Abstract
Human embryonic stem cells (hESCs) are potential therapeutic tools and models of human development. With a growing interest in primary cilia in signal transduction pathways that are crucial for embryological development and tissue differentiation and interest in mechanisms regulating human hESC differentiation, demonstrating the existence of primary cilia and the localization of signaling components in undifferentiated hESCs establishes a mechanistic basis for the regulation of hESC differentiation. Using electron microscopy (EM), immunofluorescence, and confocal microscopies, we show that primary cilia are present in three undifferentiated hESC lines. EM reveals the characteristic 9 + 0 axoneme. The number and length of cilia increase after serum starvation. Important components of the hedgehog (Hh) pathway, including smoothened, patched 1 (Ptc1), and Gli1 and 2, are present in the cilia. Stimulation of the pathway results in the concerted movement of Ptc1 out of, and smoothened into, the primary cilium as well as up-regulation of GLI1 and PTC1. These findings show that hESCs contain primary cilia associated with working Hh machinery.
Introduction
Driving human embryonic stem cells (hESCs) along specific differentiation pathways remains a significant challenge for translational medicine and the development of hESC therapies. During early embryology, signaling pathways, such as hedgehog (Hh) and Wnt, are critical for human development (Corbit et al., 2005; Haycraft et al., 2005; Huangfu and Anderson, 2005; Liu et al., 2005; May et al., 2005) and, recently, have been shown to be mediated by the primary cilium (for reviews see Michaud and Yoder, 2006; Singla and Reiter, 2006; Christensen et al., 2007; Satir and Christensen, 2007). Therefore, in the search for mechanisms regulating hESC differentiation, it is vital to first establish the existence of primary cilia and the localization of signaling components in undifferentiated hESCs.
Primary cilia are single, generally nonmotile, cilia with a 9 + 0 axoneme, differing from the 9 + 2 arrangement of motile cilia. Primary cilia are implicated as key cellular sensory structures involved in signal transduction and coordination of intra- and intercellular signaling pathways (for reviews see Michaud and Yoder, 2006; Singla and Reiter, 2006; Christensen et al., 2007; Satir and Christensen, 2007). Signaling in primary cilia is thought to be initiated by receptors positioned within the cilium and relayed through transcription factors, which may become activated directly in the cilium or in the cell body via basal body scaffold proteins. Specific growth factor receptors in the primary cilium, such as PDGF receptor-α, enable the cell to respond differentially to ligands and to initiate cell division (Schneider et al., 2005).
Mutations giving rise to defective primary cilia or improper placement of signaling molecules within the cilium result in a plethora of clinical manifestations (Pazour, 2004; Badano et al., 2006). These include obesity, rod–cone dystrophy, renal abnormalities, polycystic kidney disease, polydactyly, genital abnormalities, learning disabilities, congenital heart disease, hearing loss, situs inversus, and Bardet-Biedl syndrome (Blacque and Leroux, 2006). In particular, mutations in genes encoding intraflagellar transport proteins impair Hh signaling and result in limb bud and neural tube defects, which are similar to those seen in Hh signaling mutations (Corbit et al., 2005; Haycraft et al., 2005; Huangfu and Anderson, 2005; Liu et al., 2005; May et al., 2005). Hh signaling is essential during embryonic development for left–right asymmetry axis, limb and heart development, and neurogenesis (Corbit et al., 2005; Haycraft et al., 2005; Huangfu and Anderson, 2005; Liu et al., 2005; May et al., 2005). In the adult, Hh signaling is involved in stem cell maintenance and tissue homeostasis.
We hypothesized that primary cilia might be found in hESCs, wherein they could play a critical role in hESC differentiation parallel to that in normal early embryogenesis. In this study, we demonstrate that primary cilia are a general feature of hESC lines and that essential signaling components of the Hh pathway are present and functional in primary cilia of undifferentiated hESCs. Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) images provide definitive evidence and reveal novel features of hESCs and their primary cilia. To date, this is the first study conclusively showing the presence of these unique organelles in hESCs by definitive confocal and electron micrographs of hESC primary cilia and by dynamic colocalization of key signaling molecules essential for early development and known to be functional in the Hh signaling pathway, as was recently demonstrated in primary cilia of cultured mouse fibroblasts (Rohatgi et al., 2007).
Results and discussion
In this study, we demonstrate that the primary cilium is a dynamic ultrastructural feature in three different lines of undifferentiated hESCs. The presence of this organelle is not limited to specific culture conditions. HESCs from H1 (male) and H9 (female) lines (approved by the National Institutes of Health; Olivier et al., 2006) were grown on matrigel without feeder cells (described in Yao et al. [2006]) with serum replacement for 6 d. Primary cilia were first identified by immunofluorescence (IF) markers of acetylated tubulin (AcTb) in both H1 and 9 hESCs after 6 d of culture in DME:F12 with serum replacement (Figs. 1 A and 2 A). Primary cilia became more prominent after starvation of hESCs by placement in DME:F12 without serum replacement for 24 h (Fig. 1 B). Another hESC line, LRB003 (female; studied in the Denmark laboratory and supported by funds independent of the National Institutes of Health; Laursen et al., 2007), was cultured in monolayers on 0.1% gelatin with conditioned medium from cultured human foreskin fibroblasts (hFF), and primary cilia were observed after 4 d as the cells entered growth arrest in confluent colonies in the culture dish (Fig. 1, D and E).
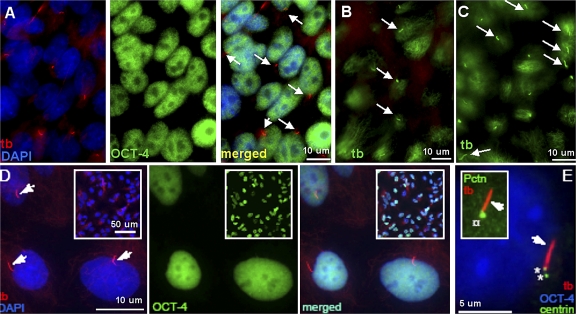
Figure 1.
Immunolabeling of primary cilia in undifferentiated hESCs. (A) Characterization of undifferentiated colonies of H1 hESCs grown on matrigel for 5 d in DME:F12 with serum replacement. Undifferentiated cells are identified by nuclear colocalization of anti–OCT-4 (OCT-4, green) and DAPI (dark blue) in the merged image (light blue). More than 97% of cells on average expressed OCT-4 in a nuclear pattern indicating their undifferentiated state. Primary cilia stained with anti-AcTb (tb, red) are indicated by arrows. (B) H1 hESCs grown on matrigel for 5 d in DME:F12 with serum replacement (i.e., unstarved), labeled with anti-AcTb (tb, green) to show primary cilia (arrows). (C) H1 hESCs grown on matrigel for the same period of time in DME:F12 without serum replacement (i.e., starved) for 24 h and labeled with anti-AcTb (tb, green). Primary cilia are indicated by arrows. (D) Characterization of undifferentiated colonies of LRB003 hESC grown on 0.1% gelatin with conditioned medium. Undifferentiated cells are identified by nuclear colocalization of anti–OCT-4 (OCT-4, green) and DAPI (dark blue) in the merged image (light blue). Anti-AcTb (tb, red) marks the primary cilia (arrows) as well as the microtubular network in the cytoplasm. Less than or equal to 95% of the cells were ciliated and positive for OCT-4. Primary cilia are not easily visualized at the low resolution images (as shown in the insets). (E) A primary cilium (tb, red, arrow) in undifferentiated LRB003 hESCs emerges from one of the centrioles (asterisks) marked with anti-centrin (centrin, green). Nuclear localization of anti–OCT-4 (OCT-4, blue) denotes that the cell has not differentiated. The inset shows anti-pericentrin (Pctn, green) marking the centrosome (¤) at the base of the primary cilium (arrow).
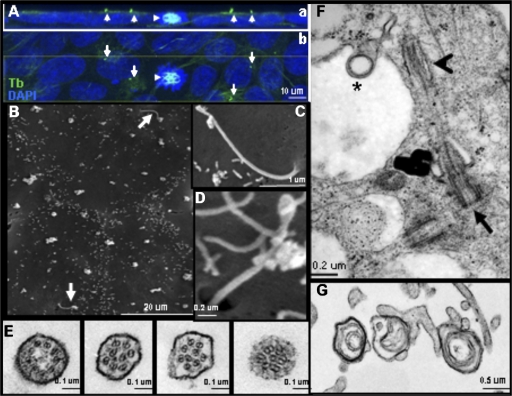
Figure 2.
Electron microscopy of hESC primary cilia. (A) Confocal microscopy of hESCs grown on matrigel in a monolayer viewed from the side (a) and the top (b), showing primary cilia (arrows) on undifferentiated hESCs but not on dividing cells (arrowhead). (B) SEM of H1 cells grown for 7 d on matrigel in N2/B27-supplemented medium and starved in DME:F12 without serum replacement for the last 24 h of growth. See Online supplemental material for details. Primary cilia (arrows) are seen on two adjacent cells among numerous smaller microvilli. (C and D) Enlarged images of primary cilia. Note paddle tips. (E) TEM cross sections of primary cilia of H1 and 9 hESCs grown for 7 d on matrigel in N2/B27-supplemented medium and starved in DME:F12 without serum replacement for the last 24 h before fixation. The first panel shows the 9 + 0 arrangement, which becomes disorganized along the ciliary length as shown on the rest of the panels. (F) TEM longitudinal section of a primary cilium (arrowhead) emerging from one of a pair of centrioles/basal bodies. The perpendicular orientation of the basal bodies suggests that the cilium arises from the mother centriole (arrow). A lamellar vesicle (asterisk) is seen budding from the cell surface. (G) TEM of lamellar-type bodies secreted by hESCs.
Confirmation that the hESCs remained undifferentiated was made by IF using the transcription factor OCT-4 (Fig. 1, A, D, and E) and stage-specific embryonic antigen 4 (not depicted). Both markers were used to assure undifferentiated hESCs. Anti-AcTb identified potential primary cilia (Fig. 1, B and C) and antibodies against tumor rejection antigen 1-85 (Tra-1-85)–marked human cells (see Fig. 3 D). After 5 d in culture, short (∼2–3 μm) AcTb extensions characteristic of primary cilia were seen on ∼33% of H1 hESCs (25 cilia/75 cells counted from five different fields of colonies on a plate; Fig. 1 B). In the remaining H1 cells, anti-AcTb was often seen as a single concentrated spot, which likely represents a very short cilium of length <1 μm. When cultures were starved in DME:F12 alone (without serum replacement) for 24 h, the AcTb extensions increased in length to ∼4–6 μm, and the number of cells with these extensions increased to ∼50% (41/80 cells, counted as the H1 hESCs; Fig. 1 C). H9 behaved essentially similarly (unpublished data).
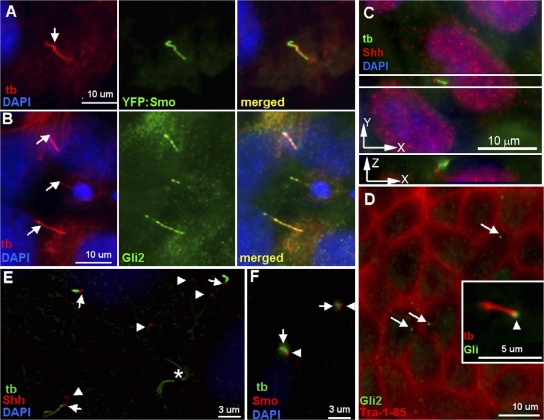
Figure 3.
Hh signaling proteins localize to hESC primary cilia. (A) Localization of YFP:Smo (green) to the primary cilium (tb, red, arrow) in LRB003 hESCs. The merged image shows colocalization. Nuclei are stained with DAPI (blue). (B) Immunolocalization of Gli2 (green) to primary cilia (tb, red, arrows) in LRB003 hESCs. The merged image shows colocalization. Nuclei are stained with DAPI (blue). Gli data from H1 and LRB003 cells were obtained with different antibodies. (C) H1 cells grown for 7 d on matrigel in N2/B27-supplemented medium and labeled for primary cilia (tb, green) and anti-SHh (SHh, red). In addition, a z series of a field showing separate SHh labeling (red) located distinctly to the side of the primary cilium (green) is depicted. (D) Similar cells labeled with anti–Tra-1-85 (red) and anti-Gli2 (Gli2, green). Arrows indicate punctuate localization of the Gli2 protein (green dots) and the inset specifically localizes the Gli2 protein (arrowhead) at one end of a primary cilium (tb, red). (E) Anti-SHh (SHh, red) localizes near the base of most primary cilia. 33/71 cells possess primary cilia (46.5%), which is consistent with Fig. 1 C. 22/33 cells with primary cilia (66%) exhibit SHh near their base (arrows). SHh also localizes to points not associated with the cilia (arrowheads). The asterisk points to the midbody of cells that have recently undergone cytokinesis. These structures are not associated with Hh signaling molecules. (F) Anti-Smo (Smo, red) localizes predominantly to the base of primary cilia. This pattern of Smo expression is similar to that observed at 0 and 1 h of SAG stimulation (see Fig. 4). Arrows point to primary cilia (tb, green) and arrowheads indicate Smo localization.
In the LRB003 hESCs, after ∼7 d in culture and independent of starvation, primary cilia with lengths of 5–10 μm emerged as solitary organelles from >90% of the confluent cells (Fig. 1 D). To show unequivocally that cilia were present on undifferentiated LRB003 hESCs, we used triple colocalization with anti-AcTb, anti–OCT-4, and either anti-centrin or anti-pericentrin (Fig. 1 E). Single primary cilia (labeled by anti-AcTb) were shown to emanate from the centrosomes (labeled by anti-pericentrin; Fig. 1 E, inset) of OCT-4–positive cells and, in particular, from one of the two centrioles (labeled by anti-centrin; Fig. 1 E), probably the mother centriole, which also functions as the ciliary basal body.
SEM of H1 and 9 hESCs (95% OCT-4 positive) revealed single AcTb-positive projections ∼4–6 μm in length and ∼0.25 μm in diameter (characteristic of primary cilia) seen at the free surfaces of the cells, which is in contrast to the many microvilli that are shorter and have a smaller diameter (Fig. 2 B). SEM also demonstrated paddle tips at the ends of some primary cilia (Fig. 2, C and D). Confocal images (Fig. 2 A) show the outward orientation of primary cilium from growth-arrested cells in a monolayer, whereas mitotic cells lack a primary cilium (Pan and Snell, 2007).
To show definitively that the structures are primary cilia, we fixed hESC cultures in situ and processed them for TEM. Some colonies were cut parallel to and just above their free surfaces to give cross-sectional views of projecting structures, and other sections were oriented through the cell bodies perpendicular to this direction to show longitudinal views of the cilia and their basal bodies. Cross sections near the apical surfaces of the cells showed axonemes, which are enclosed by a unit membrane (Fig. 2 E). The 9 + 0 pattern can be clearly observed in cross sections close to the basal body, as are a disarray of nine doublets, including 8 + 1, 6 + 1, and other patterns (Fig. 2 E), either from the same cell but at different sections along the length of the cilia approaching the tip or in different cells at varying stages of ciliary growth. One centriole pair can also be observed close to the cell surface with a primary cilium growing from one of the centrioles (Fig. 2 F), which has become the ciliary basal body. Primary cilia often emerge from a concavity in the cell, which may be interpreted as a small depression in the cell's apical surface as shown in the SEM (Fig. 2 D) and TEM (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200706028/DC1) images. Rarely (in <1% of observed cells), two primary cilia originate within one cell (unpublished data). A rich array of polysomes and cytoplasmic microtubules, running parallel to the apical surface, are seen near the basal body (Fig. S1 A). Immediately below this level, a ciliary rootlet emerging from the basal body and microfilament bands of the adherens junctions of the confluent hESCs can be found (Fig. S2). In addition, lamellar-type vesicles are observed both intracellularly and extracellularly, adherent to the hESC surface (Fig. 2, F and G; and Fig. S1 B).
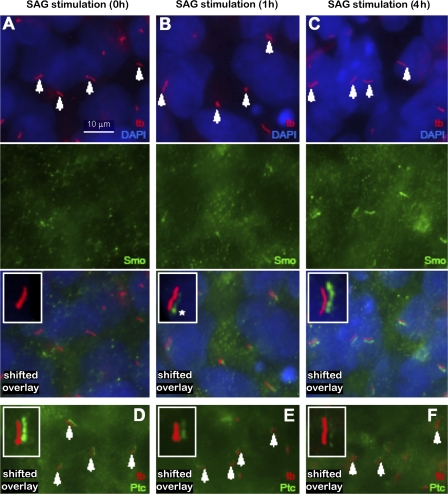
Next, we examined whether components of the Hh signaling system were present and functional in the hESC primary cilia. It has been reported previously that the sonic Hh (SHh) receptors patched (Ptc) and smoothened (Smo) and their downstream effectors Gli1, 2, and 3 are expressed in hESCs (Rho et al., 2006). In various cells, upon binding of SHh to its receptor Ptc, Smo is activated, which is followed by the processing and activation of Gli transcription factors that enter the nucleus to control differential processes during early and late embryogenesis. Smo was previously reported to be a constituent of nodal cilia, Madin Darby canine kidney cell cilia, and other primary cilia (Corbit et al., 2005; May et al., 2005), and Gli2 was found at the tip of mesenchymal primary cilia during limb formation (Haycraft et al., 2005). Time-dependent studies in mammalian differentiated cells support a model in which SHh triggers the removal of Ptc from the primary cilium, permitting Smo to enter the cilium and initiating signaling (Rohatgi et al., 2007). We therefore tested whether Smo, Ptc, and Gli2 are present in hESC primary cilia, and we followed the movement of Smo and Ptc in and out of the cilium upon stimulation by Smo agonist (SAG). The use of SAG to induce activation of SHh signaling has been established by Chen et al. (2002). In transfected LRB003 hESCs, YFP:Smo strongly and almost exclusively localizes to the primary cilium (Fig. 3 A). The ciliary staining of YFP:Smo was remarkably higher than that of anti-Smo (Fig. 4 A) because of overexpression of Smo from the construct. Furthermore, with a Gli2-specific antibody, we show that Gli2 strongly localizes in a punctuate pattern along the entire length of primary cilia but is absent in the nucleus of these cells (Fig. 3 B). Also, in H1 and 9 hESCs, anti-Gli2 localizes to the primary cilia (Fig. 3, B and D), whereas Smo localizes to the base of ∼3/4 of the cells with primary cilia (Fig. 3 F). In addition, by fluorescence immunolocalization, small amounts of SHh can be localized near the base of the cilia, which is clearly located to the side of the primary cilium (Fig. 3 C, z series) in ∼2/3 of the ciliated H1 cells (Fig. 3, C and E). In LRB003 cells, upon stimulation with SAG, the ciliary level of Smo starts to increase beginning at 1 h (Fig. 4 B) as compared with 0 h (Fig. 4 A). This is followed by a major accumulation of Smo along the length of the cilium at 4 h of SAG treatment (Fig. 4 C). This infers that translocation of Smo along the cilium is initiated by the docking of Smo at the base of the cilium. The opposite pattern of translocation can be seen for Ptc, which leaves the cilium upon SAG stimulation (Fig. 4, D–F). This movement of Hh components into and out of the cilium (Fig. 4), along with the z series showing SHh located to the side of the primary cilium (Fig. 3 C), eliminates the possibility of nonspecific antibody binding to the centrosome in light of the fact that centrosomes never migrate up the cilium. Experiments similar to that described by Orozco et al. (1999) are planned for the direct viewing of intracellular and ciliary transport of intraflagellar transport motor proteins and their Hh cargoes in hESCs that would establish mechanisms of trafficking. Knockdown experiments, for example, using siRNA of KIF3A, would be informative and are presently underway.
Figure 4.
Translocation of Smo and Ptc in and out of the primary cilium after SAG stimulation. (A) Localization of anti-Smo (green) to the primary cilium of LRB003 cells (tb, red, arrows) at 0 h of SAG treatment. (B and C) Localization of anti-Smo to the primary cilium (arrows) at 1 and 4 h, respectively. The insets in A–C show high resolution (shifted overlays) of Smo (green) in the primary cilium (red). The asterisk indicates the base of the cilium. (D) Localization of anti-Ptc (green) to the primary cilium (tb, red, arrows) at 0 h of SAG treatment. (E and F) Localization of anti-Ptc at 1 and 4 h, respectively, to primary cilium (arrows). The insets in D–F show high resolution images (shifted overlays) of Ptc (green) in the primary cilium (red). Nuclei were stained with DAPI.
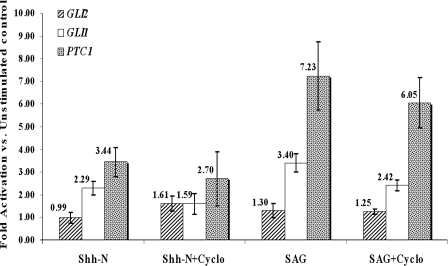
The addition of 5 μM SHh or 10 μg/ml SAG to H1 hESCs for 18 h up-regulated GLI1 (approximately twofold) and PTC1 (approximately fivefold) mRNA levels compared with baseline levels of these components without exogenous ligand stimulation, as determined by real-time PCR with GAPDH as an internal control (Fig. 5). As expected, GLI2 mRNA was essentially nonresponsive. Cyclopamine, a Smo inhibitor (Lipinski et al., 2006), modestly inhibited the up-regulation in the presence of inducers under the conditions used (Fig. 5). GLI2 mRNA was not affected. These data are consistent with the dynamics of the Hh signaling machinery, as described by Rohatgi et al. (2007), in differentiated cells and, together with the localization studies of Hh signaling proteins, support the conclusion that Hh signaling proceeds through hESC primary cilia. Whether or not the SHh ligand is produced by the hESC and whether the function of the signal is to maintain the cells undifferentiated or act as a precursor to differentiation remains to be determined.
Figure 5.
Up-regulation of mRNAs for Hh components in H1 HESCs by SHhN or SAG induction as determined by real-time PCR (Materials and methods). A comparison of the relative increase in mRNA compared with baseline levels (without stimulation) for the respective components is shown. This experiment was reproduced twice with essentially the same qualitative results, and the mean values are shown as numbers above each bar with the corresponding standard error bars, which represent the standard error of the mean. Each experiment used three pooled samples to obtain sufficient RNA for the technique.
The presence of the extracellular lamellar bodies in undifferentiated hESCs may also be related to Hh or other signaling pathways. Similar vesicles have been reported to be involved in signaling in association with cells with nodal or primary cilia in several embryonic tissues. It would be interesting if the lamellar vesicles seen here are indeed akin to nodal vesicular parcels containing SHh signals, as described in early embryonic nodal cells (Tanaka et al., 2005; Hirokawa et al., 2006), or to prominin-1–containing particles of dividing neuroepithelial cells of the developing mammalian central nervous system (Dubreuil et al., 2007). The content of the H1 lamellar vesicles remains to be investigated further.
In summary, definitive confocal and transmission electron micrographs, coupled with SEM and IF microscopy, conclusively demonstrate the presence of primary cilia with many known features in hESCs. For a detailed review of primary cilia ultrastructure in differentiated cells, see Satir and Christensen (2007). Because Hh signaling pathways of embryological development and patterning operate via primary cilia, it is perhaps not surprising to find Ptc, Smo, and Gli2 localized and potentially functional within the hESC primary cilia. Whether SHh is released in cultures before or after differentiation of hESCs is unclear, and whether important receptors of other signaling pathways, such as Wnt (Gerdes et al., 2007; Pan and Thomson, 2007), are localized in hESC primary cilia remains to be determined.
Collectively, our results suggest that primary cilia may be involved in the regulation and coordination of the first steps of hESC differentiation and/or the maintenance of the undifferentiated state/self-renewal. Because hESCs hold promise for the treatment of many diseases and provide an excellent system for studying mechanisms involved in early human development, these findings provide the groundwork to determine specific aspects of early differentiation controlled by the machinery of primary cilia. This knowledge may ultimately reveal pathways for manipulation of hESC differentiation into specific cell and tissue lineages.
Materials and methods
Cell cultures (Albert Einstein College of Medicine)
hESCs from H1 and 9 lines (National Institutes of Health approved) were maintained in a humidified incubator at 37°C with an atmosphere consisting of 6% CO2, 7% O2, and 87% N2 and were grown on matrigel (BD Biosciences) without feeder cells in DME nutrient mixture F12 (Ham; Invitrogen) with l-glutamine and 15 mM Hepes, supplemented with the serum replacements N2 (chemically defined supplement containing 1000 mg/liter human transferrin, 50 mg/liter insulin recombinant full chain, 0.6 mg/liter progesterone, 161 mg/liter putrescine, and 173 mg/liter selenite; Invitrogen) as 100× concentrate of Bottenstein's N2 formulation (Bottenstein, 1985) and B27 (50× serum supplement designed for the long-term viability of hippocampal and other neurons of the central nervous system; Invitrogen), in addition to 20 ng/ml of basic FGF (R&D Systems), BSA fraction V, 1% nonessential amino acids, 50 U/ml penicillin, 50 ng/ml streptomycin, 1 mM l-glutamine, and 1-thioglycerol added for 6 d (as described in detail in Yao et al. [2006]) and observed by phase microscopy using an inverted light microscope (CK40; Olympus). To passage the cells, differentiated cells were scraped in PBS under a binocular magnifier with a Pasteur pipette scraper (elongated and twisted using heat), treated with prewarmed collagenase type IV for 5 min to detach the hESC colonies, aspirated, concentrated using a macrocentrifuge (Eppendorf), and either plated on 6-well tissue culture plates (Thermo Fisher Scientific) coated with 1:4 matrigel for propagation, on gamma-irradiated 35-mm glass-bottom microwell dishes (MatTek Cultureware) covered with 1:4 matrigel for IF, or on carbon-coated glass coverslips on the bottom of each well of a 6-well plate covered with 1:4 matrigel for TEM or SEM. The cultures were monitored microscopically and at day 6 were either maintained for an additional day in the same supplemented medium or starved in plain DME:F12 for 24 h. The cells were then prepared for IF microscopy using the protocol described in IF Microscopy (Albert Einstein College of Medicine).
The LRB003 cell line (not approved by the National Institutes of Health) was studied, as described in the next section, in the Copenhagen laboratory and solely supported by Danish funding agencies (see Acknowledgments).
Cell cultures (Copenhagen)
The hESC line LRB003 (Laursen et al., 2007) was initially cultured on 35-mm dishes (Thermo Fisher Scientific) coated with 0.1% gelatin (Sigma-Aldrich) on a confluent layer of mitotically inactivated hFF (Line #CCD-1112Sk; American Type Culture Collection). The hESC culture medium consisted of the following: knockout DME, 15% knockout serum replacement, 2 mM GlutaMAX, nonessential amino acids, 50 U/ml penicillin, 50 ng/ml streptomycin, and 0.1 mM β-mercaptoethanol (Invitrogen); and 4 ng/ml basic FGF (R&D Systems). Cells were maintained in a humidified incubator at 37°C with an atmosphere consisting of 6% CO2, 7% O2, and 87% N2. After 5–7 d of incubation, hESCs were passaged using trypsin (Invitrogen) for experimental culturing conditions in a feeder-free environment. The cells were plated on 16-well glass slides (Thermo Fisher Scientific) coated with 0.1% gelatin (BD Biosciences) in the absence of hFF. The conditioned media used consisted of hFF supernatant and hESC culture media (1:1).
IF microscopy (Albert Einstein College of Medicine)
hESCs from H1 and 9 cell lines were washed in Dulbecco's PBS without calcium and magnesium (Mediatech, Inc.) at RT, and then fixed in 3.7% paraformaldehyde in PBS for 15 min. They were then rinsed three times with PBS, incubated in 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 10 min, and blocked with 2% BSA in PBS for 1 h at RT or overnight at 4°C, and primary antibodies (monoclonal anti-AcTb mouse anti–human IgG2b [Sigma-Aldrich]; purified polyclonal anti–γ-tubulin rabbit anti–human IgG [BioLegend]; purified polyclonal anti-zinc finger protein Gli2 rabbit anti–human IgG [Aviva Systems Biology]; monoclonal anti–Tra-1-85 mouse anti–human IgG1 [Millipore]; and PE-conjugated monoclonal anti–stage-specific embryonic antigen 4 mouse anti–human IgG3 [R&D Systems]) were added in 1:300 dilution in blocking buffer for 1 h at RT or overnight at 4°C. Rabbit anti–human Oct-3/4 polyclonal IgG (Santa Cruz Biotechnology, Inc.), rabbit anti–human Smo polyclonal IgG (Santa Cruz Biotechnology, Inc.), and rabbit anti–human SHh antibody polyclonal IgG (Cell Signaling Technology) were used at dilutions of 1:100 in blocking buffer and incubated overnight at 4°C. The cells were then washed three times in PBS with 5-min incubations between washes. The secondary antibodies Cy3-conjugated AffiniPure goat anti–mouse IgG (H + L) and Cy5-conjugated AffiniPure goat anti–rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories) were added at 1:400 dilution in blocking buffer and incubated for 1 h at RT in the dark. All appropriate controls were done for the IF experiments described. Negative controls consisted of cells incubated with secondary antibody only. The cells were then washed again three times in PBS with 5-min incubations between washes and taken for IF imaging or stored at 4°C. The cells were incubated in DAPI (1:1,000 dilution) for 15 min in PBS before imaging. IF imaging was performed on an inverted (IX70; Olympus) and a confocal microscope (described in detail in Confocal microscopy; TCS SP2 AOBS; Leica) and viewed at a final magnification of 600 using CY3 (red) and 5 (far red) fluorescence filters. A cooled charge-coupled device camera (Sensicam QE; Sony) and IP Laboratory software (BD Biosciences) were used to capture the images, whereas ImageJ (National Institutes of Health) and Photoshop CS2 version 9.0.2 (Adobe) were used to view and analyze the data.
IF microscopy (Copenhagen)
After 1 wk of incubation, hESCs on 16-well glass slides were washed once with PBS (136.89 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, and 1.7 mM KH2HPO4) and then fixed with 4% paraformaldehyde for 20 min. After three 5-min washes with PBS, the wells were permeabilized with 0.1% Triton X-100 for 20 min. After three 5-min washes with PBS, the wells were blocked with 4% FBS for 45 min. Wells were incubated overnight at 4°C in the following primary antibodies: monoclonal mouse anti-AcTb at 1:10,000; polyclonal goat anti-pericentrin, polyclonal goat anti-centrin, polyclonal rabbit anti-Gli2, polyclonal rabbit anti–OCT-4, polyclonal rabbit anti-Ptc (Santa Cruz Biotechnology, Inc.) at 1:200; and polyclonal rabbit anti-Smo (MBL International) at 1:200. The next day, cells were washed five times with PBS and allowed to stand 5 min, followed by three more quick washes with PBS. The cells were incubated 1 h with the following secondary antibodies: Alexa Fluor488–conjugated goat anti–rabbit IgG, Alexa Fluor488–conjugated donkey anti–goat IgG, and Alexa Fluor568–conjugated goat or rabbit anti–mouse IgG (1:600; Invitrogen); and coumarin/aminomethylcoumarin acetate–conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories). Secondary antibody incubation was occasionally followed by DAPI incubation. Cells were visualized on a microscope (Eclipse E600; Nikon) with EPI-FL3 filters and a cooled charge-coupled device camera (MagnaFire; Optronics), and digital images were processed using Photoshop.
Confocal microscopy (Albert Einstein College of Medicine)
Images were collected with a confocal microscope (TCS SP2 AOBS) with 60× oil immersion optics. Laser lines at 488, 543, and 633 nm for excitation of DAPI, Cy3, and Cy5, respectively, were provided by an Ar laser and a HeNe laser. Detection ranges were set to eliminate crosstalk between fluorophores.
SAG stimulation (Copenhagen)
Confluent cultures of LRB003 cells were incubated in the presence of 1 μM SAG (Qbiogene) for 0, 1, and 4 h, followed by IF microscopy analysis with rabbit anti-Smo and anti-Ptc. Primary cilia were visualized with anti-AcTb and nuclei with DAPI. All images were taken with equivalent time exposures.
Exposure of H1 hESCs to SHhN, SAG, and cyclopamine (Albert Einstein College of Medicine)
Recombinant human SHh (C24II), amino terminal peptide (SHhN; R&D Systems), and SAG were dissolved in PBS containing 0.1% BSA. Cyclopamine (Toronto Research Chemicals) was dissolved in 95% ethanol. SHhN, SAG, and cyclopamine, in medium containing 0.5% serum, were applied to H1 hESCs in culture (in triplicate) at concentrations of 5 μM, 10 μg/ml, and 1 μM, respectively, for 18 h. The exposure time and concentrations used were derived from Lipinski et al. (2006).
RNA purification from H1 hESC lines and real-time quantitative PCR (Albert Einstein College of Medicine)
After 18 h, RNA was extracted from three pooled wells of H1 hESCs after stimulation, pretreated with DNase, and further purified by RNeasy columns (QIAGEN). cDNA synthesis was performed using the SuperScript III double-stranded cDNA synthesis kit (Invitrogen) on a Mastercycler Gradient (Eppendorf). Single-stranded cDNA was cleaned on a QIAquick PCR purification kit (QIAGEN) and 50 ng was used for the PCR quantification. Gene expression was assayed by quantitative real-time RT-PCR using TaqMan gene expression master mix (Applied Biosystems) and TaqMan gene expression assay primer and probe sets (Applied Biosystems) of PTCH1 (Assay ID, Hs00181117_m1), GLI1 (Hs00171790_m1), and GLI2 (Hs00257977_m1) on the iCycler (Applied Biosystems) and normalized using the internal control gene human GAPDH (FAM/MGB Probe, non–primer limited; Applied Biosystems), which was used as the endogenous reference in the H1 hESC line assays. Each sample was run twice in triplicate. PCR reactions were run for 40 cycles. The log-linear phase of amplification was monitored to obtain threshold cycle values. The comparative threshold cycle method was used to determine levels of expression. Absence of primer dimers was verified by running the PCR product on a 1.5% agarose gel.
Transfection of cells (Copenhagen)
800 ng of pSmo:YFP (provided by P. Beachy, Stanford University School of Medicine, Stanford, CA) was mixed with FuGene6 (Roche) at 6:1 at RT for 45 min. Afterward, 16 μl of the mix was aliquoted to each well containing hESC colonies in 100 μl knockout DME. After 3 h at 37°C, the medium was replaced with conditioned medium and incubated at 37°C for an additional 48 h. The cells were fixed and permeabilized, and anti-AcTb was added at 1:10,000 and visualized with Alexa Fluor568–conjugated rabbit anti–mouse IgG along with DAPI staining.
TEM (Albert Einstein College of Medicine)
The carbon-coated matrigel-covered samples with hESC colonies of H1 and 9 cells, respectively, were fixed with 2.5% glutaraldehyde and 0.5% tannic acid in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide followed by 2% uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (Ladd Research Industries). Ultrathin sections were cut on a Ultracut UCT (Reichert), stained with uranyl acetate followed by lead citrate, and viewed on a transmission electron microscope (1200EX; JEOL) at 80 kV.
SEM (Albert Einstein College of Medicine)
The carbon-coated matrigel-covered samples with hESC colonies of H1 and 9 cells, respectively, were fixed in 2.5% glutaraldehyde, 0.1 M sodium cacodylate, 0.2 M sucrose, and 5 mM MgCl2, pH 7.4, dehydrated through a graded series of ethanol, critical point dried using liquid CO2 in a critical point drier (Samdri 795; Tousimis), sputter coated with gold-palladium in a sputter coater (Vacuum Desk-2; Denton), and examined in a scanning electron microscope (JSM6400; JEOL) using an accelerating voltage of 10 kV.
Online supplemental material
Fig. S1 shows a transmission electron micrograph of an hESC showing the details of lamellar vesicles with a ciliary necklace around a forming primary cilia. Fig. S2 shows a transmission electron micrograph of a section taken just below the cell cortex with a centriole/basal body showing a ciliary rootlet and microfilament bands at the adherens junctions of confluent H9 hESCs. Online supplementary material is available at http://www.jcb.org/cgi/content/full/jcb.200706028/DC1.
Supplemental Material
Acknowledgments
We thank Aaron Bell for his expert assistance with preliminary IF and EM data collection and Frank Macaluso, Michael Cammer, Juan Jimenez, and Leslie Gunther of the Albert Einstein College of Medicine Analytical Imaging Facility for their expert technical assistance. The Copenhagen team thanks Roberto Oliveri for his expert assistance with culturing LRB003 hESCs.
This work was supported in part by grants from the National Institutes of Health (NIDDK DK 41296, DK41918, and NIAAA AA008769 to P. Satir and DK064123 to R.E. Hirsch) and National Institute of General Medical Sciences (GM075037 to E.E. Bouhassira). It must be emphasized that National Institutes of Health funds were only used for experiments with the H1 and 9 hESC cell lines that are listed on the approved National Institutes of Health registry eligible for federal funding support. Enko Kiprilov is a doctoral candidate in the Albert Einstein College of Medicine medical scientist training program and is supported in part by The National Institutes of Health (grant T32-GM007288). Romain Desprat is a doctoral candidate in the Sue Golding Division of the Albert Einstein College of Medicine. The studies by the team at the University of Copenhagen were supported in part by Rigshospital Science Foundation (A.G. Byskov and C.Y. Anderson), The Lundbeck Foundation (grant numbers 150/05 and R9-A969 to S.T. Christensen, A.G. Byskov, and A. Awan), and funds from the University of Copenhagen (S.T. Christensen and C.A. Clement).
E.N. Kiprilov and A. Awan contributed equally to this paper.
Abbreviations used in this paper: AcTb, acetylated tubulin; hESC, human embryonic stem cells; hFF, human foreskin fibroblast; Hh, hedgehog; IF, immunofluorescence; Ptc1, patched 1; SAG, Smo agonist; SEM, scanning electron microscopy; SHh, sonic hedgehog; Smo, smoothened; TEM, transmission electron microscopy; Tra-1-85, tumor rejection antigen 1-85.
References
- Badano, J.L., N. Mitsuma, P.L. Beales, and N. Katsanis. 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7:125–148. [DOI] [PubMed] [Google Scholar]
- Blacque, O.E., and M.R. Leroux. 2006. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell. Mol. Life Sci. 63:2145–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottenstein, J. 1985. Growth and differentiation of neural cells in defined media. In Cell Cultures in the Neurosciences. J. Bottenstein and G. Sato, editors. Plenum Press, New York. 3–43.
- Chen, J.K., J. Taipale, K.E. Young, T. Maiti, and P.A. Beachy. 2002. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA. 99:14071–14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.T., L.B. Pedersen, L. Schneider, and P. Satir. 2007. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 8:97–109. [DOI] [PubMed] [Google Scholar]
- Corbit, K.C., P. Aansted, V. Singla, A.R. Norman, D.Y.R. Stanier, and J.F. Reiter. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. [DOI] [PubMed] [Google Scholar]
- Dubreuil, V., A.M. Marzesco, D. Corbeil, W.B. Huttner, and M. Wilsch-Bräuninger. 2007. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J. Cell Biol. 176:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, J.M., Y. Liu, N.A. Zaghloul, C.C. Leitch, S.S. Lawson, M. Kato, P.A. Beachy, P.L. Beales, G.N. DeMartino, S. Fisher, et al. 2007. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39:1350–1360. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., Y. Tanaka, Y. Okada, and S. Takeda. 2006. Nodal flow and the generation of left-right asymmetry. Cell. 125:33–45. [DOI] [PubMed] [Google Scholar]
- Haycraft, C.J., B. Banzs, Y. Aydin-Son, Q. Zhang, E.J. Michaud, and B.K. Yoder. 2005. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., and K.V. Anderson. 2005. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA. 102:11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen, S.B., K. Møllgård, C. Olesen, R.S. Oliveri, C.B. Bøchner, A.G. Byskov, A.N. Andersen, P.E. Høyer, N. Tommerup, and C.Y. Andersen. 2007. Regional differences in expression of specific markers for human embryonic stem cells. Reprod. Biomed. Online. 15:89–98. [DOI] [PubMed] [Google Scholar]
- Lipinski, R.J., J.J. Gipp, J. Zhang, J.D. Doles, and W. Bushman. 2006. Unique and complimentary activities of the Gli transcription factors in Hedgehog signaling. Exp. Cell Res. 312:1925–1938. [DOI] [PubMed] [Google Scholar]
- Liu, A., B. Wang, and L.A. Niswander. 2005. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 132:3103–3111. [DOI] [PubMed] [Google Scholar]
- May, S.R., A.M. Ashique, M. Karien, B. Wang, Y. Shen, K. Zarbilis, J. Reiter, J. Ericson, and A.S. Peterson. 2005. Loss of retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 287:378–389. [DOI] [PubMed] [Google Scholar]
- Michaud, E.J., and B.K. Yoder. 2006. The primary cilium in cell signaling and cancer. Cancer Res. 66:6463–6467. [DOI] [PubMed] [Google Scholar]
- Olivier, E.N., C. Qiu, M. Velho, R.E. Hirsch, and E.E. Bouhassira. 2006. Large-scale production of embryonic red blood cells from human embryonic stem cells. Exp. Hematol. 34:1635–1642. [DOI] [PubMed] [Google Scholar]
- Orozco, J.T., K.P. Wedaman, D. Signor, H. Brown, L. Rose, and J.M. Scholey. 1999. Movement of motor and cargo along cilia. Nature. 398:674. [DOI] [PubMed] [Google Scholar]
- Pan, G., and J.A. Thomson. 2007. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17:42–49. [DOI] [PubMed] [Google Scholar]
- Pan, J., and W. Snell. 2007. The primary cilium: keeper of the key to cell division. Cell. 129:1255–1257. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J. 2004. Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease. J. Am. Soc. Nephrol. 15:2528–2536. [DOI] [PubMed] [Google Scholar]
- Rohatgi, R., L. Milenkovic, and M. Scott. 2007. Patched1 regulates Hedgehog signaling at the primary cilium. Science. 317:372–376. [DOI] [PubMed] [Google Scholar]
- Rho, J.Y., K. Yu, J.S. Han, J.I. Chae, D.B. Koo, H.S. Yoon, S.Y. Moon, K.K. Lee, and Y.M. Han. 2006. Transcriptional profiling of the developmentally important signaling pathways in human embryonic stem cells. Hum. Reprod. 21:405–412. [DOI] [PubMed] [Google Scholar]
- Satir, P., and S.T. Christensen. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69:377–400. [DOI] [PubMed] [Google Scholar]
- Schneider, L., C.A. Clement, S.C. Teilmann, G.J. Pazour, E.K. Hoffmann, P. Satir, and S.T. Christensen. 2005. PDGFRα signaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 15:1861–1866. [DOI] [PubMed] [Google Scholar]
- Singla, V., and J.F. Reiter. 2006. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 313:629–633. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Y. Okada, and N. Hirokawa. 2005. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 435:172–177. [DOI] [PubMed] [Google Scholar]
- Yao, S., S. Chen, J. Clark, E. Hao, G.M. Beattie, A. Hayek, and S. Ding. 2006. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc. Natl. Acad. Sci. USA. 103:6907–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.