Abstract
The skeleton is one of the most important features for the reconstruction of vertebrate phylogeny but few data are available to understand its molecular origin. In mammals the Runt genes are central regulators of skeletogenesis. Runx2 was shown to be essential for osteoblast differentiation, tooth development, and bone formation. Both Runx2 and Runx3 are essential for chondrocyte maturation. Furthermore, Runx2 directly regulates Indian hedgehog expression, a master coordinator of skeletal development. To clarify the correlation of Runt gene evolution and the emergence of cartilage and bone in vertebrates, we cloned the Runt genes from hagfish as representative of jawless fish (MgRunxA, MgRunxB) and from dogfish as representative of jawed cartilaginous fish (ScRunx1–3). According to our phylogenetic reconstruction the stem species of chordates harboured a single Runt gene and thereafter Runt locus duplications occurred during early vertebrate evolution. All newly isolated Runt genes were expressed in cartilage according to quantitative PCR. In situ hybridisation confirmed high MgRunxA expression in hard cartilage of hagfish. In dogfish ScRunx2 and ScRunx3 were expressed in embryonal cartilage whereas all three Runt genes were detected in teeth and placoid scales. In cephalochordates (lancelets) Runt, Hedgehog and SoxE were strongly expressed in the gill bars and expression of Runt and Hedgehog was found in endo- as well as ectodermal cells. Furthermore we demonstrate that the lancelet Runt protein binds to Runt binding sites in the lancelet Hedgehog promoter and regulates its activity. Together, these results suggest that Runt and Hedgehog were part of a core gene network for cartilage formation, which was already active in the gill bars of the common ancestor of cephalochordates and vertebrates and diversified after Runt duplications had occurred during vertebrate evolution. The similarities in expression patterns of Runt genes support the view that teeth and placoid scales evolved from a homologous developmental module.
Author Summary
Important molecular mechanisms underlying mammalian skeletogenesis have been described but knowledge about the evolutionary origin of these gene networks is limited. The Runt gene family (Runx1–3) is of extraordinary importance for skeletogenesis. Runx2 deficient mice completely lack bone. Runx2 and Runx3 are essential for cartilage development and Runx2 regulates the key factor Indian hedgehog, which coordinates skeletogenesis. Here, we reconstructed Runt gene evolution in correlation to skeletal evolution. By analyzing lancelets, one of the closest living relatives of vertebrates, we revealed that the single Runt and Hedgehog family founder genes were co-expressed in primitive skeletal elements of the chordate stem species. Interestingly, at this stage the Runt and Hedgehog pathways were already directly linked to one another. Furthermore we isolated two Runt genes from a representative of jawless cartilaginous fish (hagfish) and three Runt genes from jawed cartilaginous fish (dogfish) which were all expressed in cartilage. The dogfish Runt genes were also found in teeth and placoid scales. This study suggests that Runt genes were involved in all ancient processes of chordate skeletogenesis. Furthermore the analysis supports the theory that most likely the gut was the tissue that originally secreted an acellular gill gut skeleton in the chordate ancestor.
Introduction
The skeleton is a hallmark of vertebrates and has been widely used over the past decades for phylogenetic analyses [1]. However, little is known about its molecular evolution.
Descriptive data are available for the matrix proteins produced by the cells that constitute the skeleton in jawless vertebrates (epitomized by hagfish and lampreys, collectively termed agnathans). Beside species specific proteins [2] they possess cartilage with type II collagen (Col2α1), which is also the characteristic matrix protein for jawed vertebrates (gnathostomes) [3],[4]. Furthermore Sox9, which directly regulates Col2α1 in mammals, was shown to be expressed in cartilage of the lamprey [3]. Interestingly SoxE (an invertebrate homolog to the mammalian Sox8/9/10) was found to be co-expressed with fibrillar collagen in the hemichordate Saccoglossus bromophenolosus [5]. The expression was found in the pharyngeal endodermal cells, which are most likely responsible for the secretion of an acellular cartilage. Such an endodermal secretion was postulated to be primarily the ancestral mode of making pharyngeal cartilage in deuterostomes [5].
Up to now no Runt gene expression has been described in skeletal elements of lancelets, agnathans and jawed cartilaginous fish in spite of the fact that Runt transcription factors (Runx1–3 synonyms: Aml1–3/Cbfa1–3/Pebp2αa–c) are central regulators of skeletal development in higher vertebrates [6],[7]. They are characterized by a highly conserved DNA binding Runt domain and the presence of two promoters [8]. Each Runt gene has two isoforms with different N-termini starting with a MASNS-like motif under the distal P1 promoter and a MRIPV sequence under the proximal Promoter P2. Furthermore the 3′ end has a conserved VWRPY-motif [8]. Runx2 is indispensable for osteogenesis as mice bearing a homozygous mutation in Runx2 completely lack bone [7], and Runx2 is together with Runx3 essential for cartilage differentiation [9],[10]. Futhermore Runx2 directly regulates the key signaling molecule Indian hedehog (Ihh), which coordinates cartilage differentiation, endochondral ossification and limb outgrowth [10]. From the three members belonging to the mammalian Hedgehog (Hh) family (Ihh, Sonic hedgehog, Desert hedgehog) also Sonic hedgehog (Shh) signaling is influenced by Runx2 during tooth morphogenesis [11]. Runx2 haploinsufficiency causes the human bone disease cleidocranial dysplasia, further substantiating its importance for skeletal development [12]. Importantly, all three mammalian Runt genes are expressed in cartilage and have been shown to play a role in the formation and differentiation of skeletal elements [6],[10],[13]. Furthermore, all Runt genes in the mouse are involved in tooth formation [14].
In contrast to the extensively studied Hox genes, which are important for patterning, Runt genes are essential for features that represent evolutionary innovations of vertebrates such as bone [1]. Such innovations result from tinkering with existing processes, from the flexibility that arises from modifications to existing gene networks, and from selective advantage provided by gene duplications or modifications [15]. As simply as this theory explains an important evolutionary process, as difficult it is to functionally analyze how the genetic networks underlying innovations like the vertebrate skeleton evolved. Based on the central role of Runt genes for skeletogenesis in higher vertebrates we hypothesized that these genes played a role in the evolution of cartilage, bone and teeth and thus might be instrumental to understand skeletal evolution in chordates. We therefore analyzed number and expression of Runt genes in hagfish (Myxine glutinosa) as a representative of jawless vertebrates, in dogfish (Scyliorhinus canicula) as a representative of cartilaginous fish and lancelets (Branchiostoma lanceolatum and B. floridae) as representatives of celphalochordates to reconstruct if Runt genes were already expressed in the developing skeleton of the chordate, vertebrate and jawed vertebrate stem species. In addition, we tested if Runt and Hh are co-expressed in lancelets and if a functional interaction between the Runt and Hh pathways might have evolved before the cellular cartilage of vertebrates evolved.
In this study we show that the stem species of chordates harboured a single Runt gene, whereas three Runt genes were present before the emergence of gnathostomes. Runt genes are expressed in developing cartilage, teeth and placoid scales of cartilaginous fish and cartilage of jawless vertebrates. In adult lancelets the Runt gene is expressed together with Hh, in the endo- and ectoderm of the gill bars. Furthermore, we demonstrate that the lancelet Runt protein can directly bind to and activate the lancelet Hh promoter. This suggests that beside SoxE and fibrillar collagen two other key factors for vertebrate skeletogenesis (Runt and Hh) were part of an ancient gene network for skeletogenesis in the gill gut stabilizing the gill bars of the common ancestor of vertebrates and lancelets approximately 700 million years ago. Our finding that the gut is an ancient Runt expression domain of deuterostomes is in accordance with the hypothesis that endodermal secretion was the ancestral mode of making pharyngeal cartilage [5].
Results
Isolation of hagfish and dogfish Runt genes
We used a PCR-based approach using cDNA as well as genomic DNA to identify Runt genes in lower vertebrates. This led to the detection of two Runt genes in hagfish (MgRunxA, MgRunxB) and three Runt genes in dogfish (ScRunx1–3). All of these newly detected Runt genes had a 3′ end with the characteristic VWRPY-motif. The two different 5′ ends of the Runt genes amplified from embryonal dogfish cDNA were homologous to the 5′ mammalian promoter variant-1 (MASNS-like) and variant-2 (MRIPV-like) motifs, respectively. In the two hagfish Runt genes amplified from adult hagfish cDNA only a single 5′ gene end was detected. According to our Blast searches against the Ensembl pre-genome sequences of lamprey (Pteromyzon marinus) the two hagfish 5′ ends represent most likely the promoter variant 2. Because of the unavailability of hagfish embryos it could not be clarified if two Runt gene promoter 1 variants are expressed during early hagfish development.
Blast searches in whole genome databases (NCBI, JGI, Ensembl) revealed that there are most likely two Runt genes in the lamprey genome, and one Runt gene in cnidarians (Nematostella vectensis), nematodes (Caenorhabditis elegans), cephalochordates (B. floridae), and tunicates (Ciona intestinalis, Oikopleura dioica) [16],[17]. We detected two Runt genes in sea urchin (Strongylocentrotus purpuratus) [18],[19], which were located on the same genomic contig, two partial Runt genes in skate (Raja eglanteria) [20], three Runt genes in mammals [6],[7] and four in pufferfish (Takifugu rubripes) [21],[22] and also four in zebrafish (Danio rerio) including a duplicated Runx2 gene [23]. In chicken (Gallus gallus) three Runt genes were found. An alignment of all newly detected Runt genes together with other deuterostome Runt genes is provided as supporting information (Figure S1) and the GeneBank accession numbers are given in the footnote.
Conserved synteny of Runt and the chloride intracellular channel (Clic) genes in human, chicken and tunicate genomes
Comparable to the human Runt loci [24], the three orthologous chicken Runt genes are followed by a Clic gene on the complementary strand. The chicken Runx1 on chromosome 1 is followed by a Clic6 homologous gene, the chicken Runx2 on chromosome 3 by a Clic5 homologous gene and the chicken Runx3 on chromosome 23 is followed by a Clic4 homologous gene. In lancelet the Runt and Clic genes are located on different scaffolds (JGI assembly vers 1.0). However, in the genome of the tunicate C. intestinalis a Clic homologous gene was found in proximity to Runt on chr_12q (JGI, Assembly vers 2.0). This strongly suggests that the entire Runt locus was triplicated during the evolution of chordates.
The last common ancestor of chordates harboured a single Runt gene
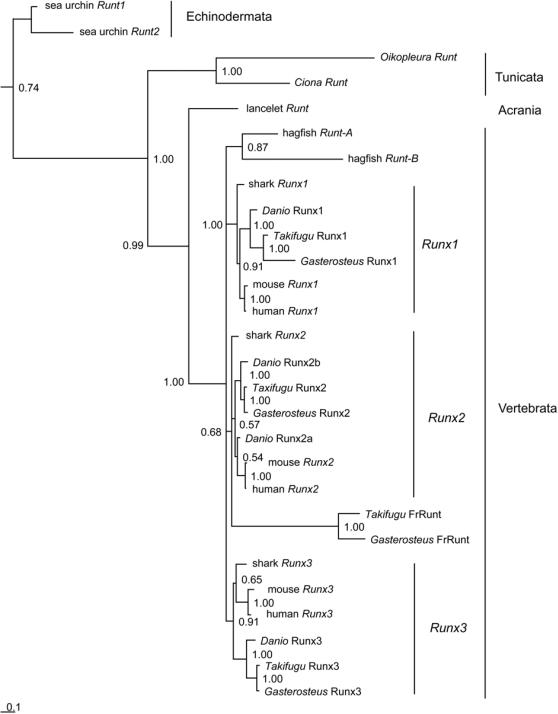
Our phylogenetic analysis (Figure 1) suggests that the stem species of chordates harboured a single Runt gene, whereas the last common ancestor of jawed vertebrates harboured three Runt genes. In addition our results indicate that the dogfish ScRunx1–3 genes are orthologous to the Amniota Runx1–3 genes. In contrast to this, the two hagfish Runt genes did not cluster with any of the three paralogous Runt genes from higher vertebrates. As outlined in Figure 2 several lineage specific Runt gene duplications have occurred: (a) in the sea urchin lineage, (b) in the stem species of bony fish and (c) probably also in hagfish. But there is a need for further data e.g. from whole genome comparison, to determine if the two hagfish Runt genes are a result of a Runt gene duplication in the stem species of vertebrates or evolved by a separate gene duplication event in the hagfish lineage.
Figure 1. Phylogenetic tree (Bayesian inference) of chordate Runt genes.
Numbers refer to branch support (Bayesian posterior probability) for the internal branches adjacent to the nodes. Sea urchin Runt genes were used to root the tree. Branch length reflects the number of substitutions per alignment site (compare scale bar).
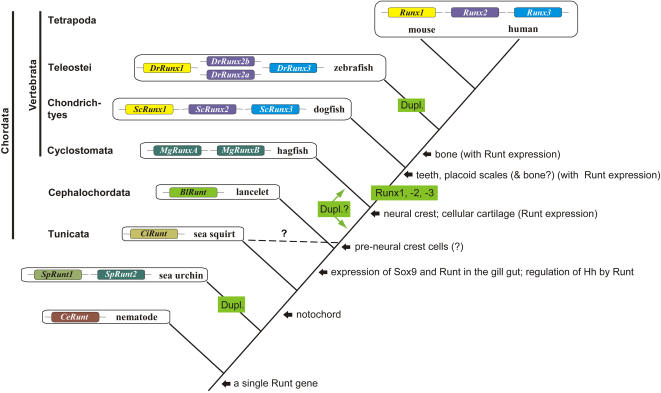
Figure 2. Overview of the Runt gene evolution in chordates.
The stepwise evolution of cartilage and bone and the most likely time intervals of Runt gene duplications (Dup) are indicated. The position of tunicates is contentious [31] which is indicated by a dashed line. In this context it is of interest that pre-neural crest cells have been observed in tunicates [60].
Runt gene expression in skeletal elements of hagfish and dogfish
To determine a possible role for Runt genes in the skeleton we asked the question if Runt genes are expressed in skeletal elements of hagfish. Using quantitative Reverse Transcriptase PCR (qRT-PCR) from dissected tissues we found that the MgRunxA gene had its highest expression in hard cartilage, followed by the gill region and soft cartilage (Figure 3). Compared to the MgRunxA gene the MgRunxB gene was only weakly expressed with the strongest expression in the gill region. In situ hybridizations confirmed the high expression of MgRunxA in hard cartilage (Figure 3B and 3C).
Figure 3. Analysis of hagfish MgRunxA and –B expression in different tissues of adult animals.
Quantification of MgRunxA and –B expression by qRT-PCR (A). Whereas MgRunxB was only weakly expressed in all tissues analyzed, MgRunxA showed a strong expression in calcified cartilage gills and soft cartilage. Expression of MgRunxA was also detected by radioactive in situ hybridisation in hard cartilage tissue (B, C). Insert of (B) is shown at higher magnification in (C) displaying the silver grains of the autoradiography emulsion indicating MgRunxA expression. B: Brain, C-h: Hard cartilage, C-s: Soft cartilage, Cho: Chorda, G: Gills, Gb: Gall bladder, G-a: Anterior gut, G-m: Midgut, G-h: Hindgut, H: Heart, L: Liver, Mu: Muscle, Sk: Skin.
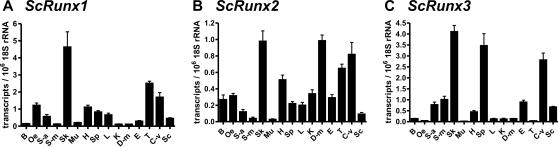
In adult dogfish the Runt genes show ubiquitous expression but it is noteworthy that all Runt genes had their third highest expression in the gill gut cartilage. For all three dogfish Runt genes the highest expression was found in the skin (Figure 4). We performed in situ hybridization to characterize the distribution of Runt expression in the skin (Figure 5A–5C). All three Runt genes were expressed in the placoid scales in the skin of dogfish embryos. ScRunx1 and ScRunx3 were expressed in the basal epidermis cells of the stratum germinativum, whereas ScRunx2 was found at the site where later the basal plate will develop. Based on the similarities between scales and teeth we performed expression analysis of Runt genes in the developing teeth of dogfish embryos. In the developing teeth the same expression pattern of ScRunx1–3 was found (Figure 5D–5F). ScRunx1 and ScRunx3 were expressed at a distal position and ScRunx2 was found at a basal position. Figure 5G shows a schematic of the different sites of Runt expression in teeth and placoid scales. In addition, Runt genes were also expressed in the developing skeleton. Runx2 expression was detected in cranial cartilage and skeletal elements of the fin whereas Runx2 and Runx3 expression was found in gill gut cartilage (Figure 6).
Figure 4. qRT-PCR results of ScRunx1–3 expression.
In dogfish the most prominent expression of all three ScRunt genes was in the skin. Also in visceral cartilage ScRunx1–3 were strongly expressed. B: Brain, C-v: Visceral cartilage, D-m: Ductus mesonephric, E: Epididymis, H: Heart, K: Kidney, L: Liver, Mu: Muscle, Oe: Oesophagus, Sc: Spinal column, Sk: Skin, S-a: Anterior stomach, S-m: Middle part of stomach, Sp: Spleen, T: Testis.
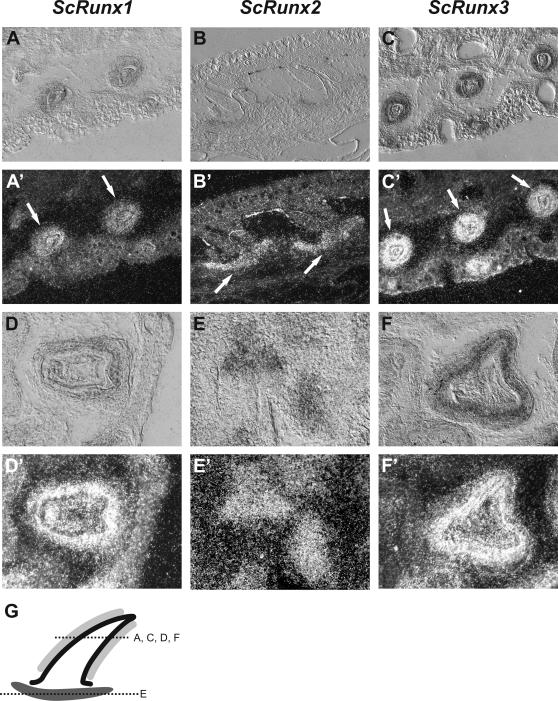
Figure 5. ScRunx1–3 expression analysis by in situ hybridization in placoid scale (A–C) and tooth development (D–F).
Bright field is given on top, dark field below. ScRunx1 (A, D) and –3 (C, F) are expressed in the basal epidermis cells of the stratum germinativum, which forms the enamel organ, whereas ScRunx2 (B, E) is found at the site of the developing basal plate. These expression patterns were identical in teeth and placoid scales. (G) Scheme of Runt expression in placoid scales and teeth with overlapping expression of ScRunx1 and –3 in the stratum germinativum (light grey) and ScRunx2 in the developing basal plate (dark grey). Dotted lines represent section planes of transverse sections in (A,C–F). Section in (B) is a longitudinal section.
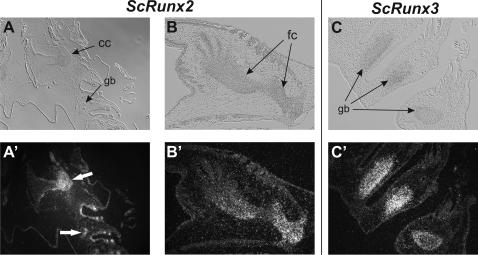
Figure 6. Expression of ScRunx2 and –3 in developing dogfish cartilage.
Expression of ScRunx2 was detected in developing cranial and gill bar cartilage (A) and in the proximal cartilage elements of the pectoral fin (B). Expression of ScRunx3 was detected in developing visceral cartilage (C). Cc: cranial cartilage, gb: gill gut cartilage, fc: fin cartilage.
Expression of Runt during lancelet (B. floridae) development in the notochord, gut and neural tube
To be able to reconstruct the Runt expression domains in the chordate stem species and to see if Runt was expressed in ancient skeletal elements such as the notochord, we analyzed Runt gene expression in lancelets, the putative sistergroup of vertebrates. Using whole mount in situ hybridization of early developmental stages (early and late gastrula) a diffuse Runt staining, indicating a maternal Runt expression, was detected, comparable to the description of maternal Runx1, –2b, and –3 expression in zebrafish. [25]–[27]. Two different probes were used, corresponding to the Runt gene variant starting with exon 1 (transcribed from the distal promoter P1) and the Runt gene variant starting with exon 2 (transcribed from the proximal promoter P2). These two probes showed overlapping staining patterns (Figure 7).
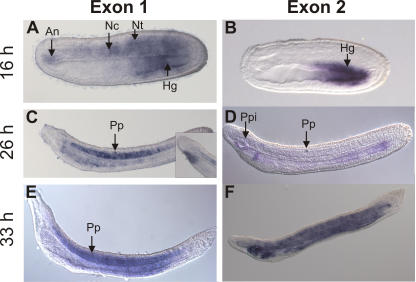
Figure 7. Runt gene expression in lancelet larvae (B. floridae).
Anterior site is located to the left and the dorsal site towards the top. Whole mount in situ hybridization at stages of 16 h (A, B), 26 h (C, D) and 33 h (E, F). A), C) and E) Runt gene exon 1 variant. B), D) and F) Runt gene exon 2 variant. Note that the primary pigment spot, indicated by an arrow, lays in the nerve chord and does not represent a Runt expression domain. An: Anterior notochord, Nt: Neural tube, Nc: Notochord, Hg: Hindgut, Pp: Primary pigment spot, Ppi: Preoral pit.
The Runt gene variant P1 was expressed at the 8 somite stage (16 h) in the posterior part of the gut, the notochord and the developing neural tube (Figure 7A). At 26 h Runt expression can be predominantly seen in the middle part of the notochord, the midgut and foregut (Figure 7C). An inconsistent staining pattern was also detected at this stage in about 50% of the larvae immediately below the preoral pit (Figure 7C insert). At 33 h the larvae showed persistent expression of the Runt exon 1 variant in the notochord and neural tube, but also in the midgut region (Figure 7E).
The Runt gene variant P2 was exclusively expressed in the hindgut at 16 h (Figure 7B). At 26 h the expression domain extended throughout the entire gut and a signal was also found in a confined region of the foregut (Figure 7D). At 33 h Runt expression was found throughout the entire larvae with the most intense signals in the tailbud and in the anterior gut region. (Figure 7F).
Runt, SoxE and Hedgehog expression in gill gut region of adult lancelet (B. lanceolatum)
Our analysis had shown that Runt genes are expressed in cartilaginous tissue of the hagfish as well as in the notochord of lancelets indicating a possible role in the development of the ancestral skeleton. We next asked the question, if Runt expression can be found in skeletal elements of adult lancelets. Based on the recent observation that adult lancelets express fibrillar collagen in their gill bars [5],[28] we hypothesized that the gill bars represent an ancestral form of cartilage regulated by similar pathways of chondrogenesis as in higher vertebrates. We showed previously that in adult lancelets only the Runt exon 2 variant is expressed [9]. As shown in Figure 8A, qRT-PCR demonstrated expression in almost all tissues, a finding that is in accordance with the broad staining pattern of the exon 2 Runt gene variant at 33 h PF (Figure 7F). However, the most intense signals in adult lancelets were found in the gill gut and the gut. Furthermore qRT-PCR showed that the lancelet SoxE gene had its highest expression and Hh its third highest expression in the gill gut region (Figure 8B and 8C). To determine where exactly Runt and Hh genes were expressed in the gill bars we performed in situ hybridization on tissue sections (Figure 8D–8G). We detected Runt and Hh gene expression in the endo- and ectodermal epithelial cells of primary and secondary gill bars (Figure 8D–8J) but not in the mesodermal coelomic cells of the primary gill bars (data not shown). Interestingly Runt and Hh were strongly coexpressed in a cell population between the endodermal epithelium with cilia and the ectodermal gland epithelium directly adjacent to both sites of the acellular matrix (arrows in Figure 8D–8G). The Hh signal was confirmed by immunohistochemistry (data not shown).
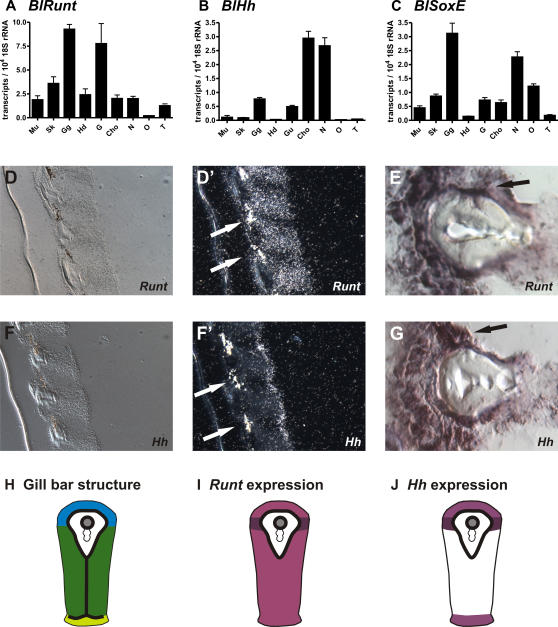
Figure 8. Analysis of Runt, SoxE and Hh gene expression in adult lancelet (B. lanceolatum).
(A–C) Quantification of Runt, SoxE and Hh expression in different tissues. (A) The strongest Runt expression is seen in the gill gut region followed by the gut and skin. (B) Hh is most strongly expressed in the chorda and neural tube followed by the gill gut and gut. (C) SoxE has its strongest expression in the gill gut and neural tube. Mu: Muscle, Sk: Skin, Gg: Gill gut, Hd: Hepatic diverticulum, G: Gut, Cho: Chorda, N: Neural tube, O: Ovaries, T: Testis. (D–G) in situ hybridization for BlRunt and BlHh show high expression in the endoderm and ecotoderm of the gill bars. (D–E) Runt expression. (F–G) Hh expression. (D, F) Bright field images. (D′, F′) Dark field images of radioactive in situ hybridizations. (E, G) Non-radioactive in situ hybridizations. High expression of BlRunt and BlHh was found in a cell population between the endodermal epithelium with cilia and the ectodermal gland epithelium directly adjacent to both sites of the acellular matrix (arrows). (H–J) Schematic drawing of Runt and Hh expression sites in secondary gill bars as shown in (D–G). (H) The gill bar tissue consists of three different single layered epithelia attached to a basal membrane - atrial epithelium (blue), lateral epithelium (dark green) and pharyngeal epithelium (light green). The basal membrane is indicated by the bold black line. The skeletal rod of secondary gill bars contains a skeletal vessel (grey filled circle) that is formed by basal membranes, and does not contain endothelial cells. (I) Runt expression is found throughout the gill bar epithelia (light purple) with strongest expression adjacent to the skeletal rods (dark purple). (J) Hh is expressed at weaker levels in the atrial and pharyngeal epithelium (light purple) and at high levels in the cell population adjacent to the skeletal rods (dark purple).
Direct regulation of lancelet Hedgehog by Runt
As both Runt and Hh showed co-expression in the gill gut region, we analyzed whether a functional relationship between both genes, as it is known for the mouse [10], exists in lancelets. Analysis of the B. floridae Hh promoter revealed several putative Runt binding sites (Figure 9A). All of them were capable of binding to B. lanceolatum Runt as shown by electrophoretic mobility shift assays (Figure 9B). To provide further evidence for a Runt dependent regulation of BfHh we cloned different fragments of the BfHh promoter into the pGL3-basic luciferase reporter vector. Both, MmRunx2 and BlRunt were able to activate the different promoter constructs (Figure 9C).
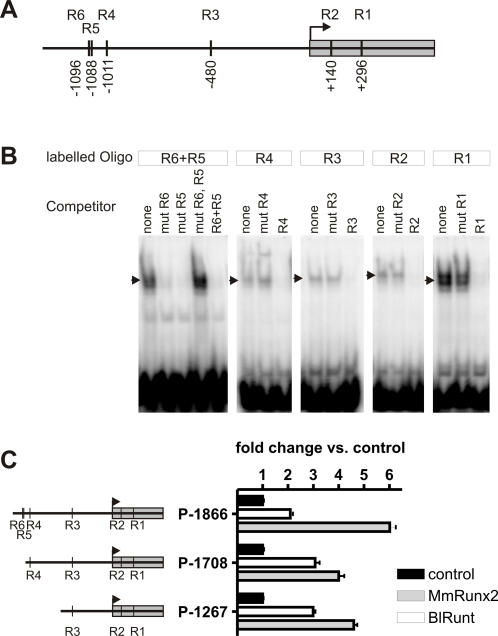
Figure 9. Runt dependent regulation of the B. floridae Hh promoter.
(A) Scheme of the BfHh promoter with putative Runt binding sites. Number and position relative to the transcription start site is given. (B) Electrophoretic mobility shift assays using oligos containing R1–R6 Runt binding sites. BlRunt can bind to each of the putative binding sites. Strongest binding is observed for the oligo with the closely adjacent binding sites R5 and R6 and for the R1 oligo. Nuclear extracts without BlRunt do not show a mobility shift of the oligos (data not shown). (C) Runt dependent activation of the BfHh promoter in NIH3T3 cells. Overexpression of either BlRunt or MmRunx2 leads to activation of the indicated promoter constructs compared to constructs co-transfected with an empty expression vector.
Discussion
Runt gene evolution in chordates
In order to get insight into the molecular mechanisms underlying the evolution of the skeleton we analyzed the evolution of the Runt gene family in various representative species. Runt genes are important regulators of neurogenesis and hematopoiesis [29],[30] and they are essential for mammalian skeletogenesis [7],[10]. Our analysis revealed that the stem species of chordates harboured most likely only a single Runt gene and as outlined in Figure 2 independent Runt duplications occurred in the clades of sea urchin (SpRunt1, SpRunt2), and bony fish (duplication of Runx2). A chordate stem species with only a single Runt gene is the most parsimonious assumption since the genomes of cnidarians, nematodes, cephalochordates and tunicates harbour also only a single Runt gene. The presence of two Runt genes in sea urchin is most likely a result of a tandem duplication, as we found both genes on a single genomic contig and they cluster together in our phylogenetic analysis (Figure 1). It was recently postulated that tunicates and not cephalochordates are the sistergroup of vertebrates [31],[32]. Focusing on our aim to reconstruct the framework for Runt gene evolution, both alternative taxonomic positions of tunicates and lancelets would be consistent with our hypothesis that the stem species of chordates harboured only a single Runt gene.
In accordance with the evidence for at least one genome wide duplication, 350 to 650 million years ago [33],[34] we detected in dogfish (as a representative of the jawed cartilaginous fish) three Runt genes, orthologous to Amniota Runx1, –2 and –3 genes, whereas only two Runt genes (MgRunxA and MgRunxB) were identified in hagfish (as a representative of jawless vertebrates). The phylogenetic tree (Figure 1) identifies the MgRunxA and MgRunxB genes as being closely related to the Runx1–3 genes. However, it is unknown if these evolved by a hagfish specific duplication or by a Runt gene duplication in the stem species of vertebrates. The phylogenetic analysis of the divergent Runt genes does not give satisfactory high support and a comparative analysis of the Runt gene loci will be needed to resolve this problem.
In the pufferfish (T. rubripes) genome, an enigmatic fourth Runt domain gene (FrRunt) was detected in addition to the orthologs of the Runx1, –2 and –3 genes, which appeared to represent either a pufferfish-specific fast evolving derivative of Runx2 or a direct descendant of the ancestral chordate Runt gene [22]. According to our data it is unlikely that the FrRunt gene represents a direct descendent of the ancestral chordate Runt gene which evolved in parallel with the vertebrate Runt genes [22] since we did not detect a FrRunt orthologous gene in tunicates, lancelets, hagfish and dogfish. Instead our phylogenetic analysis (Figure 1) and a comparison of the genomic environment of the FrRunt locus with the genomes of other bony fish (supporting information Figure S2) suggests that the FrRunt gene represents a fast evolving Runx2 orthologous gene. Such an accelerated evolution within duplicated genes is a common phenomenon [35].
Our findings that beside the human [24] also the chick and tunicate (C. intestinalis) Runt genes are followed by Clic genes together with the evidence that the FrRunt gene represents a fast evolving Runx2 gene suggests that during chordate evolution the entire Runt locus was triplicated.
Runt genes and the evolution of cartilage and bone in vertebrates
Cartilage has evolved multiple times in metazoa [1]. Here we focus on the vertebrate cellular cartilage expressing Col2α1 as the predominant matrix protein. Differentiation of this cartilage is regulated by a molecular network including Sox9, a transcription factor that directly regulates Col2α1 expression [36]. Furthermore Sox9 is a target of PTH related protein (PTHrP) that controls chondrocyte differentiation through a negative feedback loop with Indian hedgehog (Ihh). Runx2 in turn directly regulates Ihh [10]. Besides Runx2 also Runx1 and Runx3 genes are expressed during murine and zebrafish cartilage formation. However Runx2 and Runx3 appear to be the most important Runt genes for skeletogenesis [6],[23].
In hagfish soft and hard cartilage can be distinguished [2] and a Col2α1-homologous protein is expressed only in soft cartilage [4]. It is unknown if a protein homologous to Col1α1 is expressed in hard cartilage as it is the case in mammalian bone. As shown in Figure 3 the hagfish MgRunxB gene is only weakly expressed in both types of cartilage. However, the MgRunxA gene had its strongest expression in hard cartilage and its third highest expression in soft cartilage (Figure 3A). We only analyzed tissues from adult hagfish of medium size (30–40 cm). The fact that hagfish grow up to a length of 80 cm suggests that the Runt gene expression in hagfish cartilage is also of importance for the growth of the skeleton.
The view that Runt genes have a conserved functional role in skeletogenesis is also supported by our finding of Runt gene expression in the developing cartilage of dogfish. We detected a strong expression in visceral cartilage for all three dogfish Runt genes by qRT-PCR (Figure 4). Furthermore we performed in situ hybridizations on dogfish embryos and found ScRunx2 to be expressed in the cartilage of the fin and together with ScRunx3 in the gill gut cartilage (Figure 6).
In lamprey (another representative of jawless vertebrates) the Col2α1 gene is expressed in cartilage along with Sox9 and PTHrP, indicating that they were already a part of the chondrogenic gene repertoire in early vertebrate evolution [3]. Our finding of dogfish and hagfish Runt expression in cartilage together with the well-known role of Runt genes in skeletogenesis, suggests that Runt genes can now be considered to be a part of the ancient molecular machinery for cartilage formation in the stem species of vertebrates.
ScRunx1–3 gene expression in teeth and placoid scales
Placoid scales are small conical structures in the skin of cartilaginous fish. We found that all three dogfish Runt genes are expressed in the developing placoid scales (Figure 5A–5C). Interestingly, the basal plate of scales and teeth is initiated by osteoblasts which continue to secrete bone matrix in a basal direction, while slightly later, the odontoblasts secrete dentine on the pulpar side on the basal plate [37]. Since ScRunx2 is expressed in the developing basal plate it is an interesting speculation that the expression of Runx2 at this site might reflect the origin of bone as a dermal tissue in early vertebrate evolution. The dermoskeleton is the first to show mineralization in vertebrate phylogeny [38]. This mineralized dermoskeleton was composed of odontodes (dermal “teeth”) supported by extensively developed bone, imposing mineralization upon the collagenous layer of the dermis [38].
In placoid scales as well as in the developing teeth ScRunx1 and ScRunx3 were expressed in the stratum germinativum, whereas ScRunx2 was found at the site where later the basal plate will develop (Figure 5A–5F). In mammals teeth develop as epithelial appendages in which sequential and reciprocal interaction between the ectoderm and underlying neural crest derived mesenchyme constitute a central developmental mechanism [1],[14]. The dental epithelial cells differentiate into ameloblasts and mesenchymal cells into odontoblasts, secreting the matrices enamel, and dentin respectively [1]. Runx2 and Runx3 expression is confined to mesenchymal tissues, whereas Runx1 was found to be restricted to epithelia [14].
According to a classical view teeth evolved secondarily from skin denticles moving into the mouth (reviewed in [39]). However, this model was recently challenged by the proposal that sets of denticles on the pharyngeal (gill) arches and not external denticles were the precursors of the organized tooth families [39]. This alternative theory was based on the observations of homologous arrays of denticle whorls occurring within the pharyngeal region of jawless fish such as the thelodont Loganellia [40]. In this model the endoderm played an important role in the patterning process involved in the production of denticles on the postbranchial lamina [39]. It was assumed that the denticles on the postbranchial lamina have been formed in the presence of an inductive endoderm as one part of the internal visceral skeleton. This would be remarkably different to the development of external denticles, which are only under the influence of an inductive ectoderm [39].
Our Runt expression pattern supports the classical view that teeth and placoid scales have a common evolutionary origin, at least on the level of the molecular pathway underlying their development. In other words, the hypothesis that teeth and placoid scales evolved from a common developmental module, which might have been shifted and extended in its expression topology [41] is supported by the striking similarity of the Runt expression patterns in teeth and placoid scales.
Conservation of molecular pathways in skeletogenesis
The gut appears to be an ancient expression domain of Runt. This expression in the chordate stem species can be reconstructed as Runt genes are expressed in the gut in representatives of the outgroup (sea urchins, nematode [16],[17] and the lancelet (this study, Figures 7 and 8). The Runt expression in the gill bars, structures that stabilize the gill gut, might be linked to the later role of Runt genes in the evolution of the pharyngeal skeleton. In zebrafish Runx3 was shown to promote cartilage formation via the endodermal expression of Runx3 in pharyngeal pouch cells [23].
However, in vertebrates most of the branchial arch cartilage, the cranial bone forming cells (osteoblasts), as well as the cells that deposit dentin (odontoblasts) are derived from the neural crest [42]. It was previously proposed that the neural crest acquired chondrogenic ability by recruiting proto-chondrogenic gene programs from the notochord, neural tube and gill gut [4], [5], [28], [43]–[45] Strikingly, we found high Hh expression together with high Runt expression in exactly these three sites indicating that the described interaction between the Runt and Hh pathways is of relevance for chordate cartilage evolution.
Whereas the homology of the gill gut in lancelets and vertebrates is well established [28] little is known about the molecular machinery necessary for development and maintenance of the skeletal-like structures of the pharyngeal gill slits in lancelets. The gill bars are stabilized by 15 nm thick filaments aligned parallel to the long axis of the rods, and are covered by a single layered epithelium, that can be morphologically distinguished into atrial, lateral and pharyngeal epithelium [46]. Gill bars gave a positive signal when stained with an antibody against type II collagen [28] indicating a cartilage-like structure, which appears to be acellular.
To get deeper insights into the molecular machinery underlying the early evolution of the skeleton we analyzed Runt and SoxE gene expression in adult lancelets. Our analyses revealed that both genes were highly expressed in the gill bar region (Figure 8). Furthermore our in situ hybridization results revealed that the lancelet Runt gene is expressed in atrial, lateral, and pharyngeal epithelium of ectodermal and endodermal origin (Figure 8D and 8E), but not in the mesodermal coelomic cells of the primary gill bars. It has recently been reported that the lancelet gills contains lymphocyte-like cells most likely located between the cells of the lateral and pharyngeal epithelia [47]. We cannot resolve these cells in our in situ hybridizations and thus cannot detect if Runt is expressed in these cells of the gill bars (Figure 8D and 8E). The finding of endodermal Runt expression supports the model in which endodermal secretion was the ancestral mode of making cartilage [5]. Since in deuterostomes the endoderm is a plesiomorphic Runt expression domain, Runt is likely to be present also in the endoderm of the gill gut in hemichordates.
Other crucial genes for mammalian skeletogenesis are Ihh and Shh. For Ihh a direct regulation by Runx2 has been shown and Runx2 influences Shh signaling in tooth development [10],[11]. Furthermore, Runt and Ihh genes are coexpressed during skeletogenesis in zebrafish [23],[25],[48]. We observed Runt expression in the midgut and foregut of lancelet larvae, similar to a recent study [49]. The exon 1 variant, however, showed additional expression in the notochord and neural tube (Figure 7A, 7C, and 7E). These expression domains were still detected in adult lancelets together with high Hh expression (Figure 8A and 8B). The observation that the single Runt and Hh genes of lancelets are co-expressed in the notochord, neural tube and in the adult lancelet gill gut (Figures 7 and 8 and reference [50]) prompted us to investigate if also the lancelet Runt protein might regulate lancelet Hh gene expression. In our Hh promoter studies the lancelet Runt protein bound directly to Runt binding sites in the lancelet Hh promoter and regulated the reporter gene driven by this promoter (Figure 9). The highest Hh expression together with Runt co-expression was found in the notochord, the neural tube, and the gill gut, all of which were previously proposed to be involved in the evolution of chordate cartilage [4],[5],[28],[44],[45]. It is thus likely, that the direct regulation of Hh by Runt was a relevant mechanism in chordate evolution. This suggests that the core gene network involved in vertebrate cartilage, bone and tooth formation was present prior to the divergence of cephalochordates and vertebrates and the duplication of the Runt and Hh genes.
Further research is needed to determine if a small cell group directly adjacent to both sites of the acellular matrix, with high Runt and Hh expression (arrows in Figure 8D–8G), is of special importance for cartilage formation in lancelets. Another interesting aspect will be to determine if a direct regulatory interaction between the Runt and Hh pathways is also present in hemichordates and whether a direct interaction between Runt and Hh pathways was maintained during vertebrate evolution in other important developmental processes, such as vertebrate hematopoiesis [29],[51].
Materials and Methods
Materials
Lancelets (B. floridae) were collected by shovel and sieve in water of 1 m in depth in Tampa Bay, Florida and in vitro fertilization, embryo culture and fixation were performed as previously described [52]. Adult B. lanceolatum were obtained from the Biologische Anstalt Helgoland. Hagfish (M. glutinosa) were collected by S.E. Material from adult dogfish (S. canicula) was obtained from the Biologische Anstalt Helgoland and dogfish embryos from the Aquazoo (Düsseldorf).
Oligonucleotides
All primers and oligonucleotides employed in our study are given as supporting information. Primers for dogfish sequences can be found in Table S1. Primers for hagfish sequences are given in Table S2, and primers for amphioxus are listed in Table S3. Oligonucleotides employed for EMSAs are given in Table S4.
Analysis of Runt gene sequence and number
Total RNA was isolated as described previously [17] from B. floridae (larvae), B.lanceolatum (adult), M. glutinosa (adult), S. canicula (embryos 4,5 cm, 6,5 cm, and 9,5 cm as well as adult animals). Runt genes were amplified by a strategy reported previously, using degenerated primers to amplify the conserved Runt domain followed by RACE PCRs to amplify the full length Runt genes [17]. The only exception was the amplification of the hagfish MgRunxB 5′ end which was obtained by inverse PCR with gene specific primers [53]
Phylogenetic analysis
Alignments were obtained with ClustalW from 28 full length Runt amino acid sequences [54]. Ambiguously aligned proportions were omitted using Gblocks ver. 0.91b [55] with following parameters: minimum number of sequences for a conserved/flanking position (15/15), maximum number of contiguous nonconserved positions (8), minimum length of a block (5), allowed gap positions (all). The phylogenetic analysis was performed using MrBayes 3.1.5 [56], employing JTT+G+I as substitution model and running eight chains for 1.000.000 generations. Trees were sampled every 1000 generations and according to a saturation curve of likelihood values the first 500 trees were discarded as burn-in. Analysis was performed with Runt sequences from O. dioica (AAS21356.1), C. intestinalis (ci0100131551, ci010013155, ciad013o19, cinc013i02 and cies003n20), B. lanceolatum and B. floridae (AAN08567.1, AAN08565.1), M. glutinosa (DQ990008, DQ990009), S. canicula (DQ990010, DQ990012, DQ990014), D. rerio (NP_571678.1, AAS02047.1, AAQ88389.1, AAO85550.1). T. rubripes (BAF36011.1, BAF36001.1, AB280005.1, NP_001092121), G. aculeatus (Ensemble Gene Id: ENSGACP00000020145, ENSGACG00000012322, ENSGACG00000011721, ENSGACG00000007301), M. musculus (EDL03777.1, BAA03485.1, EDL29993.1) and H. sapiens (NP_001001890.1, EAX04278.1, NP_004341.1), while using the sea urchin Runt genes from S. purpuratus (U41512.2, XM_776533.1) as an outgroup.
In situ hybridizations
Whole mount in situ hybridizations with lancelet larvae were performed as previously described [43]. Radioactive in situ hybridizations on paraffin embedded tissue sections were performed as reported in [57] with the exception of using lower hybridization and washing temperatures of 50°C, and using 0,2× SSC instead of 2× SSC for washing of B. lanceolatum tissue sections. Non-radioactive in situ hybridization on cryo-sections of B. lanceolatum was carried out using the GenePaint System [58]. Probes for MmIhh and ScRunx3 were used as hybridization controls for B. lanceolatum.
Expression profiling of Runt genes in M. glutinosa, S. canicula and B. lanceolatum by qRT-PCR
QRT-PCR was performed on an ABIPrism 7900HT Cycler (Applied Biosystems, Forster City, USA) using SYBR Green PCR Master Mix (Applied Biosystems). TaqMan Reverse Transcription Reagents (Applied Biosystems) were used to synthesize the cDNA and primers were generated using the Primer Express software (Applied Biosystems). Quantification was performed using the standard curve method with dilutions of plasmids containing the sequence to be amplified in a known copy number as a standard. For the analysis of SoxE expression by qRT-PCR first a SoxE cDNA fragment was amplified by employing primers which were designed according to a SoxE sequence of B. floridae. Expression of target genes was normalized using 18S rRNA as reference.
Immunohistology
For immunohistochemistry on paraffin sections citrate antigen retrieval was performed. Anti-human Ihh antibody (Santa Cruz) was applied 1∶50 over night. Secondary antibody (biotinylated anti-goat, Sigma-Aldrich) was applied 1∶500 for one hour. Subsequent staining was performed with the Vectastain ABC kit from Vector laboratories according to the manufacturers′ instructions.
EMSA
Electrophoretic mobility shift assays for putative binding sites were performed as described in [59] with nuclear extracts from chicken DF-1 cells infected with a RCAS-virus expressing the Runt cDNA from B. lanceolatum. Specific binding was confirmed with a labeled oligo containing the putative binding site and using either wild type oligos or oligos with mutated binding sites as competitors.
Luciferase reporter assays
PCR amplified fragments of the B. floridae Hh promoter (AC150424) were cloned into the pGL3-basic reporter vector. NIH3T3 cells were transfected in 24-well plates with the reporter constructs (250 ng per well) together with an expression vector containing either the cDNA for BlRunt or MmRunx2 or an empty vector as control (100 ng per well). 5 ng per well of pRL-CMV were co-transfected for normalization. Cells were lysed with 100 µl passive lysis buffer (Dual Luciferase Assay Kit; Promega, Madison, USA). 5 µl of the lysate were measured using the Dual-Glo Luciferase Assay Kit (Promega) with 25 µl of the assay reagents each. Measurements were performed on a 1450 MicroBeta Scintillation and Luminescence Counter (Perkin Elmer, Waltham, USA). The result of a representative experiment is shown which was confirmed five times independently.
Data deposition
The sequences reported in this paper have been deposited in the GenBank databases. Dogfish: MASNS-like-promoter variant 1, ScRunx1 Acc-Nr DQ990011, ScRunx2 DQ990013, ScRunx3 DQ990015 and MRIPV-like-motifs promoter variant 2, ScRunx1 DQ990010, ScRunx2 DQ990012, ScRunx3 DQ990014. Hagfish: MgRunxA DQ990008, MgRunxB DQ990009. Lancelet: SoxE EF051347.
Supporting Information
Alignment used for Phylogenetic Analysis. Alignment (ClustalW, BioEdit: http://www.mbio.ncsu.edu/BioEdit/bioedit.html) of newly detected Runt genes in hagfish (MgRunxA and B, DQ990008, DQ990009) and dogfish (ScRunx1-3, DQ990010, DQ990012, DQ990014) with other deuterostome Runt genes. The conserved sequence blocks used for the phylogenetic analysis are underlined with #. Parameters used with Gblocks 0.91b were: Minimum number of sequences for a conserved / flanking position: 15/15; Maximum number of contiguous nonconserved positions: 8; minimum length of a block: 5; allowed gap positions: all. 338 (52%) of the original 645 alignment positions were used in the phylogenetic analysis.
Abbreviations: B.l.: Branchiostoma lanceolatum; C.i.: Ciona intestinalis; D.r.: Danio rerio; G.a.: Gasterosteus aculeatus; H.s.: Homo sapiens; M.m.: Mus musculus; M.g.: Myxine glutinosa; O.d.: Oikopleura dioica; S.p.: Strongylocentrotus purpuratus; S.c.: Scyliorhinus canicula; T.r.: Takifugu rubripes.
(0.09 MB DOC)
Synteny Analysis.
A search for cross-species conserved gene orders was performed as previously described [1]. We compared a larger contig of the FrRunt locus (Ensemble: Scaffold 39) than previously analyzed (Ensemble: Scaffold 835[2]) to the zebrafish genome and detected a synteny region between the 3′ genomic region of the FrRunt gene and chromosome 1 of zebrafish comprising Fstl1 and Gja5 (A). Furthermore we detected in the stickleback (G. aculeatus) genome a FrRunt orthologous gene with a genomic environment almost identical to the FrRunt gene locus (B). The gene orthologous to Clic 5 located 3′ of Runx2a in the zebrafish genome was found by Blast searches on group 1 in the stickleback genome. Together these results suggest that a translocation between a region of the 3′ end of the FrRunt locus and chromosome 1 had occurred in the common stem species of pufferfish and stickleback.
(0.05 MB DOC)
Dogfish Primers.
Primers employed to amplify and analyze the expression of Runt genes in dogfish. PA: Primary amplification, RA: Reamplification.
(0.08 MB DOC)
Hagfish Primers.
Primers employed to detect Runt genes and analyze Runt gene expression in hagfish. PA: Primary amplification, RA: Reamplification.
(0.07 MB DOC)
Amphioxus Primers.
Primers employed to analyze Sox9, Hedgehog and Runt genes in lancelets.
(0.08 MB DOC)
EMSA Oligos.
Oligos employed for the electrophoretic mobility shift assays.
(0.07 MB DOC)
Acknowledgments
We thank T. Bartolomaeus (FU Berlin), P. Seemann (MPI for Molecular Genetics, Berlin), W. Gettmann, H. Bosch (Aquazoo Düsseldorf), M. Kruess (Biologische Anstalt Helgoland) for support and discussion and K. Seitz and PN Robinson for their editorial help.
Footnotes
The authors have declared that no competing interests exist.
No external funding was granted for this study.
References
- 1.Hall BK. Bones and Cartilage, Developmental and Evolutionary Skeletal Biology. London: Elsevier Academic Press; 2005. p. 760. [Google Scholar]
- 2.Robson P, Wright GM, Keeley FW. Distinct non-collagen based cartilages comprising the endoskeleton of the Atlantic hagfish, Myxine glutinosa. Anat Embryol (Berl) 2000;202:281–290. doi: 10.1007/s004290000113. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G, Miyamoto MM, Cohn MJ. Lamprey type II collagen and Sox9 reveal an ancient origin of the vertebrate collagenous skeleton. Proc Natl Acad Sci U S A. 2006;103:3180–3185. doi: 10.1073/pnas.0508313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang G, Cohn MJ. Hagfish and lancelet fibrillar collagens reveal that type II collagen-based cartilage evolved in stem vertebrates. Proc Natl Acad Sci U S A. 2006;103:16829–16833. doi: 10.1073/pnas.0605630103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rychel AL, Swalla BJ. Development and evolution of chordate cartilage. J Exp Zoolog B Mol Dev Evol. 2007;308:325–335. doi: 10.1002/jez.b.21157. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida CA, Komori T. Role of Runx proteins in chondrogenesis. Crit Rev Eukaryot Gene Expr. 2005;15:243–254. doi: 10.1615/critreveukargeneexpr.v15.i3.60. [DOI] [PubMed] [Google Scholar]
- 7.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 8.Rennert J, Coffman JA, Mushegian AR, Robertson AJ. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–63. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XP, Aberg T, James MJ, Levanon D, Groner Y, et al. Runx2 (Cbfa1) inhibits Shh signaling in the lower but not upper molars of mouse embryos and prevents the budding of putative successional teeth. J Dent Res. 2005;84:138–143. doi: 10.1177/154405910508400206. [DOI] [PubMed] [Google Scholar]
- 12.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 13.Saltman LH, Javed A, Ribadeneyra J, Hussain S, Young DW, et al. Organization of transcriptional regulatory machinery in osteoclast nuclei: Compartmentalization of Runx1. J Cell Physiol. 2005 doi: 10.1002/jcp.20329. [DOI] [PubMed] [Google Scholar]
- 14.Yamashiro T, Aberg T, Levanon D, Groner Y, Thesleff I. Expression of Runx1, –2 and –3 during tooth, palate and craniofacial bone development. Mech Dev. 2002;119(Suppl 1):S107–S110. doi: 10.1016/s0925-4773(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 15.Hall BK. Consideration of the neural crest and its skeletal derivatives in the context of novelty/innovation. J Exp Zoolog B Mol Dev Evol. 2005;304:548–557. doi: 10.1002/jez.b.21057. [DOI] [PubMed] [Google Scholar]
- 16.Nam S, Jin YH, Li QL, Lee KY, Jeong GB, et al. Expression pattern, regulation, and biological role of runt domain transcription factor, run, in Caenorhabditis elegans. Mol Cell Biol. 2002;22:547–554. doi: 10.1128/MCB.22.2.547-554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stricker S, Poustka AJ, Wiecha U, Stiege A, Hecht J, et al. A single amphioxus and sea urchin runt-gene suggests that runt-gene duplications occurred in early chordate evolution. Dev Comp Immunol. 2003;27:673–684. doi: 10.1016/s0145-305x(03)00037-5. [DOI] [PubMed] [Google Scholar]
- 18.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Guerra A, Aze A, Morales J, Mulner-Lorillon O, Cosson B, et al. The genomic repertoire for cell cycle control and DNA metabolism in S. purpuratus. Dev Biol. 2006;300:238–251. doi: 10.1016/j.ydbio.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Anderson MK, Pant R, Miracle AL, Sun X, Luer CA, et al. Evolutionary origins of lymphocytes: ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J Immunol. 2004;172:5851–5860. doi: 10.4049/jimmunol.172.10.5851. [DOI] [PubMed] [Google Scholar]
- 21.Ng CE, Osato M, Tay BH, Venkatesh B, Ito Y. cDNA cloning of Runx family genes from the pufferfish (Fugu rubripes). Gene. 2007;399:162–173. doi: 10.1016/j.gene.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Glusman G, Kaur A, Hood L, Rowen L. An enigmatic fourth runt domain gene in the fugu genome: ancestral gene loss versus accelerated evolution. BMC Evol Biol. 2004;4:43. doi: 10.1186/1471-2148-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flores MV, Lam EY, Crosier P, Crosier K. A hierarchy of Runx transcription factors modulate the onset of chondrogenesis in craniofacial endochondral bones in zebrafish. Dev Dyn. 2006;235:3166–3176. doi: 10.1002/dvdy.20957. [DOI] [PubMed] [Google Scholar]
- 24.Strippoli P, D'Addabbo P, Lenzi L, Giannone S, Canaider S, et al. Segmental paralogy in the human genome: a large-scale triplication on 1p, 6p, and 21q. Mamm Genome. 2002;13:456–462. doi: 10.1007/s00335-001-2157-0. [DOI] [PubMed] [Google Scholar]
- 25.Flores MV, Tsang VW, Hu W, Kalev-Zylinska M, Postlethwait J, et al. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr Patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- 27.Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Chau JY, et al. Runx3 is required for hematopoietic development in zebrafish. Dev Dyn. 2003;228:323–336. doi: 10.1002/dvdy.10388. [DOI] [PubMed] [Google Scholar]
- 28.Rychel AL, Smith SE, Shimamoto HT, Swalla BJ. Evolution and Development of the Chordates: Collagen and Pharyngeal Cartilage. Mol Biol Evol. 2006;23:541–549. doi: 10.1093/molbev/msj055. [DOI] [PubMed] [Google Scholar]
- 29.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 30.Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, et al. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 32.Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 33.McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 34.Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications – the adventure of a hypothesis. Trends Genet. 2005;21:559–567. doi: 10.1016/j.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Steinke D, Salzburger W, Braasch I, Meyer A. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics. 2006;7:20. doi: 10.1186/1471-2164-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9(Suppl A):S69–S75. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- 37.Reif WE. Development of Dentition and Dermal Skeleton in Embryonic Scyliorhinus canicula. Journal of Morphology. 1980:275–288. doi: 10.1002/jmor.1051660303. [DOI] [PubMed] [Google Scholar]
- 38.Donoghue PC, Sansom IJ, Downs JP. Early evolution of vertebrate skeletal tissues and cellular interactions, and the canalization of skeletal development. J Exp Zoolog B Mol Dev Evol. 2006;306:278–294. doi: 10.1002/jez.b.21090. [DOI] [PubMed] [Google Scholar]
- 39.Johanson Z, Smith M. Origin and evolution of gnathostome dentitions: a question of teeth and pharyngeal denticles in placoderms. Biol Rev. 2005;80:303–345. doi: 10.1017/s1464793104006682. [DOI] [PubMed] [Google Scholar]
- 40.Johanson Z, Smith MM. Placoderm fishes, pharyngeal denticles, and the vertebrate dentition. J Morphol. 2003;257:289–307. doi: 10.1002/jmor.10124. [DOI] [PubMed] [Google Scholar]
- 41.Donoghue PC, Sansom IJ. Origin and early evolution of vertebrate skeletonization. Microsc Res Tech. 2002;59:352–372. doi: 10.1002/jemt.10217. [DOI] [PubMed] [Google Scholar]
- 42.Hall BK. The neural crest in development and evolution. New York: Springer; 1999. p. 314. [Google Scholar]
- 43.Meulemans D, Bronner-Fraser M. Central role of gene cooption in neural crest evolution. J Exp Zoolog B Mol Dev Evol. 2005;304:298–303. doi: 10.1002/jez.b.21047. [DOI] [PubMed] [Google Scholar]
- 44.Meulemans D, McCauley D, Bronner-Fraser M. Id expression in amphioxus and lamprey highlights the role of gene cooption during neural crest evolution. Dev Biol. 2003;264:430–442. doi: 10.1016/j.ydbio.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Baker CV, Bronner-Fraser M. The origins of the neural crest. Part II: an evolutionary perspective. Mech Dev. 1997;69:13–29. doi: 10.1016/s0925-4773(97)00129-9. [DOI] [PubMed] [Google Scholar]
- 46.Rähr H. Ultrastructure of gill bars of Branchiostoma lanceolatum with special reference to gill skeleton and blood vessels (Cephalochordata). Zoomorphology. 1982;99:167–180. [Google Scholar]
- 47.Huang G, Xie X, Han Y, Fan L, Chen J, et al. The identification of lymphocyte-like cells and lymphoid-related genes in amphioxus indicates the twilight for the emergency of adaptive immune system. PLoS ONE. 2007;2:e206. doi: 10.1371/journal.pone.0000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avaron F, Hoffman L, Guay D, Akimenko MA. Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev Dyn. 2006;235:478–489. doi: 10.1002/dvdy.20619. [DOI] [PubMed] [Google Scholar]
- 49.Meulemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS ONE. 2007;2:e787. doi: 10.1371/journal.pone.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimeld SM. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev Genes Evol. 1999;209:40–47. doi: 10.1007/s004270050225. [DOI] [PubMed] [Google Scholar]
- 51.Dyer MA, Farrington SM, Mohn D, Munday JR, Baron MH. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development. 2001;128:1717–1730. doi: 10.1242/dev.128.10.1717. [DOI] [PubMed] [Google Scholar]
- 52.Holland ND, Holland LZ. Embryos and larvae of invertebrate deuterostomes. In: Stern CD, Holland PWH, editors. Essential developmental biology: a practical approach. Oxford: IRL Press; 1993. pp. 21–32. [Google Scholar]
- 53.Triglia T, Peterson MG, Kemp DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 55.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 56.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 57.Albrecht AN, Schwabe GC, Stricker S, Boddrich A, Wanker EE, et al. The synpolydactyly homolog (spdh) mutation in the mouse – a defect in patterning and growth of limb cartilage elements. Mech Dev. 2002;112:53–67. doi: 10.1016/s0925-4773(01)00639-6. [DOI] [PubMed] [Google Scholar]
- 58.Reymond A, Marigo V, Yaylaoglu MB, Leoni A, Ucla C, et al. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002;420:582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- 59.Stock M, Schafer H, Stricker S, Gross G, Mundlos S, et al. Expression of galectin-3 in skeletal tissues is controlled by Runx2. J Biol Chem. 2003;278:17360–17367. doi: 10.1074/jbc.M207631200. [DOI] [PubMed] [Google Scholar]
- 60.Jeffery WR. Ascidian neural crest-like cells: phylogenetic distribution, relationship to larval complexity, and pigment cell fate. J Exp Zoolog B Mol Dev Evol. 2006;306:470–480. doi: 10.1002/jez.b.21109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment used for Phylogenetic Analysis. Alignment (ClustalW, BioEdit: http://www.mbio.ncsu.edu/BioEdit/bioedit.html) of newly detected Runt genes in hagfish (MgRunxA and B, DQ990008, DQ990009) and dogfish (ScRunx1-3, DQ990010, DQ990012, DQ990014) with other deuterostome Runt genes. The conserved sequence blocks used for the phylogenetic analysis are underlined with #. Parameters used with Gblocks 0.91b were: Minimum number of sequences for a conserved / flanking position: 15/15; Maximum number of contiguous nonconserved positions: 8; minimum length of a block: 5; allowed gap positions: all. 338 (52%) of the original 645 alignment positions were used in the phylogenetic analysis.
Abbreviations: B.l.: Branchiostoma lanceolatum; C.i.: Ciona intestinalis; D.r.: Danio rerio; G.a.: Gasterosteus aculeatus; H.s.: Homo sapiens; M.m.: Mus musculus; M.g.: Myxine glutinosa; O.d.: Oikopleura dioica; S.p.: Strongylocentrotus purpuratus; S.c.: Scyliorhinus canicula; T.r.: Takifugu rubripes.
(0.09 MB DOC)
Synteny Analysis.
A search for cross-species conserved gene orders was performed as previously described [1]. We compared a larger contig of the FrRunt locus (Ensemble: Scaffold 39) than previously analyzed (Ensemble: Scaffold 835[2]) to the zebrafish genome and detected a synteny region between the 3′ genomic region of the FrRunt gene and chromosome 1 of zebrafish comprising Fstl1 and Gja5 (A). Furthermore we detected in the stickleback (G. aculeatus) genome a FrRunt orthologous gene with a genomic environment almost identical to the FrRunt gene locus (B). The gene orthologous to Clic 5 located 3′ of Runx2a in the zebrafish genome was found by Blast searches on group 1 in the stickleback genome. Together these results suggest that a translocation between a region of the 3′ end of the FrRunt locus and chromosome 1 had occurred in the common stem species of pufferfish and stickleback.
(0.05 MB DOC)
Dogfish Primers.
Primers employed to amplify and analyze the expression of Runt genes in dogfish. PA: Primary amplification, RA: Reamplification.
(0.08 MB DOC)
Hagfish Primers.
Primers employed to detect Runt genes and analyze Runt gene expression in hagfish. PA: Primary amplification, RA: Reamplification.
(0.07 MB DOC)
Amphioxus Primers.
Primers employed to analyze Sox9, Hedgehog and Runt genes in lancelets.
(0.08 MB DOC)
EMSA Oligos.
Oligos employed for the electrophoretic mobility shift assays.
(0.07 MB DOC)