INTRODUCTION
Here we describe a simple and cost-effective PCR-based method to introduce deletions, point mutations, frame-shift, or truncations with very high efficiency. The scope and additional applications of the method are discussed.
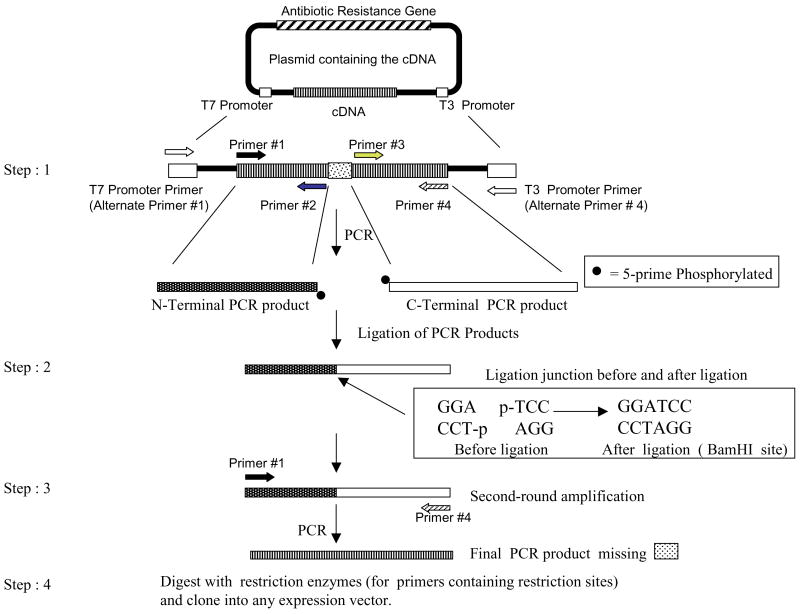
Site-directed mutagenesis involves the introduction of mutations at the DNA level to alter the primary amino acid sequence of proteins (1,2). To approach the desired level of reliability in generating mutants, several methods have been described, some of which are technically demanding and/or require special reagents (1–6). Megaprimer-based methods for site-directed mutagenesis utilize three oligonucleotide primers (4) or only one mutagenic primer (5) in two rounds of PCR. However, when longer fragments are mutagenized, the efficiency of the methods decreases dramatically and the number of side products increases. Inverse PCR can generate mutations with two primers but solely utilizes plasmid DNA templates (6). We describe a highly efficient, cost-effective, and reliable method to generate mutations along the entire length of the cDNA. The strategy involves the following steps. (i) Four primers (e.g., Figure 1, primers #1 #4) amplify the cDNA in the first-round PCR. Primer pairs (#1 and #2) and (#3 and #4) generate the N- and C-terminal PCR products, respectively. Primers #2 and #3 are purchased as 5′ phosphorylated primers. Primers #1 and #4 are unphosphorylated. (ii) T4 DNA ligase-mediated joining of the PCR products immediately following the first-round PCR. (iii) Second-round amplification of the ligated product using primers #1 and #4. (iv) Restriction digestion of the PCR product (from step 3) and cloning into any vector of choice. In our case, primer #2 contained the desired mutation (S65L, S65W, or R167Q). The PCRs contained the following components: 5.0 ng template DNA, 2.5 μL forward primers (0.05 μg/μL, 10× stock), and 2.5 μL reverse primers (0.05 μg/μL, 10× stock), 2.5 μL 2 mM dNTP, 0.5 μL 100 mM MgSO4, 2.5 μL 10× Pfu buffer (Stratagene, La Jolla, CA, USA), 0.5 μL Pfu DNA polymerase (5 U/μL; Stratagene), and sterile water to 25 μL. Standard PCR cycling conditions were used (1 cycle of 94°C for 4 min, 54°C for 1 min, 72°C for 1 min; 28 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 7 min). One micro-liter (approximately 100 ng) of each PCR product was directly used for blunt-end ligation [1 h at room temperature, 1 μL 10× buffer (New England Biolabs, Beverly, MA, USA), 1 μL each of N- and C-terminal PCR products, 0.5 μL T4 DNA ligase (100,000 U/mL), and 6.5 μL sterile water]. One micro-liter (approximately 10 ng) of the ligation reaction was used for the second-round PCR under the conditions described above. Final PCR products were digested with restriction enzymes and gel-purified using the Zymoclean Gel RNA Recovery Kit (Zymo Research, Orange, CA, USA). Fifty nanograms of insert were ligated to compatible sites in 50 ng mammalian expression vector, pEGFP-C1, and transformed into chemically competent bacteria.
Figure 1. Strategy for site-directed mutagenesis.
Step 1. Introduction of a deletion shown as an example. Note that gene-specific primers or alternate primers (T7 or T3 promoter primers) are used to achieve a minimum size of a 114-bp product. Step 2. Ligation of the PCR products. Step 3. Amplification of the ligated product to generate cDNA containing the desired mutation. Step 4. Restriction digestion of the final PCR product and ligation into the expression vector.
Pfu DNA polymerase is used to ensure blunt ends and avoid second-site mutations. Initially, gel-purified PCR products from step 1 were used for ligation (step 2). Subsequently, we found that PCR products could be directly used without removing PCR primers (Figure 2). Moreover, Pfu buffer was found to be compatible with ligase buffer; thus, PCR products can be directly ligated without removing the Pfu buffer. The minimum size of the PCR product amplified was 114 bp (Figure 2, lane 15). Thus, if mutations close to translation initiation or termination sites are needed, alternate primers complementary to the vector (Figure 1, T7 or T3 promoter primers) or the untranslated regions (UTRs) of the cDNA can be used to amplify at least a 114-bp PCR product. In the second round of PCR, primers are designed to only amplify the open reading frame (ORF).
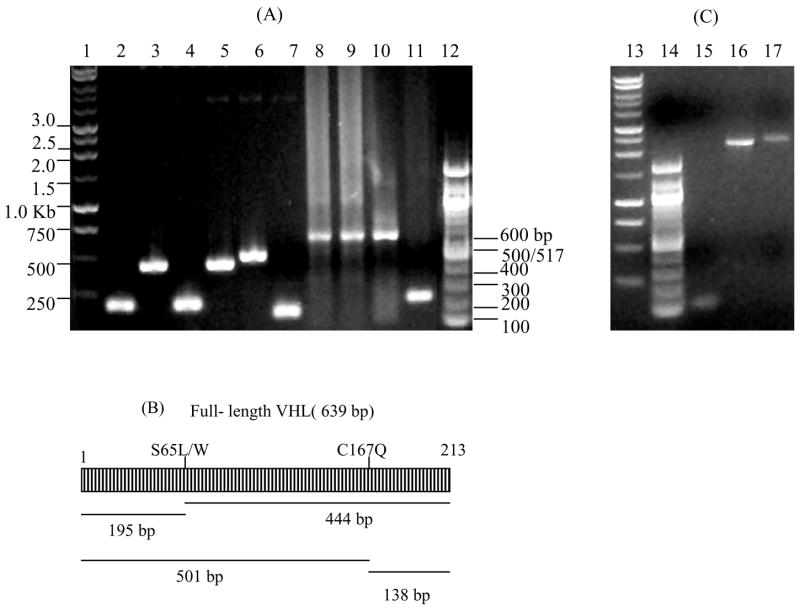
Figure 2. Applications of the strategy.
(A) Generation of point mutants and a deletion of the VHL protein. Lanes 1 and 13, 1 kb DNA ladder (Promega, Madison, WI, USA); lanes 12 and 14, 100-bp DNA ladder (New England Biolabs); lanes 2, 4, and 6, N-terminal PCR products of VHL mutants S65L, S65W, or R167Q, respectively; lanes 3, 5, and 7, C-terminal PCR products of VHL mutants S65L, S65W, or R167Q, respectively; lanes 8 10, full-length S65L, S65W, and R167Q point mutants after second-round PCR; lane 11, simultaneous introduction of a point mutation and deletion of amino acids 66 166 by joining S65L N-terminal PCR product (lane 2) with 167 213 C-terminal PCR product (lane 7). (B) Expected PCR products for VHL. (C) Fusion of membrane localization signal (mem) of integrin alpha-3 (114 bp, lane 15) to EGFP-integrin UTR (2.2 kb, lane 16) to create mem-EGFP-integrin 3′ UTR (lane 17). VHL, Von Hippel-Lindau; EGFP, enhanced green fluorescent protein; UTR, untranslated region.
Using our method, we cloned a number of mutants (examples shown in Figure 2) of the Von Hippel-Lindau (VHL) protein in a single day. The procedure can also be adapted to generate novel fusion proteins containing distinct sequence elements. For example, we generated fusions containing the membrane localization signal of integrin alpha-3 and a reporter [enhanced green fluorescent protein (EGFP)] containing the 3′ UTR of integrin alpha-3 (Figure 2C) to study messenger RNA (mRNA) transport and protein localization of the reporter fusion. All the constructs were confirmed by sequencing.
The strategy has several advantages. (i) The method is cost-effective and can be completed in a single day. (ii) The amplified PCR product is significantly smaller, allowing direct manipulation of larger cDNAs. (iii) Any vector, DNA fragment, or PCR product containing the cDNA can be the starting template. (iv) Small epitope tags can be fused in-frame to the protein of interest (see use of alternate primers). (v) Sufficient quantities of the final PCR product are available for cloning into multiple vectors, thus reducing the time and effort spent on subcloning. (vi) As opposed to the ligase-independent overlap-extension PCR method (7), “mixing and matching” various N-terminal and C-terminal PCR products (Figure 2A, lane 11) generates a higher number of deletion mutants with fewer step 1 PCRs. This is advantageous where a large number of random deletions are required; for example, in mapping protein-protein interaction domains. (vii) The strategy is technically very simple and involves no intermediate purification steps.
Acknowledgments
This work is supported by a MUSC-DOD Phase VII (Geocenters) grant to V.D.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
References
- 1.Braman J, editor. In Vitro Mutagenesis Protocols. 2. Humana Press; Totowa, NJ: 2002. [Google Scholar]
- 2.Ishii TM, Zerr P, Xia XM, Bond CT, Maylie J, Adelman JP. Site-directed mutagenesis. Methods Enzymol. 1998;293:53–71. doi: 10.1016/s0076-6879(98)93007-5. [DOI] [PubMed] [Google Scholar]
- 3.Chiu J, March PE, Lee R, Tillett D. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 5.Nagy ZB, Felfoldi F, Tamas L, Puskas LG. A one-tube, two-step polymerase chain reaction-based site-directed mutagenesis method with simple identification of the mutated product. Anal Biochem. 2004;324:301–303. doi: 10.1016/j.ab.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Stemmer WP, Morris SK. Enzymatic inverse PCR: a restriction site independent, single fragment method for high-efficiency, site-directed mutagenesis. BioTechniques. 1992;13:214–220. [PubMed] [Google Scholar]
- 7.Lee J, Lee HJ, Shin MK, Ryu WS. Versatile PCR-mediated insertion or deletion mutagenesis. BioTechniques. 2004;36:398–400. doi: 10.2144/04363BM04. [DOI] [PubMed] [Google Scholar]