Abstract
Production of Ran–guanosine triphosphate (GTP) around chromosomes induces local nucleation and plus end stabilization of microtubules (MTs). The nuclear protein TPX2 is required for RanGTP-dependent MT nucleation. To find the MT stabilizer, we affinity purify nuclear localization signal (NLS)–containing proteins from Xenopus laevis egg extracts. This NLS protein fraction contains the MT stabilization activity. After further purification, we used mass spectrometry to identify proteins in active fractions, including cyclin-dependent kinase 11 (Cdk11). Cdk11 localizes on spindle poles and MTs in Xenopus culture cells and egg extracts. Recombinant Cdk11 demonstrates RanGTP-dependent MT stabilization activity, whereas a kinase-dead mutant does not. Inactivation of Cdk11 in egg extracts blocks RanGTP-dependent MT stabilization and dramatically decreases the spindle assembly rate. Simultaneous depletion of TPX2 completely inhibits centrosome-dependent spindle assembly. Our results indicate that Cdk11 is responsible for RanGTP-dependent MT stabilization around chromosomes and that this local stabilization is essential for normal rates of spindle assembly and spindle function.
Introduction
The GTP-bound form of the small GTPase Ran is essential for spindle assembly. Ran's guanine nucleotide exchange factor, RCC1, is enriched on chromosomes, whereas Ran–GTPase-activating protein is evenly distributed in the cytoplasm, generating a gradient of RanGTP around chromosomes (Kalab et al., 2002, 2006; Caudron et al., 2005). RanGTP controls both microtubule (MT) nucleation (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Zhang et al., 1999) and plus end stabilization (Carazo-Salas et al., 2001; Wilde et al., 2001). MT nucleation is triggered by the RanGTP-dependent release of TPX2 from importins in the proximity of chromosomes (Gruss et al., 2001). However, the mechanism by which RanGTP induces local MT plus end stabilization remained unknown. In this study, we purify the RanGTP-dependent stabilization factor and identify it as Cdk11. We show that Cdk11 localizes to the spindle in the Xenopus laevis system and demonstrate that inactivation of Cdk11 in extracts results in abnormal spindle assembly.
Results and discussion
Purification of the RanGTP-dependent MT stabilization activity
We had previously demonstrated that in TPX2-depleted metaphase (M-phase) extracts, RanQ69L (a mutant of Ran that cannot hydrolyze GTP) did not induce free MT nucleation but still mediated MT stabilization (Gruss et al., 2002). This indicated that RanGTP stabilizes MTs in a TPX2-independent manner. In wild-type M-phase extracts containing TPX2, however, MT stabilization initially increased with RanQ69L concentration but then decreased (Gruss et al., 2002). Because the high concentration of RanQ69L induced numerous ectopic, extracentrosomal asters, we reasoned that the activity responsible for MT stabilization was diluted among the numerous asters, resulting in no visible effect on aster size. To avoid this problem, we identified a polyclonal anti-TPX2 antibody that blocked MT nucleation (Fig. S1 B, available at http://www.jcb.org/cgi/content/full/jcb.200706189/DC1). In the presence of this antibody, higher concentrations of RanQ69L stabilized MTs, whereas RanT24N (a mutant that cannot bind GTP) did not (Fig. S1 C).
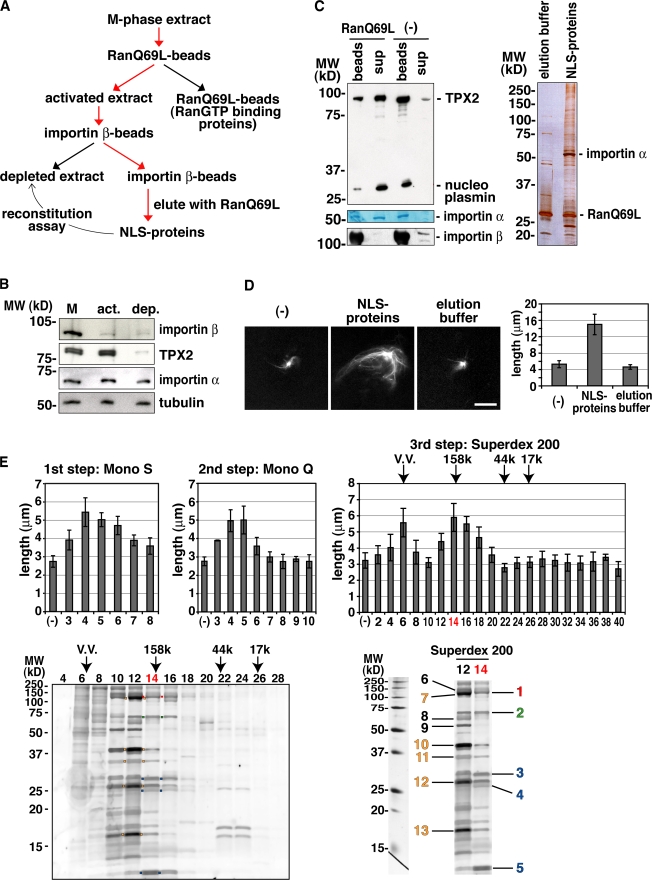
The MT nucleation factor TPX2 is inhibited through interaction between its NLS sequence and the importin α/β heterodimer and is activated when RanGTP dissociates it from the importins (Gruss et al., 2001). In several independent assays, we found that the importin α/β heterodimer also specifically inhibits the MT stabilization activity in egg extracts (Fig. S1 D and not depicted). Based on these findings, we designed a purification strategy to identify the MT stabilization factor (Fig. 1 A). An M-phase extract was first treated with RanQ69L beads to release endogenous NLS proteins from importin α/β. RanGTP-binding proteins such as importin β were then removed from the extract (Fig. 1, A and B; Nachury et al., 2001). The resulting extract (activated extract) was then applied to importin β beads. NLS proteins like TPX2 were bound to these beads and, thereby, were efficiently depleted (Fig. 1, A and B). As expected, importin α was partially removed (Fig. 1 B). NLS proteins were then eluted with RanQ69L in the presence of 500 mM NaCl from the importin β beads (Fig. 1, A and C). When the eluted fraction (NLS proteins) was added back to the depleted extract supplemented with centrosomes and the anti-TPX2 antibody, it generated larger centrosomal asters, whereas the elution buffer did not (Fig. 1, A and D).
Figure 1.
Purification of the RanGTP-dependent MT stabilization activity from Xenopus egg extract. (A) Purification strategy. (B) Immunoblot of the extract outlined in A. M, M-phase extract; act, activated extract; dep, depleted extract. The labeled proteins were detected by specific antibodies. (C) Elution of NLS proteins and endogenous importin α from the importin β column by RanQ69L and 500 mM NaCl. (left) After incubation, supernatant (sup) and beads were analyzed by immunoblotting (TPX2, nucleoplasmin, and importin β) or Coomassie staining (importin α). (right) Silver staining of proteins in elution buffer or the NLS protein fraction. Note that a major band (importin α) and various other bands (expected NLS proteins) were detected in addition to RanQ69L. (D) MT stabilization activity detected in the NLS protein fraction. The NLS protein fraction or elution buffer was incubated in the depleted extract containing centrosomes, anti-TPX2 antibodies, and Cy3-labeled tubulin (left). The MT length of the centrosomal asters was quantified (right), as shown in Fig. S1 A (available at http://www.jcb.org/cgi/content/full/jcb.200706189/DC1). (E) Purification of the MT stabilization activity from the NLS protein fraction. First step, Mono S; second step, Mono Q; third step, Superdex 200. Molecular mass standards (kD) are indicated. VV, void volume. The MT stabilization activity of purified fractions was assayed as described in Fig. 1 D. (bottom left) SDS-PAGE and Cypro ruby staining of the Superdex 200 fractions. Red boxes, Cdk11; green boxes, DKC1, NOP5, and XNOP56; blue boxes, small nuclear RNP proteins U2A′, U2B″, and Sm proteins; orange boxes, the processing of precursor complex. (bottom right) Proteins identified by mass spectrometry from the active stabilization fractions (Table I). The colored numbers correspond to the colored boxes in the bottom left panel. (D and E) Error bars represent SD. n > 20 asters; n > 3 experiments. Bar, 20 μm.
On the basis of this reconstitution assay, we further purified the MT stabilization activity on Mono S, Mono Q, and Superdex 200 columns (Fig. 1 E). We analyzed the Superdex 200 fractions by SDS-PAGE, and visible bands in the active fractions were identified by mass spectrometry (Fig. 1 E). All proteins identified were known nuclear proteins (Fig. 1 E and Table I). In the most active fraction (fraction 14), we found three different protein complexes, including Xenopus Cdk11 (Fig. 1 E, band 1 [red]), the human orthologue of which had been reported to be required for spindle assembly in HeLa cells (Petretti et al., 2006). Cdk11 RNAi led to the formation of short or monopolar spindles and to mitotic arrest (Petretti et al., 2006). We also identified Cyclin-L1 (Fig. 1 E, band 8), a known cyclin partner of Cdk11, in the active fractions (Berke et al., 2001).
Table I. Proteins identified by mass spectrometry from the Superdex 200 fractions (Fig. 1 E).
| No. | Protein name | Gene name | GI accession no. |
|---|---|---|---|
| 1 | Cdk11a | MGC80275 | gi|50414818 |
| 2 | DKC1a | NAb | gi|71051901 |
| NOP5a | MGC78950 | gi|50414537 | |
| XNop56 | XNop56 | gi|14799394 | |
| 3 | U2 small nuclear RNP polypeptide A′a | MGC81833 | gi|51950159 |
| 4 | U2 small nuclear RNP polypeptide B″a | LOC495019 | gi|54035240 |
| 5 | Sm protein Ea | MGC80249 | gi|49119142 |
| 6 | ISWI | ISWI | gi|49899007 |
| 7 | POP1a | NAb | gi|37243880 |
| 8 | Cyclin-L1 | ccnl1 | gi|80477781 |
| 9 | Importin α2 subunit | kpna2 | gi|1079301 |
| 10 | Ribonuclease P 40-kD subunita | MGC81617 | gi|51703458 |
| 11 | Ribonuclease P protein subunit p38a | MGC81745 | gi|51950085 |
| 12 | Ribonuclease P protein subunit p30a | LOC494740 | gi|52354780 |
| POP4a | MGC82042 | gi|49257840 | |
| 13 | POP5a | LOC779510 | gi|110645356 |
NA, not applicable. Note that only nuclear proteins were identified.
The protein name has not been assigned. We used the name of the human homologue.
No gene name available. The sequences were derived from the Unigene database.
Cdk11 is the RanGTP-dependent MT stabilization factor
We cloned Xenopus Cdk11 and expressed the protein in insect cells (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200706189/DC1). The purified recombinant Cdk11 kinase showed MT stabilization activity in the depleted extracts, whereas a catalytically inactive version of the protein (Solomon et al., 1992) did not (Fig. S2, A and B). The stabilization activity induced by recombinant Cdk11 was inhibited by importin β, and the inhibition effect was reversed by the further addition of RanQ69L (Fig. S2 C). Importin β and RanQ69L had no effect on aster size in the absence of Cdk11 (unpublished data).
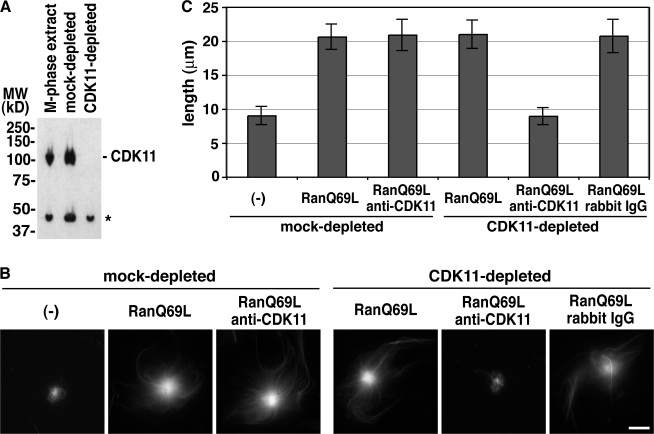
To determine whether Cdk11 was essential for RanGTP-dependent stabilization of MTs in nontreated Xenopus egg extracts, we prepared antibodies against full-length Xenopus Cdk11 (Fig. 2 A) and used them to specifically deplete the protein from the extracts. The depletion seemed to work efficiently and specifically as judged by immunoblot analysis (Fig. 2 A), but Cdk11 depletion alone had no measurable effect on the RanGTP-dependent aster size increase (Fig. 2 B). However, because Cdk11 was likely to act catalytically, it was still possible that residual amounts of Cdk11 caused MT stabilization in response to RanGTP. To examine this possibility, we added anti–full-length Cdk11 antibodies to a Cdk11-depleted extract and tested the effect of RanQ69L addition on MT length. No RanQ69L-dependent MT stabilization was observed under such conditions (Fig. 2, B and C). The addition of anti-Cdk11 antibodies to mock-depleted extracts did not block RanQ69L-dependent MT stabilization, nor did the addition of control rabbit IgG to Cdk11-depleted extracts (Fig. 2, B and C). These results indicated that Cdk11 is indeed required for RanGTP-dependent MT stabilization.
Figure 2.
Cdk11 is responsible for RanGTP-dependent MT stabilization in M-phase extracts. (A) Depletion of Cdk11 from M-phase extracts. An M-phase extract was immunodepleted using control or anti–full-length Cdk11 antibodies, and the depletion efficiency was evaluated by immunoblotting. The asterisk indicates a cross-reacting band. (B) RanGTP-dependent MT stabilization assay. The mock- or Cdk11-depleted extracts were incubated with centrosomes, anti-TPX2 antibodies, and Cy3-labeled tubulin in the presence or absence of 12 μM RanQ69L, 0.22 mg/ml anti-Cdk11 antibodies, or rabbit IgG. (C) Quantification of the MT length assayed in B as described in Fig. S1 A (available at http://www.jcb.org/cgi/content/full/jcb.200706189/DC1). Error bars represent SD. n > 20 asters. This experiment was reproduced three times. Bar, 20 μm.
Cdk11 localizes at spindle poles and on spindle MTs
The identification of Cdk11 as a RanGTP-dependent MT stabilization factor suggested that it could localize to mitotic MTs. Cdk11 has been shown to localize to mitotic centrosomes in HeLa cells by immunofluorescence (Petretti et al., 2006). The Cdk11 antibody used recognized two isoforms of Cdk11 in the immunoblot, full-length Cdk11 and a shorter variant, p58. p58 is translated from the Cdk11 mRNA through an internal ribosomal entry site and contains the C-terminal kinase domain of Cdk11 but lacks the N terminus, including the NLS sequences (Cornelis et al., 2000). We prepared specific antibodies that distinguished Xenopus Cdk11 and p58 (Fig. S2, A and C) and examined the localization of both proteins by immunofluorescence. In Xenopus XL177 cells, Cdk11 was detected at spindle poles and MTs in addition to the mitotic cytoplasm (Fig. S2 B). p58 was detected only throughout the cytoplasm (Fig. S2 B). In spindles assembled in Xenopus egg extracts, Cdk11 was also detected at spindle poles, whereas p58 was not (Fig. S2 D). Furthermore, recombinant GFP-fused Cdk11 but not p58 bound at spindle poles in egg extracts (Fig. S2, E and F). These results indicated that in contrast to p58, Cdk11 localizes to mitotic centrosomes and MTs.
Cdk11 is essential for normal spindle assembly
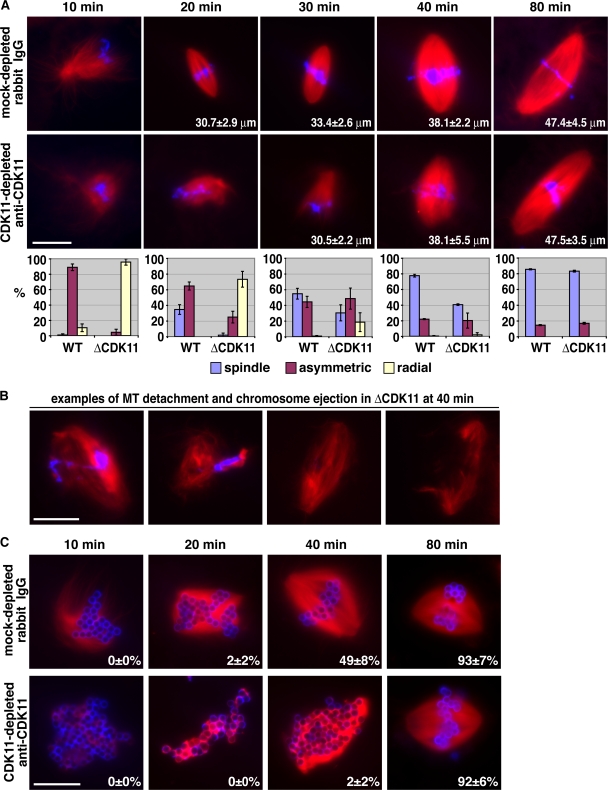
To address the physiological significance of RanGTP/Cdk11-dependent MT stabilization, we examined the effect of Cdk11 inhibition on spindle assembly in Xenopus egg extracts. After Cdk11 depletion and addition of anti-Cdk11 antibodies, apparently normal spindles eventually formed around sperm nuclei, but they formed much slower than in mock-depleted extracts (Fig. 3 A). In Cdk11-inactivated extracts, the initial asymmetrical MT growth toward chromosomes was strongly inhibited (Fig. 3 A). At later time points (20 and 30 min), asymmetrical MT organization and spindle assembly began (Fig. 3 A). The spindle structures that formed were frequently unstable with detached MTs and ejected chromosomes (Fig. 3 B). These defects were not detected in control conditions, including mock-depleted/rabbit IgG, mock-depleted/anti-Cdk11 antibody, or Cdk11-depleted/rabbit IgG extracts (Fig. 3 A and not depicted). Moreover, when we added back recombinant Cdk11 to a Cdk11-depleted extract together with anti-Cdk11 antibody, normal spindle assembly was restored, confirming the specificity of the inhibition (unpublished data). These results indicated that Cdk11 is important for the initial asymmetrical growth of centrosomal MTs toward chromosomes, MT–chromosome interactions, and normal spindle assembly rate.
Figure 3.
Cdk11-dependent MT stabilization is required for normal spindle assembly rate. (A and C) Representative structures observed in mock- or Cdk11-depleted extracts supplemented with 0.44 mg/ml rabbit IgG or anti-Cdk11 antibodies, respectively. Cycled spindles were assembled in the presence of Cy3-labeled tubulin (red), fixed at the indicated time points by squashing, and stained with Hoechst 33342 (blue). These experiments were reproduced three times. (A) Sperm spindle assembly. The numbers inside images represent the mean length of spindles. n > 30 spindles. The graphs below the images represent the percentage of structures observed according to the code indicated. n > 50 structures; n > 2 experiments. Error bars represent SD. WT, wild type. (B) Examples of abnormal sperm spindle structures observed in a Cdk11-inactivated extract at 40 min. They represent 36 ± 5% of all structures observed (47 ± 3% of spindles) and were not incorporated in the graph in A. (C) DNA bead spindle assembly. The numbers inside the images represent the percentages of bipolar spindles observed over the total number of structures counted. n > 50 structures; n > 2 experiments. Bars, 20 μm.
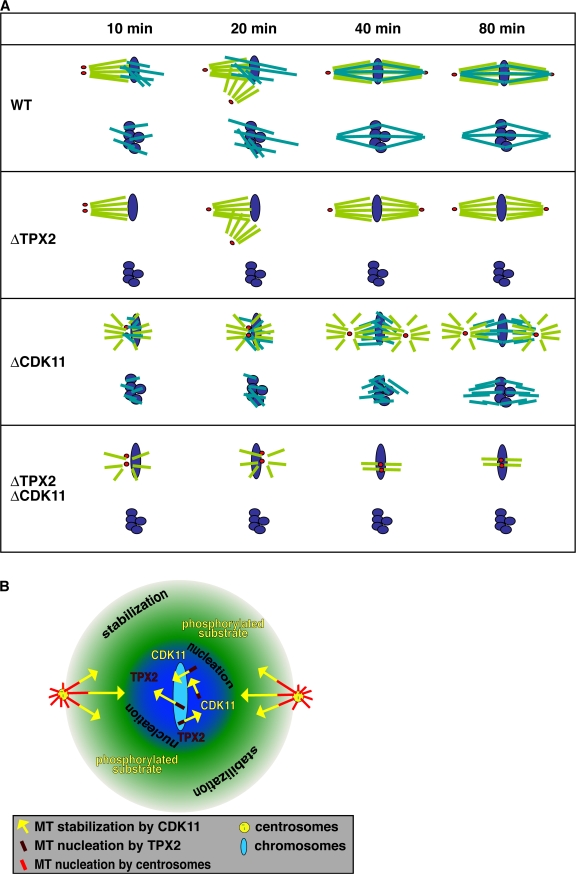
The aforementioned experiments suggested that Cdk11 inactivation primarily affected centrosomal MTs and that under such conditions, spindles arose mostly by self-organization of TPX2-dependent chromosomal MTs. Therefore, we examined the effect of Cdk11 inactivation on chromatin bead–induced spindle assembly. Chromatin bead spindles also assembled slower in Cdk11-inactivated extracts than in controls. Few fully formed spindles (2%) were first visible after 40 min, whereas in control experiments, 49% of the structures recorded at this time were bipolar spindles (Fig. 3 C). Both the number and length of MTs nucleated around chromatin beads before 40 min were dramatically reduced compared with the control (Fig. 3 C). On Cdk11 inhibition, spindles formed between 40 and 80 min from a mass of disorganized, very short MTs. These results indicated that Cdk11 is required for the elongation of chromosome-induced MTs and that this is an important parameter determining the rate of spindle assembly. Therefore, Cdk11 regulates the length of both centrosomal and chromosomal MTs in the vicinity of chromosomes. In the absence of CDK11, MTs are shorter, and, as a result, spindle self-organization takes longer.
The centrosomal spindle assembly pathway requires CDK11 activity
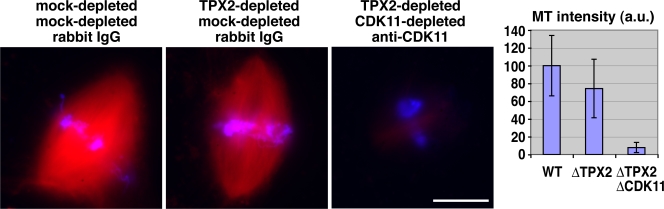
In the absence of TPX2, DNA bead spindles do not form because no MTs are nucleated (Gruss et al., 2001). In the presence of centrosomes, spindles do form around sperm nuclei in the absence of TPX2, although they contain fewer MTs (Fig. 4; Wittmann et al., 2000; Gruss et al., 2002; Caudron et al., 2005). This suggested that the stabilization of centrosomal MTs by Cdk11 might be indispensable for such centrosome-dependent spindle assembly. Indeed, when both Cdk11 and TPX2 were inactivated, no spindles formed around sperm nuclei (Fig. 4).
Figure 4.
Cdk11 is essential for spindle assembly in the absence of TPX2. Representative spindle structures observed under the indicated conditions. Cycled sperm spindles were assembled in the presence of Cy3-tubulin (red), fixed at 80 min, centrifuged onto coverslips, and stained with Hoechst 33342 (blue). MT intensity in spindles was quantified using a macro. Error bars represent SD. n > 50 spindles. This experiment was reproduced three times. WT, wild type. Bar, 20 μm.
Cdk11 was previously reported to be involved in transcription and pre-mRNA splicing (Trembley et al., 2002; Hu et al., 2003). Cdk11 knockout in mice resulted in early embryonic lethality. Cells within the embryos exhibited both proliferative defects and mitotic arrest followed by apoptosis. Consistently, Cdk11 RNAi in HeLa cells induced abnormal spindle assembly, mitotic arrest by checkpoint activation, and cell death (Petretti et al., 2006; Hu et al., 2007). In those knockout and RNAi experiments, both isoforms of Cdk11 (full-length Cdk11 and p58) were down-regulated because they are translated from the same mRNA (Cornelis et al., 2000).
In this study, we found Cdk11 but not p58 in the fractions containing the stabilization activity (Table I). This is reasonable because only Cdk11 bears NLS sequences. Xenopus Cdk11 but not p58 localizes to spindle poles and MTs in cultured cells and egg extracts. Moreover, the antibody used for our depletion experiments efficiently recognizes Cdk11 but not p58. Therefore, the phenotypes we report in egg extracts are caused by the lack of full-length Cdk11 protein. In our hands, immunodepletion of Cdk11 alone was insufficient to eliminate its function. Although Cdk11 seemed efficiently depleted from M-phase extracts, it is difficult to estimate how much remained and to determine how much Cdk11 is actually required to maintain spindle assembly. The inactivation results nevertheless appear specific and are validated by several controls. They are also further supported by the strong and specific phenotype caused by the simultaneous inactivation of Cdk11 and TPX2.
The most obvious consequence of Cdk11 inactivation in egg extracts is a significant delay in spindle assembly associated with the generation of abnormally short MT bundles during the early phases of spindle assembly. The slow-forming spindles probably self-organize from short MTs that are constantly nucleated close to chromosomes by TPX2 and aligned by motors (Fig. 5 A; Burbank et al., 2007). Also in Cdk11-inactivated extracts, we observed the frequent ejection of chromosomes during the assembly of spindles (Fig. 3 B). This indicates that Cdk11-dependent MT stabilization is required to allow stable MT attachment to chromosomes. This idea is consistent with the previous report that Cdk11 depletion in HeLa cells led to mitotic checkpoint activation and suggested improper MT–kinetochore attachment (Petretti et al., 2006).
Figure 5.
Schematic interpretation of the effects of Cdk11 on spindle MTs. (A) Spindle assembly around sperm nuclei and DNA beads. Sperm spindles: in wild-type (WT) extracts, both centrosomal and chromosomal MTs are stabilized by Cdk11 and contribute to spindle assembly. Initially, centrosomal MTs grow asymmetrically toward chromosomes through a Cdk11-dependent MT stabilization. In the absence of TPX2, centrosomal MTs still grow asymmetrically and form a bipolar spindle. In the absence of Cdk11, centrosomal MTs are not stabilized but interact with short chromosomal MTs. The MT populations become organized by cross-linking motors. When both TPX2 and Cdk11 are inactivated, there are no chromosomal MTs, and centrosomal MTs are too short and dynamic to be organized by cross-linking motors. Red dots, centrosomes; purple ovals, chromosomes; light green, centrosomal MTs; blue green, TPX2-dependent chromosomal MTs. DNA bead spindles: in wild-type extract, spindles form exclusively from TPX2 nucleated chromosomal MTs. In the absence of TPX2, no MTs are formed. In the absence of Cdk11, short and dynamic MTs assemble and spindle assembly is slowed down, but cross-linking motors finally organize them into a normal spindle containing short MTs. When both TPX2 and Cdk11 are inactivated, no MTs are nucleated. Purple, DNA beads; blue green, TPX2-dependent chromosomal MTs. (B) Hypothetical Cdk11 stabilization gradient. Cdk11 is released from importins and activated around chromosomes approximately over the same area as other NLS molecules like TPX2. However, its phosphorylated substrate that stabilizes MTs can diffuse further away before being dephosphorylated by the global opposing phosphatase. Yellow arrows indicate the increased length of MTs caused by the local activity of Cdk11.
In extracts in which TPX2 and Cdk11 were inactivated, sperm spindles did not form at all. This suggests that centrosomal MTs, even though they are constantly nucleated, are too short and dynamic to self-organize into proper spindles and that they have to be locally stabilized by the chromosome-dependent factor, Cdk11 (Fig. 5 A). This is interesting given that chromosomal MTs alone can assemble spindles in the absence of Cdk11, albeit slowly (Fig. 5 A). Thus, Cdk11 is essential for centrosomal MTs but is not necessary for chromosomal MTs to self-organize into spindles.
Recombinant Cdk11 has MT stabilization activity in the depleted extracts. However, it is necessary to add ∼1 μM of this protein to achieve maximum stabilization. This amount is significantly higher than the estimated Cdk11 amount in the purified fractions and endogenous Cdk11 concentration in M-phase extracts (∼200 nM; unpublished data). This suggests that the recombinant protein is not fully active, possibly as a result of the lack of one or more components from the purified fractions. It is possible that cyclin L regulates Cdk11 in spindle assembly, but this remains to be investigated. The function of Cdk11 depends on its kinase activity, suggesting that a Cdk11 substrate must be involved in the regulation of MT dynamics. Because a fraction of Cdk11 localizes to spindle MTs and centrosomes, it may bind to MTs through interaction with its substrate. Moreover, the Cdk11 released from importins by RanGTP around chromosomes supposedly forms a soluble gradient of active kinase congruent with the free NLS protein gradient (Fig. 5 B; Caudron et al., 2005). The phosphorylated Cdk11 substrate may form a second, more extended gradient than that of the active Cdk11 itself because of its own diffusion before its dephosphorylation by a phosphatase (Fig. 5 B; Bastiaens et al., 2006; Kholodenko, 2006).
In summary, the physiological role of the RanGTP-dependent activation of Cdk11 around chromosomes is to stabilize both centrosomal and chromosomal MT plus ends locally (Fig. 5 B). This is required for a spindle assembly rate compatible with successful completion of a cell cycle.
Materials and methods
Xenopus egg extracts, spindle assembly, immunodepletion, and antibody addition
Xenopus M-phase egg extracts (M-phase extracts; cytostatic factor arrested) were prepared as described previously (Murray, 1991). Cycled spindle assembly, immunodepletion, and antibody addition were performed as described previously (Wittmann et al., 2000; Hannak and Heald, 2006). In antibody addition experiments, rabbit IgG (Sigma-Aldrich) or anti–full-length Cdk11 antibody was added to extracts at 0.2–0.5 mg/ml.
RanGTP-dependent MT stabilization assay
A standard assay reaction contained 10 μl of M-phase extract, 1 μM Cy3-labeled tubulin, 0.15 mg/ml anti–full-length TPX2 antibody, and 2,000 isolated centrosomes per microliter in the presence or absence of 12–32 μM RanQ69L-GTP. Samples were incubated at 20°C for 30 min, fixed with 1 ml 0.25% glutaraldehyde, 10% glycerol, and 0.1% Triton X-100 in BRB80 (80 mM K-Pipes, 1 mM MgCl2, and 1 mM EGTA, pH 6.8), and spun down onto 12-mm round coverslips through a cushion of 25% glycerol in BRB80. The coverslips were postfixed with cold methanol for 10 min at −20°C, washed with PBS, and mounted on slides. Images were acquired using a microscope (Axiovert 200M; Carl Zeiss, Inc.), a plan-Neofluar 40× NA 1.3 oil objective lens (Carl Zeiss, Inc.), a Cy3 emission filter, a camera (AxioCam HRm; Carl Zeiss, Inc.), and AxioVision software (Carl Zeiss, Inc.). The mean MT length of centrosomal asters was quantified using a macro written in Matlab (The MathWorks; Fig. S1).
Preparation of recombinant proteins and affinity beads
His-RanQ69L, His-RanT24N, His–importin β, His–importin α, and His-ED mutant of importin α were expressed in bacteria and purified with talon beads (BD Biosciences). Loading of GTP on RanQ69L was described previously (Weis et al., 1996). Cyclin B Δ90 was prepared as described previously (Glotzer et al., 1991).
Saturating amounts of z tag-RanQ69L or GST–importin β bacterial lysate were incubated with IgG Sepharose or glutathione Sepharose, respectively. The beads were washed with cytostatic factor extract buffer (CSF-XB; 10 mM K-Hepes, 100 mM KCl, 3 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 50 mM sucrose, pH 7.7). To prepare GST–importin β/α or GST–importin β/ED beads, excess purified recombinant His–importin α or His-ED mutant was incubated with the GST–importin β lysate at 4°C for 1 h. Then, the mixture was incubated with glutathione Sepharose at 4°C for 1 h. The beads were washed with CSF-XB.
Preparation of the NLS protein fraction and the depleted extract
M-phase extracts with 10 μg/ml cyclin B Δ90 were incubated with a 40% wet bead volume of z-RanQ69L beads in a Mobicol column (MoBiTec) at 4°C for 1 h with rotation. The beads were collected by centrifugation, and the remaining extract (activated extract) was incubated with a 120% wet bead volume of GST–importin β beads at 4°C for 1 h with rotation. After centrifugation, the extract (depleted extract) was frozen in 50-μl aliquots in liquid nitrogen and stored at −80°C. The GST–importin β beads were washed five times with wash buffer (CSF-XB, 100 mM KF, 80 mM β-glycerophosphate, 0.1 mM sodium vanadate, 0.03% digitonin, and 1 mM DTT) and eluted with elution buffer (20 μM His-RanQ69L-GTP, 500 mM NaCl, 1 mM GTP, 1 mM ATP, and 10% glycerol in the wash buffer) at 4°C overnight with rotation. The NLS protein fraction was recovered as the supernatant by centrifugation and dialyzed to buffer A (CSF-XB, 10% glycerol, and 1 mM DTT). The fraction was directly used for experiments or stored in aliquots at −80°C.
Purification of MT stabilization activity from the NLS protein fraction
40 ml of the NLS protein fraction prepared from 100 ml of M-phase extracts was applied to a 1-ml Mono S column. The bound proteins were eluted with 10 ml of a 100–1,000-mM KCl gradient in buffer A and fractionated into 10 fractions. The MT stabilization activity of each fraction, which had been dialyzed to buffer A, was assayed in the depleted extract supplemented with centrosomes, anti-TPX2 antibodies, and Cy3-labeled tubulin at 20°C for 30 min. The pooled Mono S active fractions (fractions 3–7; 5 ml; ∼300 mM KCl) were adjusted to 350 mM KCl and applied to a 0.1-ml Mono Q column. The bound proteins were eluted with 1 ml of a 350–1,000-mM KCl gradient in buffer A and fractionated into 10 fractions. The pooled Mono Q active fractions (fractions 3–6; 0.4 ml; ∼400 mM KCl) were concentrated by Centricon 10 (Millipore), applied to a 2.4-ml Superdex 200 column, eluted with buffer A + 100 mM KCl, and fractionated (40 μl). The Superdex 200 fractions were separated on SDS-PAGE, and the gel was stained with Cypro ruby. Selected bands in the active fractions were cut and digested with trypsin (Shevchenko et al., 1996). The resulting peptide mixture was analyzed by matrix-assisted laser desorption/ionization time of flight (Uraflex; Bruker Daltonics) and liquid chromatography tandem mass spectrometry (liquid chromatography, Eksigent NanoLC_1D; mass spectrometry, Qstar Pulsar I; Applied Biosystems). Proteins were identified using the Mascot search tool.
Cloning and expression of Xenopus Cdk11
A cDNA clone (IMAGE:5073384; RZPD) covering the complete Xenopus Cdk11 cDNA was amplified by PCR and cloned into NcoI–XhoI sites of pFastBac HTa (Invitrogen). Baculoviruses were prepared according to the manufacturer's instructions. Recombinant Cdk11 was expressed in Sf9 insect cells and purified on Talon beads and the Mono Q column. The K460R mutant of Cdk11, in which lysine 460 was replaced with arginine, was generated by site-directed mutagenesis using PfuTurbo DNA polymerase (Stratagene). The K460R mutant, N-terminal Cdk11 (1–349 aa), and p58 Cdk11 (350–788 aa) were constructed, expressed, and purified as described for Cdk11.
Antibodies
Rabbit polyclonal antibodies against Xenopus full-length TPX2, full-length Cdk11 (1–788 aa), N-terminal Cdk11 (1–349 aa), and p58 Cdk11 (350–788 aa) were prepared and purified against recombinant proteins according to a standard protocol. Antinucleoplasmin monoclonal antibody was purchased from Iowa University.
Online supplemental material
Fig. S1 shows the quantification method to determine astral MT length used in the purification assay, identification of the TPX2 antibody that blocks MT nucleation, and specific inhibition of the MT stabilization activity by the importin α/β heterodimer. Fig. S2 shows that recombinant Cdk11 has RanGTP-dependent MT stabilization activity but that a kinase-dead mutant does not. Fig. S3 shows the localization of Cdk11 at spindle poles and on spindle MTs in Xenopus tissue culture cells and egg extracts. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706189/DC1.
Supplemental Material
Acknowledgments
We thank F. Nedelec for writing the Matlab macros used for aster and spindle analysis, Y. Yoneda for a GST–mouse importin β plasmid, M. Glotzer for a cyclin B Δ90 plasmid, I. Vernos for sharing reagents, S. Kandels-Lewis for help with Cdk11 cloning, and members of the Karsenti laboratory for discussion. We also thank Carl Zeiss for continuous support of the European Molecular Biology Laboratory Advanced Light Microscopy Facility.
H. Yokoyama was supported by the Japan Society for the Promotion of Science and by European Molecular Biology Laboratory postdoctoral fellowships.
M. Caudron's present address is Deutsches Krebsforschungszentrum B066, 69120 Heidelberg, Germany.
Abbreviations used in this paper: CSF-XB, cytostatic factor extract buffer; M-phase, metaphase; MT, microtubule.
References
- Bastiaens, P., M. Caudron, P. Niethammer, and E. Karsenti. 2006. Gradients in the self-organization of the mitotic spindle. Trends Cell Biol. 16:125–134. [DOI] [PubMed] [Google Scholar]
- Berke, J.D., V. Sgambato, P.P. Zhu, B. Lavoie, M. Vincent, M. Krause, and S.E. Hyman. 2001. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron. 32:277–287. [DOI] [PubMed] [Google Scholar]
- Burbank, K.S., T.J. Mitchison, and D.S. Fisher. 2007. Slide-and-cluster models for spindle assembly. Curr. Biol. 17:1373–1383. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., G. Guarguaglini, O.J. Gruss, A. Segref, E. Karsenti, and I.W. Mattaj. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., O.J. Gruss, I.W. Mattaj, and E. Karsenti. 2001. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3:228–234. [DOI] [PubMed] [Google Scholar]
- Caudron, M., G. Bunt, P. Bastiaens, and E. Karsenti. 2005. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 309:1373–1376. [DOI] [PubMed] [Google Scholar]
- Cornelis, S., Y. Bruynooghe, G. Denecker, S. Van Huffel, S. Tinton, and R. Beyaert. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell. 5:597–605. [DOI] [PubMed] [Google Scholar]
- Glotzer, M., A.W. Murray, and M.W. Kirschner. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., R.E. Carazo-Salas, C.A. Schatz, G. Guarguaglini, J. Kast, M. Wilm, N. Le Bot, I. Vernos, E. Karsenti, and I.W. Mattaj. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin on TPX2 activity. Cell. 104:83–93. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., M. Wittmann, H. Yokoyama, R. Pepperkok, T. Kufer, H. Sillje, E. Karsenti, I.W. Mattaj, and I. Vernos. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4:871–879. [DOI] [PubMed] [Google Scholar]
- Hannak, E., and R. Heald. 2006. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1:2305–2314. [DOI] [PubMed] [Google Scholar]
- Hu, D., A. Mayeda, J.H. Trembley, J.M. Lahti, and V.J. Kidd. 2003. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 278:8623–8629. [DOI] [PubMed] [Google Scholar]
- Hu, D., M. Valentine, V.J. Kidd, and J.M. Lahti. 2007. CDK11(p58) is required for the maintenance of sister chromatid cohesion. J. Cell Sci. 120:2424–2434. [DOI] [PubMed] [Google Scholar]
- Kalab, P., R.T. Pu, and M. Dasso. 1999. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9:481–484. [DOI] [PubMed] [Google Scholar]
- Kalab, P., K. Weis, and R. Heald. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 295:2452–2456. [DOI] [PubMed] [Google Scholar]
- Kalab, P., A. Pralle, E.Y. Isacoff, R. Heald, and K. Weis. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 440:697–701. [DOI] [PubMed] [Google Scholar]
- Kholodenko, B.N. 2006. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 7:165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A.W. 1991. Cell cycle extracts. In Xenopus laevis: Practical Uses in Cell and Molecular Biology. Vol. 36. B.K. Kay and H.B. Peng, editors. Academic Press, San Diego. 581–605.
- Nachury, M.V., T.J. Maresca, W.C. Salmon, C.M. Waterman-Storer, R. Heald, and K. Weis. 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106. [DOI] [PubMed] [Google Scholar]
- Ohba, T., M. Nakamura, H. Nishitani, and T. Nishimoto. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 284:1356–1358. [DOI] [PubMed] [Google Scholar]
- Petretti, C., M. Savoian, E. Montembault, D.M. Glover, C. Prigent, and R. Giet. 2006. The PITSLRE/CDK11p58 protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO Rep. 7:418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- Solomon, M.J., T. Lee, and M.W. Kirschner. 1992. Role of phosphorylation in p34cdc2 activation: identification of an. Mol. Biol. Cell. 3:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley, J.H., D. Hu, L.C. Hsu, C.Y. Yeung, C. Slaughter, J.M. Lahti, and V.J. Kidd. 2002. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem. 277:2589–2596. [DOI] [PubMed] [Google Scholar]
- Weis, K., C. Dingwall, and A.I. Lamond. 1996. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Wilde, A., and Y. Zheng. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 284:1359–1362. [DOI] [PubMed] [Google Scholar]
- Wilde, A., S.B. Lizarraga, L. Zhang, C. Wiese, N.R. Gliksman, C.E. Walczak, and Y. Zheng. 2001. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3:221–227. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., M. Wilm, E. Karsenti, and I. Vernos. 2000. TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149:1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., M. Hughes, and P.R. Clarke. 1999. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J. Cell Sci. 112:2453–2461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.