Abstract
It has been widely reported that the small GTP-binding protein Rap1 has an anti-Ras and anti-mitogenic activity. Thus, it is generally accepted that a normal physiological role of Rap1 proteins is to antagonize Ras mitogenic signals, presumably by forming nonproductive complexes with proteins that are typically effectors or modulators of Ras. Rap1 is activated by signals that raise intracellular levels of cAMP, a molecule that has long been known to exert both inhibitory and stimulatory effects on cell growth. We have now tested the intriguing hypothesis that Rap1 could have mitogenic effects in systems in which cAMP stimulates cell proliferation. The result of experiments addressing this possibility revealed that Rap1 has full oncogenic potential. Expression of Rap1 in these cells results in a decreased doubling time, an increased saturation density, and an unusual anchorage-dependent morphological transformation. Most significantly, however, Rap1-expressing cells formed tumors when injected into nude mice. Thus, we propose that the view that holds Rap1 as an antimitogenic protein should be restricted and conclude that Rap1 is a conditional oncoprotein.
Rap proteins (Ras proximate) (1) are GTPases that belong to the Ras superfamily of G proteins (2), and were originally identified by GTPγS35 binding (3), low stringency hybridization by using Ras probes (4), and by expression cloning as a Ras revertant clone (5). Although Rap shares only ≈50% overall sequence identity with Ras, their effector domain regions are indistinguishable (6), raising the possibility that both proteins share similar or, perhaps, antagonistic functions. The identical effector domains at the biochemical level and the ability of Rap1 to revert Ras-mediated cellular transformation at the biological level led to the current notion that Rap1 is an antimitogenic protein that functions by antagonizing Ras action (7). This view was supported by findings showing that the interaction of Rap with typical Ras modulators (8) and effector proteins (9) does not result in their activation (10). Thus, a simple model emerged, explaining Rap’s antimitogenic action by its ability to form nonproductive complexes with Ras effector molecules.
However, Rap’s function does not appear to be restricted solely to its antimitogenic action. Although numerous reports have linked Rap to inhibition of Ras-dependent signaling (11), there are some observations suggesting that Rap1 proteins might exert a positive role in the control of cell growth. Microinjection of Rap1b protein into Swiss 3T3 fibroblasts promoted, in the presence of insulin, a GTP-dependent G1/S transition (12). Another line of evidence came from studies in a human genetic syndrome characterized by the development of tumors in a variety of tissues, known as tuberous sclerosis. This disease was recently associated with the loss of tuberin, a protein with Rap1-GAP activity (13), potentially leading to a constitutive activation of Rap1 in those tumors. Although these results suggest a potential mitogenic/oncogenic role for Rap1 proteins, it falls short of directly implicating Rap in cell division and tumorigenesis; while Rap increased DNA synthesis in Swiss 3T3 cells, it is unknown if it indeed leads to mitogenesis, and the lack of GAP activity in tuberous sclerosis may not be restricted only to Rap (14). Therefore, it is of primary importance to develop models that permit a coherent treatment of the intriguing possibility that Rap is endowed with oncogenic capabilities.
It has long been known that cAMP, like Rap1, exerts both stimulatory and inhibitory effects on cell growth (15), even though the biological effectors still remain elusive. Rap1 is activated by signals that raise intracellular levels of cAMP (16), suggesting that Rap may be intimately linked with some of the cAMP biological effects. In addition, cAMP can induce neuronal differentiation in PC12 cells, and this process is dependent on Rap1 (17). Thus, like cAMP, Rap’s biological action appears complex in that, depending on the signaling program of the cell, Rap1 may either inhibit or stimulate cell growth and even promote cell differentiation. The apparent parallel between the effects of Rap1 and cAMP on cell growth allows important predictions regarding the duality in the role of Rap1 in cell growth control. One prediction would be that Rap’s mitogenic and oncogenic potential could be revealed if expressed in a system in which mitogenic effects of cAMP have been described. Thus, we began to study the effects of Rap1 on the growth of Swiss 3T3 fibroblasts, a system in which cAMP is known to be a positive regulator of cell growth (18). In this report we show that expression of exogenous wild-type Rap1b in Swiss 3T3 cells, is sufficient to reveal the full oncogenic potential of Rap1. These cells presented a decreased doubling time, an increased saturation density and an intriguing and unusual anchorage-dependent morphological transformation. Most importantly, in vivo, Rap-expressing cells were tumorigenic when injected into nude mice. To our knowledge, the results described here represent the first observations directly linking Rap proteins to tumorigenesis.
EXPERIMENTAL PROCEDURES
Establishment of Cell Lines.
Stable cell lines expressing hemagglutinin (HA)-tagged wild type Rap1b were generated by transfection into Swiss 3T3 cells, utilizing the same plasmids and protocols described previously (16). Unlike in NIH 3T3 cells, and for unknown reasons, the system did not show any isopropyl β-d-thiogalactoside inducibility when expressed in Swiss 3T3 cells. Therefore, two independent clones constitutively expressing HA-tagged wild-type Rap1b (pL7-3-24 and pL7-3-50) were isolated and used throughout this study to avoid clonal variability. The isolation of stable Swiss 3T3 cell lines expressing dominant negative, constitutive active and phosphorylation-deficient Rap1b constructs under a tetracyclin-regulatable expression system is now in progress.
Growth Curves, Soft-Agar, and Tumor Formation Assays.
Cells were maintained in DMEM/10% fetal bovine serum containing penicillin, streptomycin, and 2 mM glutamine in 5% CO2 at 37°C. For growth curves, cells were plated in six-well dishes (Falcon; 50,000 cells per well in triplicates). Medium was changed daily and cells were counted after trypsinization. For potential autocrine activity, cell culture inserts were used (Falcon, 0.45 μm). Cell lines to be tested for release were grown on the inserts (50,000 per insert), and their effects on control pL7-Hy cells plated at the bottom (50,000 per well) were evaluated daily by counting after trypsinization. For soft agar assays, cells were resuspended in 0.3% agar (Difco) (5,000 cells per well in six-well dishes) in serum-containing medium and set on top of 1% agar layer. After 3 weeks of incubation, colonies (>50 cells) were scored under the microscope.
Thymidine Incorporation Assays.
Cells were plated into 96-well plates (10,000 per well). Next day cells were made quiescent by serum starvation in DMEM/0.2% BSA for 20 h. After 16 h of agonist stimulation, cells were labeled with [methyl-3H]thymidine (Amersham; 1 μM, 1 μCi/ml; 1 Ci = 37 GBq), and 24 h later samples were collected by using a cell harvester. Filters were dried and analyzed by scintillation counting.
BrdUrd Labeling.
To monitor cells traversing S phase by BrdUrd incorporation assays, cells were grown to 90% confluency on glass coverslips, and made quiescent by serum starvation in DMEM/0.2% BSA for 18–20 h. After agonist stimulation, cells were pulse-labeled for 2 h with 100 μM BrdUrd (Sigma) at different time intervals. At the end of the labeling period, cells were fixed in 4% paraformaldehyde for 5 min and permeabilized with 0.5% Triton X-100 for another 5 min After washing, incorporated BrdUrd was detected by indirect immunofluorescence. Samples were incubated for 30 min at 37°C with sheep anti-BrdUrd antibody (Biodesign International; dilution, 1/100 in PBS/2% BSA) in the presence of DNase (Promega; 10 units/ml). After extensive washes in PBS/0.1% Tween 20, samples were incubated for 30 min at 37°C with fluorescein isothiocyanate-conjugated goat-anti sheep serum (Sigma; dilution, 1/150 in PBS/2% BSA) containing 0.2 μg/ml 4′,6-diamidino-2-phenylindole (Sigma). After extensive washes in PBS-0.1% Tween/20, samples were mounted in PermaFluor and viewed by epifluorescence at low magnification (10×).
RESULTS AND DISCUSSION
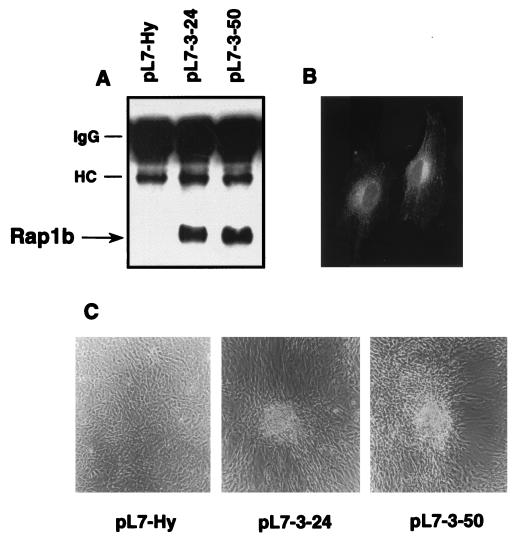
HA-tagged Rap1b-expressing cells were generated by stable transfection into Swiss 3T3 cells. Independent clones were isolated and two of them (pL7-3-24 and pL7-3-50), positive for HA-Rap expression, were further characterized (Fig. 1). As reported before for native Rap1 (19), HA-tagged Rap1b showed a perinuclear localization (Fig. 1B). At a subconfluent stage, Rap1b cells were slightly bigger, flatter and more spread compared with control cells transfected with empty vector (pL7-Hy). However, when Rap1b-expressing cells reached confluency, they began to form structures resembling transformed foci, while control cells remained as a single monolayer (Fig. 1C). The kinetics of formation of these structures were relatively fast; at 1 day postconfluency, cells showed an apparent increase in cell-cell contact at the expense of an apparent decrease in cell-substratum interaction. Thus, the morphological changes observed seem to differ from the classical cell doubling-dependent focus-forming activity characteristic of the transformed phenotype.
Figure 1.
Establishment of wild-type Rap1b-expressing Swiss 3T3 cell lines. (A) Expression of HA-tagged Rap1b as evaluated by immunoprecipitation-coupled blotting techniques by using a monoclonal anti-HA antibody (HA.11, Babco, Richmond, CA). Samples were analyzed on nonreducing SDS/PAGE with lysates from control (pL7-Hy) and two independent Rap1b-expressing clones (pL7-3-24 and pL7-3-50). HC and IgG indicate the position of heavy chain and whole Ig, respectively. (B) pL7-3-50 cells were analyzed by indirect immunofluorescence with the HA.11 antibody (dilution, 1/150). HA-tagged Rap1b shows a perinuclear localization, as reported for endogenous Rap proteins. (C) Morphological changes induced by wild-type Rap1b. Cells plated on six-well dishes were photographed under low magnification 1 day after reaching confluency. Typical fields with foci-like structures are shown here for Rap1b-expressing cells, as compared with a single monolayer observed in control cells.
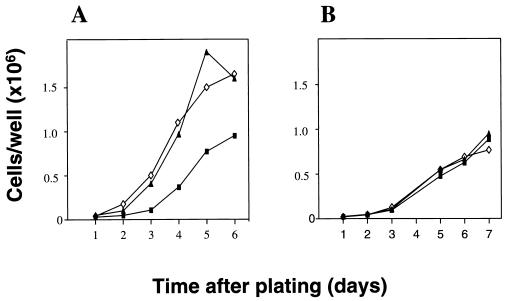
Rap1b-expressing cells showed a decreased doubling time and displayed a 1.5–2.0-fold increase in saturation density on growth curve assays (Fig. 2A). The distribution of Rap1b into vesicle-like structures (Fig. 1B) would suggest the possibility that Rap1b might be mediating the release of potential autocrine growth factors, responsible for the growth differences observed. To assess this possibility, the growth curves were repeated utilizing cell culture inserts. If Rap1b-expressing cells grown on the upper inserts were able to release diffusable mitogens, it should modify the curve profiles on control cells plated in the lower wells. As observed in Fig. 2B, no significant differences were observed in the growth curves. Though we cannot rule out an effect on a potential membrane-bound juxtacrine activity, these results suggest that the observed growth advantage is not mediated by the release of diffusable growth factors into the medium. Therefore, in spite of the known antimitogenic properties associated with Rap1 proteins, wild-type Rap1b is able to exert a positive mitogenic effect when expressed in the cAMP-responsive cell line Swiss 3T3.
Figure 2.
Growth properties of Rap1b-expressing Swiss 3T3 cells. (A) Growth curves of control (pL7-Hy, ▪) and Rap1b-expressing (pL-3-24, ◊; and pL-7-3-50, ▴) cells in serum-containing medium. Cells were plated on six-well dishes (50,000 per well, per triplicate) and counted daily after trypsinization. Results are expressed as average of triplicates (variation <5%). (B) Cell culture inserts (Falcon; 0.45-μm pore size) containing 50,000 cells of each cell line were placed on top of wells containing 50,000 pL7-Hy cells. Plates were treated as before and bottom wells (control cells) were counted daily to test for potential autocrine activity.
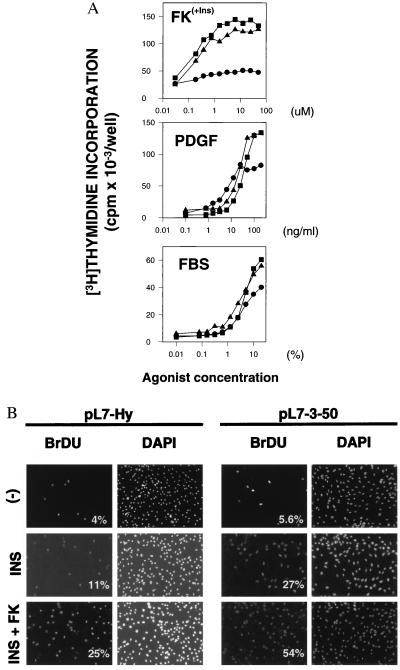
The ability of Rap1 to promote S phase entry was evaluated by [3H]thymidine incorporation. Dose-response curves for different mitogens were analyzed on pL7-3-24, pL7-3-50 and pL7-Hy control cells, and the results are shown in Fig. 3A. Rap1b-expressing cells clearly showed an increase in the maximal response, supporting the hypothesis of Rap1b’s involvement in the mitogenic response. Interestingly, the magnitude of the stimulatory effect was not the same for all the agonists tested, with a range forskolin > platelet-derived growth factor > fetal bovine serum observed. No difference in the EC50 values was observed, correlating with the observations described above (Fig. 2B), namely, that the mitogenic properties were not mediated by the release of diffusable growth factors into the medium. The possibility that changes in substrate uptake and/or specific activity could be responsible for the differences observed was excluded by BrdUrd pulse-labeling experiments. The results showed that, upon stimulation of quiescent cells with the appropriate mitogens, the number of nuclei incorporating BrdUrd in the population of cells expressing Rap1b doubled when compared with control cells (Fig. 3B). Thus, the observed mitogenic effects of Rap1b are clearly linked to its ability to promote S phase entry.
Figure 3.
Rap1b-expressing cells show an increased ability to traverse S phase. (A) [3H]Thymidine incorporation assays indicate that Rap1b- expressing cells (pL7-3-24, ▪ and pL7-3-50, ▴) show an increase in the maximal response, as compared with control (pL7-Hy, •) cells. Dose-responses were performed for fetal bovine serum (FBS), platelet-derived growth factor (PDGF) and forskolin (FK). Forskolin dose-response was performed in the presence of a constant amount of insulin (Ins, 1 μg/ml) and 3-isobutyl-1-methylxanthine (100 μM). (B) Nuclear incorporation of BrdUrd was assayed by indirect immunofluorescence (32). Cells were left untreated (−), or stimulated with insulin (INS, 1 μg/ml) or insulin plus forskolin (INS+FK, 1 μg/ml and 10 μM, respectively) in the presence of 100 μM 3-isobutyl-1-methylxanthine. Total nuclei was visualized by 4′,6-diamidino-2-phenylindole staining, and the results expressed as % BrdUrd/4′,6-diamidino-2-phenylindole. Experiments were done in duplicates and three or four independent fields per sample analyzed and expressed as averages (variation <10%).
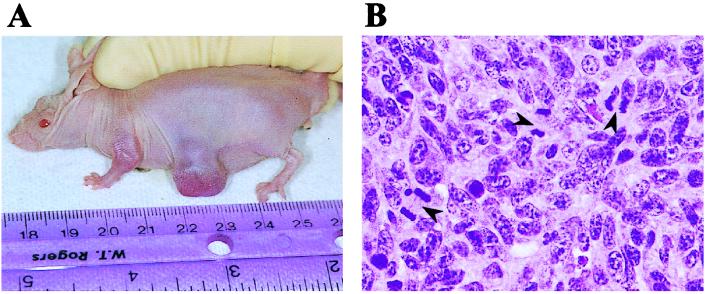
One of the best in vitro correlates of tumorigenesis is anchorage-independence of growth, assayed as colony formation in soft agar (20). Given the morphological changes and growth properties observed in the Rap1b-expressing cells, we anticipated that they would display an anchorage-independent growth when assayed in soft agar. To our surprise, none of the various cell lines tested were able to form colonies in soft agar (Table 1). This implies that despite the morphological transformation observed in Rap-expressing cells, they retained the anchorage-dependent growth characteristic of the parental cells. While the lack of anchorage-independence would seem to argue against a possible oncogenic role for Rap, the striking morphological transformation as well as the increased mitogenicity associated with Rap expression prompted us to analyze a possible tumorigenic property of Rap in vivo. Surprisingly, Rap1b-expressing cells formed tumors after subcutaneous injection into nude mice (Fig. 4). After inoculation of the cells, tumors appeared with a latency of approximately 3–4 weeks and reached sizes of 1–1.5 cm after 2 months (Table 1 and Fig. 4A). Rap tumors were invariably localized, without any signs of invasion, metastases or hypervascularization. Pathology showed cell pleomorphism, including giant cells with enlarged hyperchromatic nuclei. Numerous mitotic figures were observed (Fig. 4B). Cells established in vitro from tumor samples were indistinguishable from the parental pL7–3-50 cells, both in apparent general morphology and exogenous HA-Rap1b expression (not shown). No tumors were observed in mice injected with control cells after ≈4 months (Table 1). These results show that increased expression of wild-type Rap1b in Swiss 3T3 cells not only exerts a positive effect on cell proliferation but it is also tumorigenic when assayed in nude mice, revealing that Rap1 proteins are endowed with oncogenic capabilities when assayed in an appropriate model system.
Table 1.
Anchorage-dependent tumorigenic properties of Rap1b-expressing cells
| Exp. | Tumor formation, animals bearing tumors/animals injected
|
Soft agar assay, number of colonies/plate
|
|||
|---|---|---|---|---|---|
| pL7-Hy | pL7-3-50 | pL7-Hy | pL7-3-24 | pL7-3-50 | |
| 1 | 0/4 | 4/4 | 1 | 0 | 1 |
| 2 | 0/2 | 4/4 | 0 | 0 | 0 |
Ralp1b-expressing cells (pL7-3-50) induced tumor formation in all animals tested, whereas no tumor was observed for control (pL7-Hy) cells after 4 months of injection. For soft agar assays, cells were plated (5,000 cells per well in six-well dishes) in serum-containing medium and set on top of a 1% agar layer. Colonies (>50 cells) were scored under the microscope after 3 weeks of incubation.
Figure 4.
Rap1b-expressing cells induce tumor formation in nude mice, (A) pL7-3-50 cells were injected into nude mice, and a tumor scored 2 months after injection is shown here. (B) A typical hematoxylin/eosin staining showing several mitotic figures is shown here (arrowheads).
Growth factors and integrin-dependent adhesion are required for normal cells to proliferate (21). The requirements for growth factors and anchorage are usually bypassed in most tumor models as manifested experimentally by growth in soft agar and low serum concentration (22). The results in this report show that Rap1b-expressing cells present an uncharacteristic growth factor-dependent, anchorage-dependent, yet tumorigenic phenotype. This indicates that wild-type Rap1b is not able to bypass adhesion-mediated and growth factor signaling events. Thus, it is possible that both Rap and adhesion signals converge downstream in the pathway or, alternatively, that Rap1b activation lies upstream the integrin pathway. Interestingly, links between actin cytoskeleton and both potential upstream (23) and downstream (24) Rap1 modulators have been reported. Further evaluation of integrin-dependent events (focal contacts, stress fibers, etc) are underway to resolve this issue. Whether the latency observed in tumor formation reflects the need for the establishment of an active extracellular matrix remains to be determined.
This report establishes that Rap, like cAMP, is able to trigger a mitogenic response both in vitro and in vivo. A common pattern emerging from these studies is that, like cAMP, Rap’s mitogenic action is only revealed in the presence of other growth factors, i.e., insulin/insulin growth factor 1. This synergistic behavior implies a collaboration between Ras-dependent and cAMP-dependent signaling events. Thus, it is tempting to hypothesize that Rap1 activation might represent a downstream event of the cAMP synergistic component. Several arguments support this notion. (i) agonists that increase cAMP are able to activate Rap1b in a phosphorylation-dependent manner (16); (ii) Rap1b-expressing cells show an increased mitogenic response to agents that raise cellular cAMP, and the magnitude of this increment is proportional to the enrichment of cAMP in the growth factor mix [Fig. 3A; the differences observed for forskolin (+insulin) are larger than those for platelet-derived growth factor or serum]; (iii) the mitogenic activity promoted by platelet-derived growth factor in Swiss 3T3 cells has been linked with an autocrine loop that requires cAMP (25, 26); (iv) unlike Ras, Rap-transformed cells do not show any decrease in growth factor requirements for growth, as evidenced in the dose-response curves (Fig. 3A). This need for a growth factor (i.e., insulin) in order for Rap1b to express its mitogenic action suggests that Rap1b might be acting solely on the cAMP synergistic component. Accordingly, Rap-expressing cells do not show any mitogenic advantage in the presence of forskolin as the only agonist, and have an 3-isobutyl-1-methylxanthine-dependent advantage in the presence of insulin alone.
Thus, we have established that a parallel exists between the effects of Rap1 and cAMP with respect to mitogenicity and that, in vivo, this parallel is translated into tumorigenicity. This notion is consistent with recent findings demonstrating that cAMP-induced neuronal differentiation in PC12 cells is Rap1-dependent (17). These examples clearly illustrate that the view that holds Rap as an anti-Ras and antimitogenic protein is not universally true. To the contrary, these results, point to a collaboration between Ras and Rap1 proteins to express a full biological response. Moreover, the observed effects of Rap1 may go beyond the transduction of the cAMP signal. As recently reported, Ca2+-mobilizing agents are able to activate Rap1b in platelets (27). Rap1b serves in vitro as a substrate for CaM-kinases, which phosphorylate the same residue that protein kinase A does (28). This suggests that Rap activation might act as an integrator signal for multifunctional kinases that are activated by different heterotrimeric G protein coupled receptors. Whether the same set of effectors are responsible for all Rap1 biological responses remains to be determined.
It is clear that any component involved in a signal transduction pathway leading to cell proliferation, cell cycle progression and/or apoptosis might carry an oncogenic potential (29). Understanding how altered signal transduction contributes to oncogenesis is of fundamental importance. As discussed, Rap proteins may be intimately linked with the genesis and/or promotion of growth of certain tumors. However, the demonstration of altered Rap1 signaling in human tumors is needed to ultimately validate the notion of Rap1 as an oncogene. Our results suggest that attention should be focused on noninvasive tumors with cAMP-responsive cell lineages. Interestingly, a high level of Rap1 expression was recently associated with certain gliomas (30), a cell lineage responsive mitogenically to cAMP (31). Regardless of its mechanism of action, Rap1b activation might represent a rate-limiting step in a signaling pathway that ultimately determines cell cycle progression. The available effectors and effector domain mutants will allow us to start dissecting the pathway to identify the different signals involved in the observed phenotypes, i.e., morphological changes, mitogenic activity, and tumorigenesis. In light of the new results, we propose that the current dogma of Rap1 as an antimitogenic protein needs to be reevaluated and that, in fact, Rap1 is a conditional oncogene.
Acknowledgments
We thank Dr. M. Ostrowski and Dr. R. Juliano for support at early stages of the project, Sarah Short and Mike Fisher for technical assistant with the nude mice, and Drs. A. Howe and S. Peterson for stimulating discussions and assistance with the manuscript. This work was supported by National Institutes of Health Grant R29 CA71649 (to D.L.A.). F.R.N. is a visiting scientist at National Institute of Environmental Health Sciences.
ABBREVIATION
- HA
hemagglutinin
References
- 1.Noda M. Biochem Biophys Acta. 1993;1155:97–109. doi: 10.1016/0304-419x(93)90024-7. [DOI] [PubMed] [Google Scholar]
- 2.Boguski M S, McCormick F. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 3.Nagata K, Itoh H, Katada T, Takenaka K, Ui M, Kaziro Y, Nozawa Y. J Biol Chem. 1989;264:17000–17005. [PubMed] [Google Scholar]
- 4.Pizon V, Lerosey I, Chardin P, Tavitian A. Nucleic Acids Res. 1988;16:7719. doi: 10.1093/nar/16.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 6.Valencia A, Chardin P, Wittinghofer A, Sander C. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 7.Zhang K, Noda M, Vass W C, Papageorge A G, Lowy D R. Science. 1990;249:162–165. doi: 10.1126/science.2115210. [DOI] [PubMed] [Google Scholar]
- 8.Hata Y, Kikuchi A, Sasaki T, Schaber M D, Gibbs J B, Takai Y. J Biol Chem. 1990;265:7104–7107. [PubMed] [Google Scholar]
- 9.Ikeda M, Koyama S, Okazaki M, Dohi K, Kikuchi A. FEBS Lett. 1995;375:37–40. doi: 10.1016/0014-5793(95)01169-f. [DOI] [PubMed] [Google Scholar]
- 10.Urano T, Emkey R, Feig L A. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 11.Cook S J, Rubinfeld B, Albert I, McCormick F. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida Y, Kawata M, Miura Y, Musha T, Sasaki T, Kikuchi A, Takai Y. Mol Cell Biol. 1992;12:3407–3414. doi: 10.1128/mcb.12.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wienecke R, Konig A, DeClue J E. J Biol Chem. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- 14.Xiao G H, Shoarinejad F, Jin F, Golemis E A, Yeung R S. J Biol Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- 15.Boynton A L, Whitfield J F. Adv Cyclic Nucleotide Res. 1983;15:193–294. [Google Scholar]
- 16.Altschuler D L, Peterson S N, Ostrowski M C, Lapetina E G. J Biol Chem. 1995;270:10373–10376. doi: 10.1074/jbc.270.18.10373. [DOI] [PubMed] [Google Scholar]
- 17.Vossler M R, Yao H, York R D, Pan M G, Rim C S, Stork P J. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 18.Rozengurt E, Legg A, Strang G, Courtenay-Luck N. Proc Natl Acad Sci USA. 1981;78:4392–4396. doi: 10.1073/pnas.78.7.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pizon V, Desjardins M, Bucci C, Parton R G, Zerial M. J Cell Sci. 1994;107:1661–1670. doi: 10.1242/jcs.107.6.1661. [DOI] [PubMed] [Google Scholar]
- 20.Freedman V H, Shin S I. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz M A. J Cell Biol. 1997;139:575–578. doi: 10.1083/jcb.139.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark G J, Cox A D, Graham S M, Der C J. Methods Enzymol. 1995;255:395–412. doi: 10.1016/s0076-6879(95)55042-9. [DOI] [PubMed] [Google Scholar]
- 23.Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, et al. Mol Cell Biol. 1995;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serebriiskii I, Estojak J, Sonoda G, Testa J R, Golemis E A. Oncogene. 1997;15:1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 25.Rozengurt E, Stroobant P, Waterfield M D, Deuel T F, Keehan M. Cell. 1983;34:265–272. doi: 10.1016/0092-8674(83)90157-5. [DOI] [PubMed] [Google Scholar]
- 26.Huang N N, Wang D J, Heppel L A. J Biol Chem. 1994;269:548–555. [PubMed] [Google Scholar]
- 27.Franke B, Akkerman J W, Bos J L. EMBO J. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahyoun N, McDonald O B, Farrell F, Lapetina E G. Proc Natl Acad Sci USA. 1991;88:2643–2647. doi: 10.1073/pnas.88.7.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 30.Gutmann D H, Saporito-Irwin S, DeClue J E, Wienecke R, Guha A. Oncogene. 1997;15:1611–1616. doi: 10.1038/sj.onc.1201314. [DOI] [PubMed] [Google Scholar]
- 31.Raff M C, Hornby-Smith A, Brockes J P. Nature (London) 1978;273:672–673. doi: 10.1038/273672a0. [DOI] [PubMed] [Google Scholar]