neither the V149I nor the S214A residue changes are expected to alter dynamics or structural stability of NAT1.
Functional analyses of these two SNPs found in the NAT1*11 allele are highly inconsistent. Recombinant studies in yeast demonstrated no change in catalytic activity, protein levels, or protein stability for a NAT1 haplotype possessing both V149I and S214A (Fretland et al., 2001; 2002). When recombinantly expressed in bacterial cells, the V149I variant alone resulted in acetylation rates of up to 2-fold higher, but with no changes in protein level or thermostability (Doll et al., 1997). When evaluated in COS-1 cells, the NAT1 haplotype possessing both V149I and S214A resulted in elevated protein levels and catalytic activity in one study (Zhu and Hein, 2007), whereas another found no difference in NAT1 protein levels or catalytic activity (de Leon et al., 2000). Studies of red blood cells and leukocytes from individuals possessing the NAT1*11 allele resulted in equivalent and lower catalytic activities, respectively, relative to individuals without the allele (Bruhn et al., 1999; Zhangwei et al., 2006).
Interpreting these functional results with observations from the NAT1 crystal structure is problematic since most of the studies evaluated the V149I and S214A substitutions together. Since V149I is a highly conserved residue change at the surface of the protein where no important molecular interactions are apparent, increased catalytic activity for mutants containing only the V149I SNP without increase in protein levels cannot be rationalized through observations of the NAT1 crystal structure. Since S214 is located adjacent to the active site and may be involved in AcCoA binding, S214A alone could play a role in the increased activity of the NAT1 protein via a mechanism that increases the steady state level of acetylated NAT1 enzyme. However, current studies do not report increased activity for S214A alone, and additional functional studies to investigate this hypothesis are needed.
R187Q (560G>A)
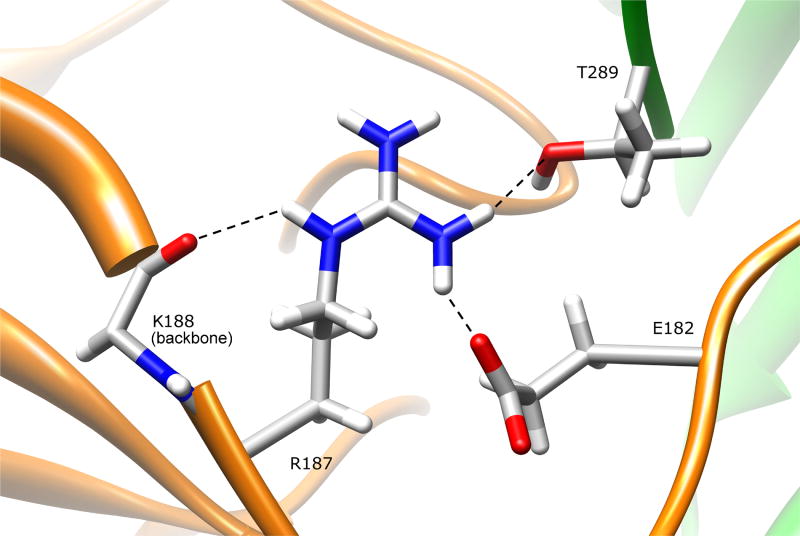
Analysis of residue R187 was not possible using the NAT1 crystal structure with PDB code 2PQT due to missing structural information for this residue. Therefore, we utilized the NAT1 F125S mutant crystal structure (PDB ID# 2IJA) to evaluate the R187Q variant. The backbone root mean square deviation (RMSD) between NAT1 structures 2IJA (chain A) and 2PQT is 0.54, indicating that these structures are very similar. The side-chain of residue R187 is 7.2% surface exposed in the domain II beta barrel, and is partially surface exposed both to the protein surface and the active site pocket (Fig. 5). The arginine side-chain hydrogen bonds to the side chain of E182 (R187:NH1-E182:OE1 2.71 Å) and backbone of K188 (R187:NE-K188:O 3.01 Å) in domain II, and to the side-chain of C-terminal residue T289 (R187:NH1-T289:OG1 3.00 Å) in domain III. These interactions help shape the active site pocket, and stabilize the conformations of both the C-terminal tail and the domain II loop. Changing this residue to glutamine will result in loss of these interactions, although the smaller glutamine residue may be capable of maintaining some of these hydrogen bonding interactions. Loss of the R187 interactions could lead to a change in the dynamics or conformations of the C-terminal tail and/or the domain II loop (165–185), resulting in destabilization of NAT1 structure. Changes in C-terminal tail conformation could also influence AcCoA binding and active site C68 acetylation. Since the active site is largely shaped by its interactions with both the C-terminal tail and the domain II loop, changes in the conformation and/or dynamics of either of these structures could influence substrate selectivity and catalytic activity.
Figure 5.
NAT1 SNP Residue Arginine 187. Residue R187 is located on the domain II beta barrel, and is partially surface exposed both on the protein surface and the active site pocket. The arginine side-chain hydrogen bonds to E182 and K188, both in domain II, and to the domain III C-terminal residue T289. Mutating this residue to a glutamine results in loss of these interactions and could change the size/shape of the active site pocket.
Functional studies of the R187Q variant in yeast demonstrated reduced catalytic activity, protein levels, and thermostability (Fretland et al., 2001; 2002). Similar results were found in mammalian cells (Zhu and Hein, 2007), with reduced Vmax and increased substrate Km. Functional studies in bacterial cells also demonstrated reduced Vmax and increased substrate Km (Hughes et al., 1998). These studies are consistent with observations of the NAT1 crystal structure, which suggest that the R187Q variant could destabilize the NAT1 structure and influence substrate binding by altering the size and shape of the active site. The study in COS-1 cells (Zhu and Hein, 2007) demonstrated that the R187Q variant did not change AcCoA Km, indicating that altered interactions with the C-terminal tail do not significantly influence AcCoA binding.
M205V (613A>G)
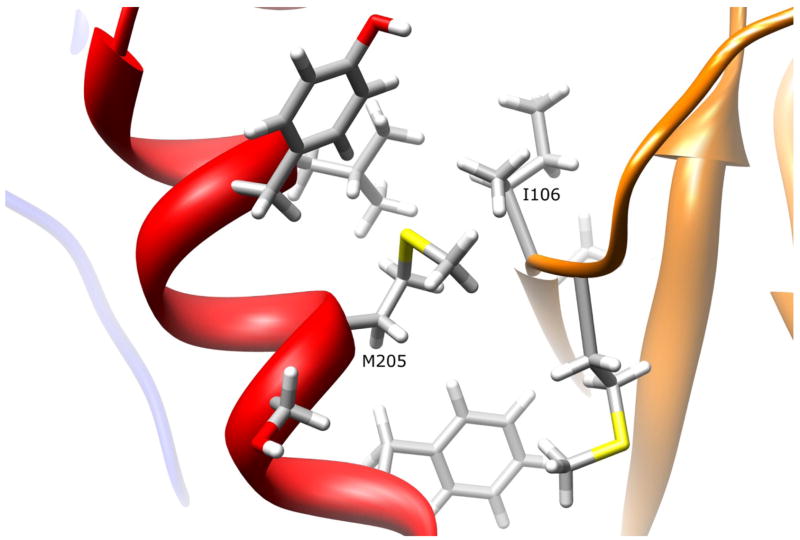
The side-chain of residue M205 is 19.2% solvent exposed in the interdomain region, which is adjacent to the active site entrance and the domain II beta barrel (Fig. 6). The side chain has no apparent interactions with surrounding residues, but is in close proximity to the backbone and side-chain of residue I106 (M205:HE1-I106:HG22 2.44 Å, M205:HG2-I106:HA 1.92 Å). Replacing the methionine with a valine residue would maintain the hydrophobicity of residue 205, and would not introduce any steric clashes with surrounding residues. Therefore, it is not expected to influence the position of catalytic triad residue H107. Since no interactions are lost and no clashes induced by replacing hydrophobic methionine with a smaller hydrophobic valine at 205, no changes in protein stability or function are expected.
Figure 6.
NAT1 SNP Residue Methionine 205. Residue M205 is partially surface exposed on the interdomain region helix. The methionine side-chain has no apparent interactions with other residues. Mutating M205 to a valine residue is not expected to influence enzyme stability or function.
Functional studies of the M205V variant in yeast (Fretland et al., 2001; 2002) and mammalian cells (Zhu and Hein, 2007) demonstrated no change in enzyme expression, catalytic activity, or thermostability. These findings are consistent with the observation that no important molecular interactions are introduced or lost in the M205V variant.
D251V (752A>T)
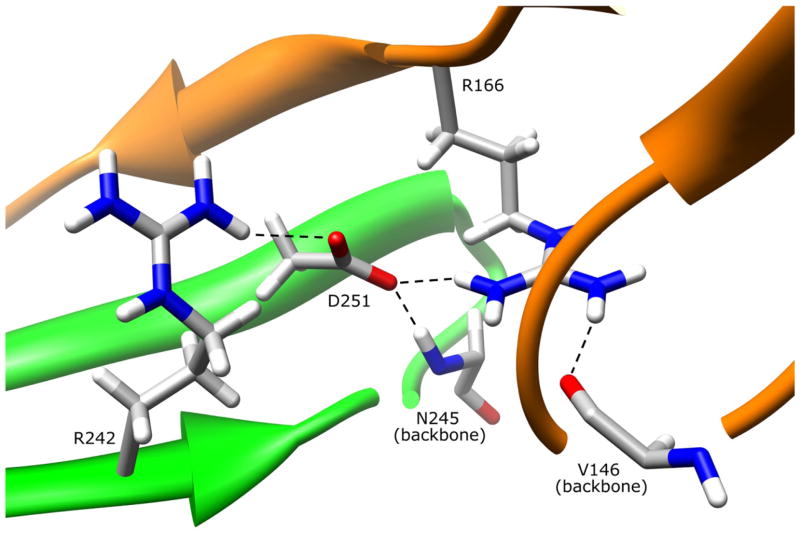
The side-chain of residue D251 is 2.1% surface exposed on the domain III beta sheet and its side-chain is oriented into the protein core (Fig. 7). The D251 side-chain hydrogen bonds to residue R166 (D251:OD1-R166:NH1 2.96 Å) on the domain II loop, to the domain III beta sheet residue R242 (D251:OD2-R242:NH1 3.16 Å), and to the backbone of N245 (D251:OD1-N245:N 2.85 Å) also in domain III. Although the interactions with R242 and N245 are not necessary for maintaining the stability of the beta sheet, hydrogen bonding with R242 may provide additional stability to the loop-stabilizing hydrogen bonding interactions between R242 and the domain II loop residue Q168. The hydrogen bond between D251 and R166 directly influences the conformation of the domain II loop, and provides support for the interaction of the domain II loop with the backbone of domain II residue V146. These and other interactions between the domain II loop and the domain III beta sheet contribute to protein stability (Walraven et al., 2007). Replacing aspartate with a hydrophobic valine residue would result in loss of all of the D251 interactions, which could affect the dynamics and conformation of the domain II loop, thereby altering protein stability.
Figure 7.
NAT1 SNP Residue Aspartate 251. Residue D251 is primarily in the protein core on the domain III beta sheet. The aspartate side-chain hydrogen bonds to R242 and the backbone of N245 in domain III. The D251 side-chain also hydrogen bonds to the domain II loop residue R166, an interaction which may stabilize R166 hydrogen bonding interaction with domain II beta barrel residue V146. These interactions are lost when D251 is mutated to a valine residue.
Functional characterization of the D251V variant in yeast (Fretland et al., 2001; 2002), COS-1 (Zhu and Hein, 2007), and HT-29 human colon adenocarcinoma cells (Butcher et al., 2004) demonstrated reduced protein levels and protein activity relative to NAT1*4 reference. Reduced thermostability was also demonstrated in yeast (Fretland et al., 2001; 2002). The HT-29 cell study reported reduced acetylation capacity for the D251V variant suggesting that acetylation status is directly correlated to enzyme degradation. However, there is no indication from the NAT1 crystal structure that the D251V residue change would directly or indirectly interfere with protein acetylation apart from its role in reducing protein stability.
E261K (781G>A)
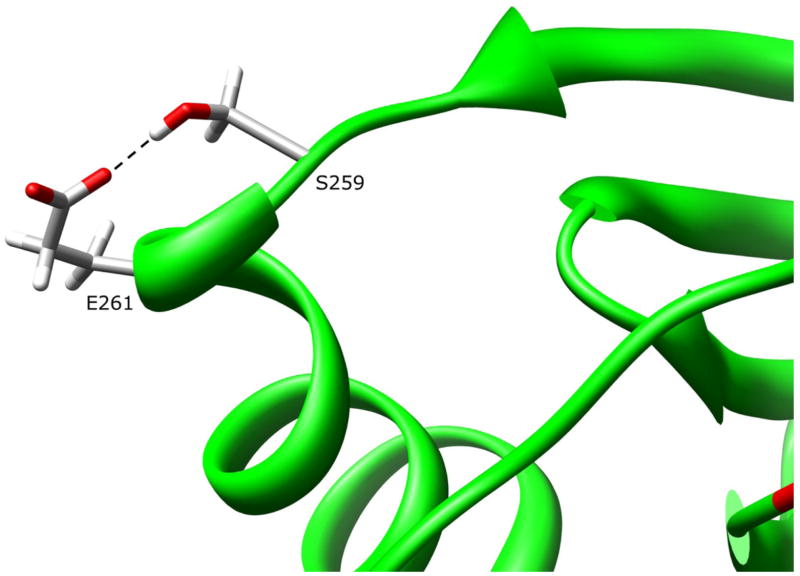
The side-chain of residue E261 is 87.5% solvent exposed on the protein surface in the domain III helix (Fig. 8). The E261 side-chain hydrogen bonds to the side chain of residue S259 (E261:OE1-S259:OG 2.69 Å) in the coil between a domain III beta sheet and helix. It is unlikely that this interaction is required for stability of the helix or the domain III tertiary structure. Although hydrogen bonding with S259 may be lost, replacing glutamate with lysine at residue 261 is not expected to cause major changes in dynamics or conformation, because the helix secondary structure is not dependent on this interaction and the domain III helix is stabilized by hydrophobic forces as described in the next section.
Figure 8.
NAT1 SNP Residue Glutamate 261. Residue E261 is surface exposed on the domain III helix. The glutamate side-chain hydrogen bonds to S259. Mutating E261 to a lysine residue is not expected to influence enzyme stability or function.
Functional studies of the E261K variant in yeast (Fretland et al., 2001; 2002), COS-1 (Zhu and Hein, 2007), and HT-29 cells (Butcher et al., 2004) demonstrated no reduction in protein levels and catalytic activity, with the yeast and HT-29 studies also demonstrating no reduction in thermostability. These data are consistent with observations from the NAT1 crystal structure that residue 261 is located on the protein surface, with a single interaction that makes no apparent contribution to the dynamics or structural conformations in that region.
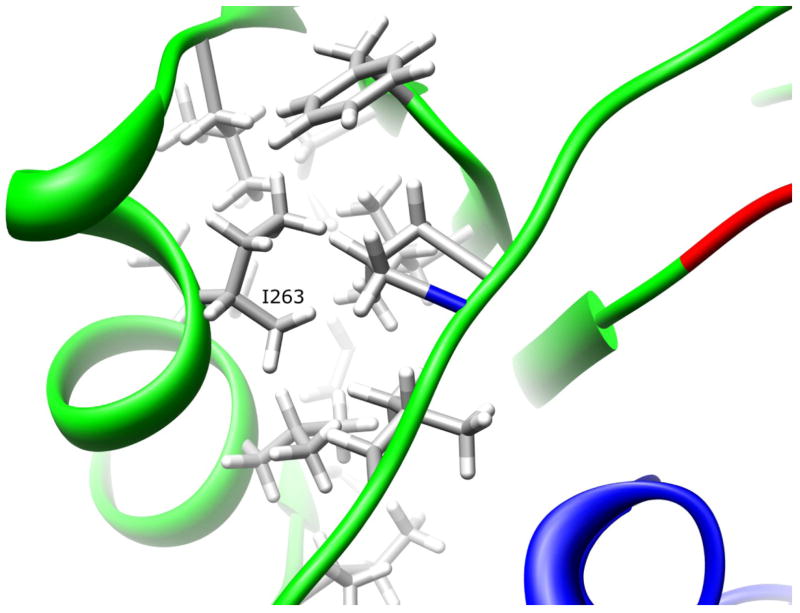
I263V (787A>G)
Residue I263 is 6.2% solvent exposed on the domain III helix (Fig. 9). The I263 side-chain is part of a hydrophobic core comprised of L234, F237, L239, L258, I263, V266, L267, I270, F271, I273, L279, and P281. This core is located at the interface between the domain III helix and beta sheet, and the C-terminal tail coil. A conservative substitution of isoleucine to the smaller valine at position 263 preserves the hydrophobic interactions without introducing steric clashes. Because the hydrophobic forces are not disturbed, this substitution is not expected to affect NAT1 structural dynamics or conformation.
Figure 9.
NAT1 SNP Residue Isoleucine 263. Residue I263 is located on the domain III helix. The isoleucine side-chain is part of a hydrophobic core that is partially responsible for the domain III tertiary structure. Mutating I263 to a valine residue is not expected to influence enzyme stability of function.
Functional studies of the I263V variant in yeast (Fretland et al., 2001; 2002) and COS-1 cells (Zhu and Hein, 2007) demonstrated no change in protein level or catalytic activity. The yeast cell studies also demonstrated no reduction in protein thermostability. These findings are consistent with observations from the NAT1 crystal structure that the domain III hydrophobic core is not disrupted by the conservative I263V residue change.
SUMMARY
The recent resolution of the NAT1 crystal structure enhances knowledge of structure-function relationships important for understanding associations of NAT1 SNPs with genetic predisposition to cancer, birth defects, and other diseases.
Acknowledgments
The studies were partially supported by National Institutes of Health grants R01-CA034627 (DWH) and T32-ES011564 (DWH to support JMW). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- Adam PJ, Berry J, Loader JA, Tyson KL, Craggs G, Smith P, De Belin J, Steers G, Pezzella F, Sachsenmeir KF, Stamps AC, Herath A, Sim E, O’Hare MJ, Harris AL, Terrett JA. Arylamine N-acetyltransferase-1 is highly expressed in breast cancers and conveys enhanced growth and resistance to etoposide in vitro. Mol Cancer Res. 2003;1:826–835. [PubMed] [Google Scholar]
- Ambrosone CB, Abrams SM, Gorlewska-Roberts K, Kadlubar FF. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch Biochem Biophys. 2007;464:169–175. doi: 10.1016/j.abb.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Bell DA, Stephens EA, Castranio T, Umbach DM, Watson M, Deakin M, Elder J, Hendrickse C, Duncan H, Strange RC. Polyadenylation polymorphism in the acetyltransferase 1 gene (NAT1) increases risk of colorectal cancer. Cancer Res. 1995;55:3537–3542. [PubMed] [Google Scholar]
- Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6:252–264. doi: 10.1186/bcr784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouvala S, Fakis G. Arylamine N-acetyltransferases: what we learn from genes and genomes. Drug Metab Rev. 2005;37:511–564. doi: 10.1080/03602530500251204. [DOI] [PubMed] [Google Scholar]
- Bruhn C, Brockmoller J, Cascorbi I, Roots I, Borchert HH. Correlation between genotype and phenotype of the human arylamine N-acetyltransferase type 1 (NAT1) Biochem Pharmacol. 1999;58:1759–1764. doi: 10.1016/s0006-2952(99)00269-5. [DOI] [PubMed] [Google Scholar]
- Butcher NJ, Arulprgasam A, Minchin RF. Proteasomal degradation of N-acetyltransferase 1 is prevented by acetylation of the active site cysteine. J Biol Chem. 2004;279:22131–22137. doi: 10.1074/jbc.M312858200. [DOI] [PubMed] [Google Scholar]
- Carmichael SL, Shaw GM, Yang W, Iovannisci DM, Lammer E. Risk of limb deficiency defects associated with NAT1, NAT2, GSTT1, GSTM1, and NOS3 genetic variants, maternal smoking, and vitamin supplement intake. Am J Med Genet A. 2006;140:1915–1922. doi: 10.1002/ajmg.a.31402. [DOI] [PubMed] [Google Scholar]
- Chen J, Stampfer MJ, Hough HL, Garcia-Closas M, Willett WC, Hennekens CH, Kelsey KT, Hunter DJ. A prospective study of N-acetyltransferase genotype, red meat intake, and risk of colorectal cancer. Cancer Res. 1998;58:3307–3311. [PubMed] [Google Scholar]
- de Leon JH, Vatsis KP, Weber WW. Characterization of naturally occurring and recombinant human N-acetyltransferase variants encoded by NAT1. Mol Pharmacol. 2000;58:288–299. doi: 10.1124/mol.58.2.288. [DOI] [PubMed] [Google Scholar]
- Doll MA, Jiang W, Deitz AC, Rustan TD, Hein DW. Identification of a novel allele at the human NAT1 acetyltransferase locus. Biochem Biophys Res Commun. 1997;233:584–591. doi: 10.1006/bbrc.1997.6501. [DOI] [PubMed] [Google Scholar]
- Dupret JM, Grant DM. Site-directed mutagenesis of recombinant human arylamine N-acetyltransferase expressed in Escherichia coli. Evidence for direct involvement of Cys68 in the catalytic mechanism of polymorphic human NAT2. J Biol Chem. 1992;267:7381–7385. [PubMed] [Google Scholar]
- Fretland AJ, Doll MA, Leff MA, Hein DW. Functional characterization of nucleotide polymorphisms in the coding region of N-acetyltransferase. Pharmacogenetics. 2001;11:511–520. doi: 10.1097/00008571-200108000-00006. [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Doll MA, Zhu Y, Smith L, Leff MA, Hein DW. Effect of nucleotide substitutions in N-acetyltransferase 1 one N-acetylation (deactivation) and O-acetylation (activation) of arylamine carcinogens: implications for cancer predisposition. Cancer Detect Prev. 2002;26:10–14. doi: 10.1016/s0361-090x(02)00005-3. [DOI] [PubMed] [Google Scholar]
- Gago-Dominguez M, Bell DA, Watson MA, Yuan JM, Castelao JE, Hein DW, Chan KK, Coetzee GA, Ross RK, Yu MC. Permanent hair dyes and bladder cancer: risk modification by cytochrome P4501A2 and N-acetyltransferases 1 and 2. Carcinogenesis. 2003;24:483–489. doi: 10.1093/carcin/24.3.483. [DOI] [PubMed] [Google Scholar]
- Gemignani F, Landi S, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Gaborieau V, Gioia-Patricola L, Bellini I, Barale R, Canzian F, Hall J, Boffetta P, Hung RJ, Brennan P. Development of lung cancer before the age of 50: the role of xenobiotic metabolizing genes. Carcinogenesis. 2007;28:1287–1293. doi: 10.1093/carcin/bgm021. [DOI] [PubMed] [Google Scholar]
- Goodfellow GH, Dupret JM, Grant DM. Identification of amino acids imparting acceptor substrate selectivity to human arylamine acetyltransferases NAT1 and NAT2. Biochem J. 2000;348:159–166. [PMC free article] [PubMed] [Google Scholar]
- Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- Hein DW, Leff MA, Ishibe N, Sinha R, Frazier HA, Doll MA, Xiao GH, Weinrich MC, Caporaso NE. Association of prostate cancer with rapid N-acetyltransferase 1 (NAT1*10) in combination with slow N-acetyltransferase 2 acetylator genotypes in a pilot case-control study. Environ Mol Mutagen. 2002;40:161–167. doi: 10.1002/em.10103. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. ‘NACCESS’, Computer Program, Department of Biochemistry and Molecular Biology. College London University; 1993. [Google Scholar]
- Hughes NC, Janezic SA, McQueen KL, Jewett MA, Castranio T, Bell DA, Grant DM. Identification and characterization of variant alleles of human acetyltransferase NAT1 with defective function using p-aminosalicylate as an in-vivo and in-vitro probe. Pharmacogenetics. 1998;8:55–66. doi: 10.1097/00008571-199802000-00008. [DOI] [PubMed] [Google Scholar]
- Ishibe N, Sinha R, Hein DW, Kulldorff M, Strickland P, Fretland AJ, Chow WH, Kadlubar FF, Lang NP, Rothman N. Genetic polymorphisms in heterocyclic amine metabolism and risk of colorectal adenomas. Pharmacogenetics. 2002;12:145–150. doi: 10.1097/00008571-200203000-00008. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Hoess K, Mitchell LE, Whitehead AS. Loss of function polymorphisms in NAT1 protect against spina bifida. Hum Genet. 2006;120:52–57. doi: 10.1007/s00439-006-0181-6. [DOI] [PubMed] [Google Scholar]
- Jiao L, Doll MA, Hein DW, Bondy ML, Hassan MM, Hixson JE, Abbruzzese JL, Li D. Haplotype of N-acetyltransferase 1 and 2 and risk of pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2379–2386. doi: 10.1158/1055-9965.EPI-06-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Inatomi H, Yang M, Kawamoto T, Matsumoto T, Bell DA. Arylamine N-acetyltransferase 1 (NAT1) and 2 (NAT2) genes and risk of urothelial transitional cell carcinoma among Japanese. Pharmacogenetics. 1999;9:401–404. doi: 10.1097/00008571-199906000-00017. [DOI] [PubMed] [Google Scholar]
- Kawamura A, Graham J, Mushtaq A, Tsiftsoglou SA, Vath GM, Hanna PE, Wagner CR, Sim E. Eukaryotic arylamine N-acetyltransferase. Investigation of substrate specificity by high-throughput screening. Biochem Pharmacol. 2005;69:347–359. doi: 10.1016/j.bcp.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Lammer EJ, Shaw GM, Iovannisci DM, Finnell RH. Periconceptional multivitamin intake during early pregnancy, genetic variation of acetyl-N-transferase 1 (NAT1), and risk for orofacial clefts. Birth Defects Res A Clin Mol Teratol. 2004a;70:846–852. doi: 10.1002/bdra.20081. [DOI] [PubMed] [Google Scholar]
- Lammer EJ, Shaw GM, Iovannisci DM, Van Waes J, Finnell RH. Maternal smoking and the risk of orofacial clefts: Susceptibility with NAT1 and NAT2 polymorphisms. Epidemiology. 2004b;15:150–156. doi: 10.1097/01.ede.0000112214.33432.cc. [DOI] [PubMed] [Google Scholar]
- Lau EY, Felton JS, Lightstone FC. Insights into the o-acetylation reaction of hydroxylated heterocyclic amines by human arylamine N-acetyltransferases: a computational study. Chem Res Toxicol. 2006;19:1182–1190. doi: 10.1021/tx0600999. [DOI] [PubMed] [Google Scholar]
- Lee KM, Park SK, Kim SU, Doll MA, Yoo KY, Ahn SH, Noh DY, Hirvonen A, Hein DW, Kang D. N-acetyltransferase (NAT1, NAT2) and glutathione S-transferase (GSTM1, GSTT1) polymorphisms in breast cancer. Cancer Lett. 2003;196:179–186. doi: 10.1016/s0304-3835(03)00311-2. [DOI] [PubMed] [Google Scholar]
- Li D, Jiao L, Li Y, Doll MA, Hein DW, Bondy ML, Evans DB, Wolff RA, Lenzi R, Pisters PW, Abbruzzese JL, Hassan MM. Polymorphisms of cytochrome P4501A2 and N-acetyltransferase genes, smoking, and risk of pancreatic cancer. Carcinogenesis. 2006;27:103–111. doi: 10.1093/carcin/bgi171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilla C, Verla-Tebit E, Risch A, Jager B, Hoffmeister M, Brenner H, Chang-Claude J. Effect of NAT1 and NAT2 genetic polymorphisms on colorectal cancer risk associated with exposure to tobacco smoke and meat consumption. Cancer Epidemiol Biomarkers Prev. 2006;15:99–107. doi: 10.1158/1055-9965.EPI-05-0618. [DOI] [PubMed] [Google Scholar]
- Liu L, Von Vett A, Zhang N, Walters KJ, Wagner CR, Hanna PE. Arylamine N-acetyltransferases: characterization of the substrate specificities and molecular interactions of environmental arylamines with human NAT1 and NAT2. Chem Res Toxicol. 2007;20:1300–1308. doi: 10.1021/tx7001614. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhang N, Zhou X, Hanna PE, Wagner CR, Koepp DM, Walters KJ. Arylamine N-acetyltransferase aggregation and constitutive ubiquitylation. J Mol Biol. 2006;361:482–492. doi: 10.1016/j.jmb.2006.06.029. [DOI] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Millikan RC, Pittman GS, Newman B, Tse CK, Selmin O, Rockhill B, Savitz D, Moorman PG, Bell DA. Cigarette smoking, N-acetyltransferases 1 and 2, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1998;7:371–378. [PubMed] [Google Scholar]
- Minchin RF, Hanna PE, Dupret JM, Wagner CR, Rodrigues-Lima F, Butcher NJ. Arylamine N-acetyltransferase I. Int J Biochem Cell Biol. 2007;39:1999–2005. doi: 10.1016/j.biocel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Morton LM, Bernstein L, Wang SS, Hein DW, Rothman N, Colt JS, Davis S, Cerhan JR, Severson RK, Welch R, Hartge P, Zahm SH. Hair dye use, genetic variation in N-acetyltransferase 1 (NAT1) and 2 (NAT2), and risk of non-Hodgkin lymphoma. Carcinogenesis. 2007;28:1759–1764. doi: 10.1093/carcin/bgm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LM, Schenk M, Hein DW, Davis S, Zahm SH, Cozen W, Cerhan JR, Hartge P, Welch R, Chanock SJ, Rothman N, Wang SS. Genetic variation in N-acetyltransferase 1 (NAT1) and 2 (NAT2) and risk of non-Hodgkin lymphoma. Pharmacogenet Genomics. 2006;16:537–545. doi: 10.1097/01.fpc.0000215071.59836.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton M, Mushtaq A, Yu TW, Wu LJ, Sinclair J, Sim E. Eubacterial arylamine N-acetyltransferases - identification and comparison of 18 members of the protein family with conserved active site cysteine, histidine and aspartate residues. Microbiology. 2001;147:1137–1147. doi: 10.1099/00221287-147-5-1137. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Lima F, Dupret JM. 3D model of human arylamine N-acetyltransferase 2: structural basis of the slow acetylator phenotype of the R64Q variant and analysis of the active-site loop. Biochem Biophys Res Commun. 2002;291:116–123. doi: 10.1006/bbrc.2002.6414. [DOI] [PubMed] [Google Scholar]
- Rovito PM, Morse PD, Spinek K, Newman N, Jones RF, Wang CY, Haas GP. Heterocyclic amines and genotype of N-acetyltransferases as risk factors for prostate cancer. Prostate Cancer Prostatic Dis. 2005;8:69–74. doi: 10.1038/sj.pcan.4500780. [DOI] [PubMed] [Google Scholar]
- Sanderson S, Salanti G, Higgins J. Joint effects of the N-acetyltransferase 1 and 2 (NAT1 and NAT2) genes and smoking on bladder carcinogenesis: a literature-based systematic HuGE review and evidence synthesis. Am J Epidemiol. 2007;166:741–751. doi: 10.1093/aje/kwm167. [DOI] [PubMed] [Google Scholar]
- Sandy J, Mushtaq A, Kawamura A, Sinclair J, Sim E, Noble M. The structure of arylamine N-acetyltransferase from Mycobacterium smegmatis--an enzyme which inactivates the anti-tubercular drug, isoniazid. J Mol Biol. 2002;318:1071–1083. doi: 10.1016/S0022-2836(02)00141-9. [DOI] [PubMed] [Google Scholar]
- Sinclair JC, Sandy J, Delgoda R, Sim E, Noble ME. Structure of arylamine N-acetyltransferase reveals a catalytic triad. Nat Struct Biol. 2000;7:560–564. doi: 10.1038/76783. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, Umbach DM, Stephens E, Castranio T, Paulson D, Robertson C, Mohler JL, Bell DA. The role of N-acetylation polymorphisms in smoking-associated bladder cancer: evidence of a gene-gene-exposure three-way interaction. Cancer Res. 1998;58:3603–3610. [PubMed] [Google Scholar]
- Wakefield L, Cornish V, Long H, Griffiths WJ, Sim E. Deletion of a xenobiotic metabolizing gene in mice affects folate metabolism. Biochem Biophys Res Comm. 2007;364:556–560. doi: 10.1016/j.bbrc.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walraven JM, Trent JO, Hein DW. Computational and experimental analyses of mammalian arylamine N-acetyltransferase structure and function. Drug Metab Dispos. 2007;35:1001–1007. doi: 10.1124/dmd.107.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikman H, Thiel S, Jager B, Schmezer P, Spiegelhalder B, Edler L, Dienemann H, Kayser K, Schulz V, Drings P, Bartsch H, Risch A. Relevance of N-acetyltransferase 1 and 2 (NAT1, NAT2) genetic polymorphisms in non-small cell lung cancer susceptibility. Pharmacogenetics. 2001;11:157–168. doi: 10.1097/00008571-200103000-00006. [DOI] [PubMed] [Google Scholar]
- Wu H, Dombrovsky L, Tempel W, Martin F, Loppnau P, Goodfellow GH, Grant DM, Plotnikov AN. Structural basis of substrate-binding specificity of human arylamine N-acetyltransferases. J Biol Chem. 2007;282:30189–30197. doi: 10.1074/jbc.M704138200. [DOI] [PubMed] [Google Scholar]
- Zhangwei X, Jianming X, Qiao M, Xinhua X. N-Acetyltransferase-1 gene polymorphisms and correlation between genotype and its activity in a central Chinese Han population. Clin Chim Acta. 2006;371:85–91. doi: 10.1016/j.cca.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Zheng W, Deitz AC, Campbell DR, Wen WQ, Cerhan JR, Sellers TA, Folsom AR, Hein DW. N-acetyltransferase 1 genetic polymorphism, cigarette smoking, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8:233–239. [PubMed] [Google Scholar]
- Zhu Y, Hein DW. Functional effects of single nucleotide polymorphisms in the coding region of human N-acetyltransferase 1. Pharmacogenomics J. 2007 doi: 10.1038/sj.tpj.6500483. [DOI] [PMC free article] [PubMed] [Google Scholar]