Abstract
The p24 family members are transmembrane proteins assembled into heteromeric complexes that continuously cycle between the ER and the Golgi apparatus. These cargo proteins were assumed to play a structural role in COPI budding because of their major presence in mammalian COPI vesicles. However, this putative function has not been proved conclusively so far. Furthermore, deletion of all eight yeast p24 family members does not produce severe transport phenotypes, suggesting that the p24 complex is not essential for COPI function. In this paper we provide direct evidence that the yeast p24 complex plays an active role in retrograde transport from Golgi to ER by facilitating the formation of COPI-coated vesicles. Therefore, our results demonstrate that p24 proteins are important for vesicle formation instead of simply being a passive traveler, supporting the model in which cargo together with a small GTPase of the ARF superfamily and coat subunits act as primer for vesicle formation.
Introduction
COPI vesicle-mediated retrograde transport from the Golgi apparatus to the endoplasmic reticulum (ER) is crucial for eukaryotic cell physiology (Semenza et al., 1990). The molecular mechanisms that drive COPI vesicle generation are not completely understood (Spang, 2002). The current model, known as priming complex model (Springer et al., 1999), proposes that Arf1p, a small GTPase, is attracted to the Golgi membrane, where it is activated by a guanine exchange factor (ArfGEF). Membrane-bound, activated Arf1p then recruits coatomer, a preassembled complex of seven subunits, which forms a priming complex with a GTPase-activating protein (ArfGAP) and a transmembrane protein (cargo or SNARE). Once enough COPI priming complexes are established, they associate laterally to form a larger, polymeric coat on the Golgi membrane. This coat polymerization leads to the deformation of the membrane and, subsequently, to the COPI vesicle generation.
In addition to the coat components, transmembrane proteins might also be important for the budding process by acting as a primer to form priming complexes (Springer et al., 1999). Specific COPI vesicle passengers, such as the members of the p24 family, have been suggested to play a role in COPI vesicle–mediated flow (Stamnes et al., 1995; Bremser et al., 1999). The p24 proteins are assembled into heteromeric complexes that continuously cycle between ER and Golgi compartments (Sohn et al., 1996; Rojo et al., 1997; Fullekrug et al., 1999; Belden and Barlowe, 2001b). In yeast, at least four members of the p24 family (Emp24p, Erv25p, Erp1p, and Erp2p) function in the p24 complex (Marzioch et al., 1999). The yeast p24 complex plays a specialized role in selective cargo recruitment into specific ER-derived vesicles (Muñiz et al., 2000, 2001). Nevertheless, this is not likely to be the only function of the p24 complex in the early secretory pathway (Elrod-Erickson and Kaiser, 1996; Bremser et al., 1999; Lavoie et al., 1999; Emery et al., 2003). The p24 proteins have been assumed to play a structural role in COPI vesicle formation because of their ability to bind COPI proteins and their major presence in mammalian COPI vesicles (Stamnes et al., 1995; Sohn et al., 1996). Indeed, mammalian p24 cytosolic tails displayed on liposomes can stimulate the formation of COPI vesicles (Bremser et al., 1999). Consistent with these findings and based on further in vitro experiments, it has been proposed that mammalian p24 proteins could inhibit GTP hydrolysis by slowing down ArfGAP activity, thereby ensuring that coatomer can remain on the membrane in order to form COPI priming complexes required for vesicle formation (Goldberg, 2000; Lanoix et al., 2001). This model predicts a critical role for p24 proteins in COPI vesicle biogenesis. However, deletion of all eight yeast p24 family members does not produce severe transport phenotypes or morphological alterations in the endomembrane system, suggesting that p24 complex is not essential for COPI function (Springer et al., 2000). Therefore, whether p24 proteins are just COPI coat-binding cargo proteins or are required for COPI coated vesicle formation itself still remains a matter of debate. Here, we provide evidence that yeast p24 complex plays an active role in COPI vesicle budding from Golgi membranes.
Results and discussion
The emp24Δ and erv25Δ mutations interact genetically with mutant alleles of genes encoding for COPI vesicle coat components
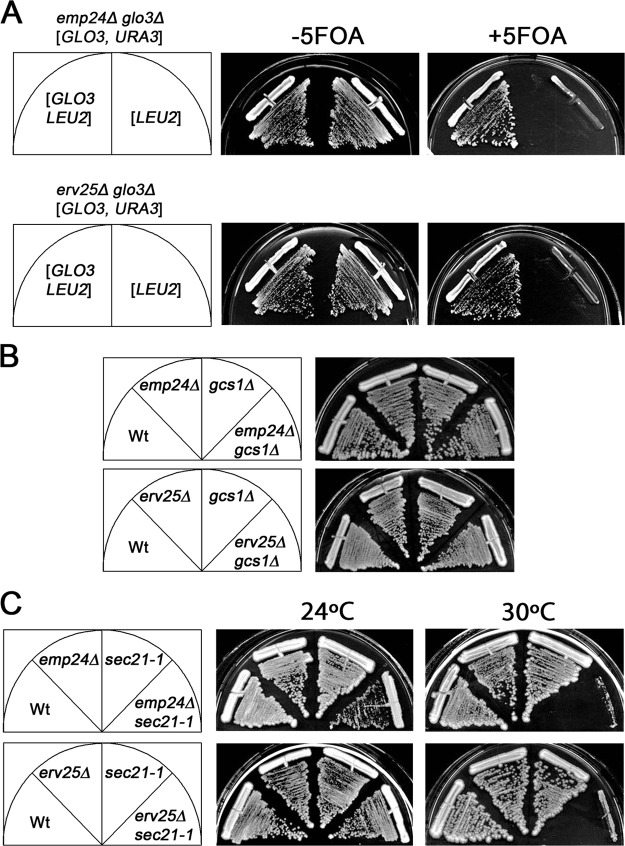
To gain more insights into the functions of the yeast p24 complex in the early secretory pathway, we performed a screen for mutations that induce synthetic lethality upon disruption of the EMP24 gene. We identified a genetic interaction between EMP24 and GLO3, a gene encoding for an ArfGAP involved in COPI vesicle formation (Lewis et al., 2004). Confirming this result, GLO3 could rescue the viability of the emp24Δ glo3Δ double mutant (Fig. 1 A). This genetic interaction is also true for other p24 members. As expected, the erv25Δ glo3Δ double mutant was inviable but could be complemented by GLO3 (Fig. 1 A). Deletion of EMP24 or ERV25 destabilizes the other three proteins of the complex, leading to a complete loss of p24 function (Marzioch et al., 1999). Thus, it is rather likely that other members of the p24 family play a similar role as Emp24p and Erv25p with respect to interaction with Glo3p.
Figure 1.
The emp24Δ and erv25Δ mutations interact genetically with mutant alleles of genes encoding for COPI vesicle coat components. (A) The emp24Δ glo3Δ strain with the URA3 based plasmid bearing GLO3 pMMY63 was transformed with LEU2-based empty vector pRS315 or GLO3 encoding plasmid pMMY67. Double transformants were replica plated at 30°C on SD-leu-ura or on 5-FOA medium to induce loss of pMMY63. (B) The emp24Δ or erv25Δ strains were crossed with a gcs1Δ strain and the resulting haploid spores were tested for growth at 30°C on YPUAD. (C) emp24Δ or erv25Δ strains were crossed with a sec21-1 strain and the resulting haploid spores were tested for growth at 24 and 30°C and grown on YPUAD.
Two partially redundant ArfGAPs operate in retrograde traffic from Golgi to ER: Glo3p and Gcs1p (Poon et al., 1999). However, neither emp24Δ nor erv25Δ showed genetic interaction with gcs1Δ (Fig. 1 B). We also found that overexpression of GCS1 was not able to restore growth of the emp24Δ glo3Δ double mutant (unpublished data). These results show that the genetic interaction between p24 coding genes and GLO3 is specific and are consistent with previous findings that Glo3p and Gcs1p have only partially overlapping functions (Poon et al., 1999; Lewis et al., 2004; Yahara et al., 2006).
Because the ArfGAP Glo3p is a component of the COPI vesicle coat through interactions with the coatomer subunits Sec21p and Sec27p (Eugster et al., 2000; Lewis et al., 2004), we wanted to investigate the genetic interaction of p24 mutations with mutations in SEC21 and SEC27. emp24Δ and erv25Δ showed a synthetic growth defect when combined with sec21-1 (Fig. 1 C) and sec27-1 (unpublished data) mutations.
Altogether, these genetic interactions between emp24Δ or erv25Δ and genes encoding COPI vesicle coat proteins involved in Golgi to ER transport strongly suggest that p24 proteins could participate in the formation of COPI vesicles.
The p24 complex is necessary for efficient trafficking from the Golgi to the ER when Glo3p GAP activity is reduced
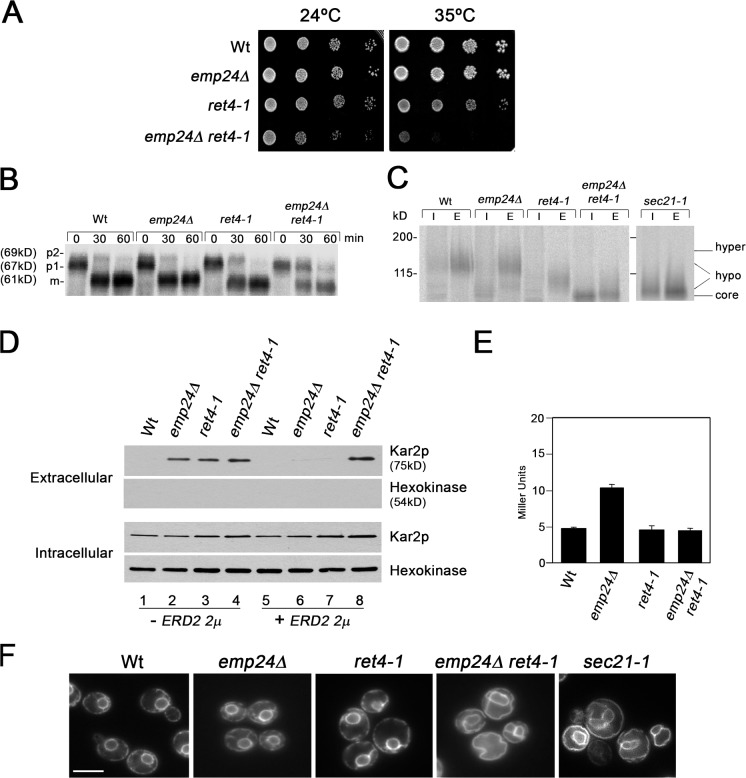
The data presented in the previous paragraph suggest that the p24 family members play a more active role in the generation of COPI-coated vesicles than previously anticipated. In the wild-type situation, this role may not be essential. However, when the ArfGAP activity provided by Glo3p is missing or compromised, p24 function in COPI vesicle generation could become vital. To investigate this possibility, we assessed the lack of p24 function in the ret4-1 mutant, a temperature-sensitive mutant allele of GLO3 (Dogic et al., 1999). The T66I mutation in ret4-1, which is localized to the GTPase-activating domain of Glo3p, reduces the GAP activity toward ARF1 in vitro. We found that emp24Δ ret4-1 mutant cells were viable at 24°C, but they grew slower than respective single mutant cells (Fig. 2 A). Furthermore, deleting EMP24 increased the temperature sensitivity of the ret4-1 allele, which is consistent with the observed genetic interaction between emp24Δ and glo3Δ (Fig. 2 A). Next, we investigated whether the emp24Δ deletion aggravates the slight defect of the ret4-1 mutant in retrograde trafficking. Because anterograde and retrograde traffic between the ER and the Golgi are interdependent, a block in retrograde transport usually has an indirect effect on anterograde transport. Thus, we analyzed the post-translational processing of several early secretory pathway cargoes such as the vacuolar hydrolase, carboxypeptidase Y (CPY) (Stevens et al., 1982), and the secreted enzyme invertase (Esmon et al., 1987) in the emp24Δ ret4-1 mutant, as an indirect way to measure defects in retrograde trafficking. Mutations in several COPI genes result in the accumulation of the immature CPY form (p1) at the ER and a general decrease in the glycosylation state of the secreted invertase (Gaynor and Emr, 1997). Likewise, the lack of p24 protein function in a ret4-1 mutant led to an accumulation of the p1 form of CPY (Fig. 2 B) and exacerbated the impairment of the invertase glycosylation and secretion in the single mutants (Fig. 2 C), which is symptomatic of a severe defect in retrograde transport.
Figure 2.
The emp24Δ ret4-1 double mutant cells exhibit COPI mutant–like phenotypes. (A) emp24Δ strain was crossed with a ret4-1 strain and the resulting haploid spores were tested for growth at 24 and 35°C on YPUAD. (B) Proliferating cells were radiolabeled for 5 min, chased for 30 min at 24°C, and lysed. CPY was immunoprecipitated, resolved by SDS-PAGE, and analyzed by PhosphoImager. ER (p1), Golgi (p2), and vacuole (m) CPY forms are indicated. (C) Low glucose–induced cells were pulse-labeled for 5 min and chased for 30 min at 24°C. Cells were converted to spheroplasts, and separated into intracellular (I) and extracellular (E) fractions from which invertase was immunoprecipitated, resolved by SDS-PAGE, and analyzed by PhosphoImager. Migration positions of core glycosylated, hypo-, and hyperglycosylated invertase are indicated. (D) Cells without (1, 2, 3, and 4) or with (5, 6, 7, and 8) ERD2 2μ plasmid (pJS209) were transferred to fresh media for 1 h at 24°C. Proteins contained in the cell culture supernatant were concentrated by TCA precipitation, resolved by SDS–PAGE, and Kar2p and Hexokinase were detected by immunoblotting. (E) β-Galactosidase assays were performed on strains harboring the reporter construct (pJC31). (F) Fluorescence images of cells expressing the ER-localized protein GFP-HDEL. Cells were grown at 24°C, except sec21-1 cells, which were grown at 24°C and then shifted 40 min at 37°C. Bar, 5 μm.
To assess more directly the defect of emp24Δ ret4-1 double mutant cells in retrograde trafficking, we determined the secretion of the ER molecular chaperone Kar2p. Most mutants that block retrograde transport secrete elevated levels of ER resident proteins with an HDEL signal, including Kar2p, into the media because the HDEL receptor Erd2p fails to retrieve the ER escaped proteins back from the Golgi (Semenza et al., 1990). Previous studies have shown that overproduction of Erd2p mitigates Kar2p secretion (Semenza et al., 1990; Belden and Barlowe, 2001a). Nevertheless, in some trafficking mutants, such as p24 mutants, the activation of the unfolded protein response (UPR) also contributes to Kar2p secretion by exceeding the capacity of the HDEL-retrieval pathway (Belden and Barlowe, 2001a). Wild-type cells did not secrete Kar2p into the media, but emp24Δ, ret4-1, and emp24Δ ret4-1 cells secreted significant amounts of Kar2p (Fig. 2 D). Interestingly, when ERD2 was overexpressed in emp24Δ and ret4-1 mutants Kar2p is not secreted into the growth media, whereas overexpression of ERD2 in emp24Δ ret4-1 cells did not reduce Kar2p secretion (Fig. 2 D). This defect in Kar2p retention might be due to an exacerbation of the UPR activation in emp24Δ ret4-1 compared with emp24Δ. To address this possibility, we measured the UPR induction from a reporter construct (pJC31) that contains the 22-bp UPRE (unfolded protein responses element) of KAR2 fused to LacZ (Cox and Walter, 1996). As shown in Fig. 2 E, β-galactosidase activity was not increased in the emp24Δ ret4-1 cells compared with the emp24Δ strain. Therefore, we assumed that Kar2p secretion in these cells is mainly caused by a severe defect in retrieval of the Erd2p receptor rather than an overloading of the HDEL-retrieval system. The observed Kar2p secretion was not due to the cell lysis, as hexokinase, a cytosolic protein, was not found in the medium (Fig. 2 D).
Because COPI mutations also cause accumulation of ER membranes (Duden et al., 1994), we examined the morphology of the ER in emp24Δ ret4-1. We visualized the ER marker GFP-HDEL by fluorescence, which highlights the nuclear envelope as well as peripheral ER elements (Rossanese et al., 2001). As previously observed (Prinz et al., 2000), GFP-HDEL showed a massive proliferation of ER membranes in a sec21-1 temperature-sensitive strain under conditions when retrograde transport is blocked (Fig. 2 F). Likewise, striking elaborations of the ER were apparent in emp24Δ ret4-1 cells, whereas the ER pattern observed in emp24Δ and ret4-1 cells was identical to that in wild-type cells. Together, these data show that emp24Δ ret4-1 double mutant cells exhibit COPI mutant–like phenotypes, supporting the idea of the involvement of the p24 complex in COPI function.
The p24 complex is required for COPI vesicle budding from Golgi membranes
Our genetic and biochemical findings suggest that the p24 complex might participate in retrograde transport from Golgi to ER by promoting the formation of COPI vesicles. We assessed directly this hypothesis by using an in vitro Golgi vesicle budding assay (Spang and Schekman, 1998). Enriched Golgi membranes devoid of ER from wild-type and emp24Δ strains were subjected to a vesicle budding assay by incubating them in the presence of GTP. Vesicles containing the COPI cargo Emp47p were produced by wild-type Golgi-enriched membranes (Fig. 3 A). In contrast, Golgi membranes from the emp24Δ mutant strain failed to form COPI vesicles (Fig. 3 A). This result indicates that the p24 complex plays an important role in COPI vesicle generation from Golgi membranes.
Figure 3.
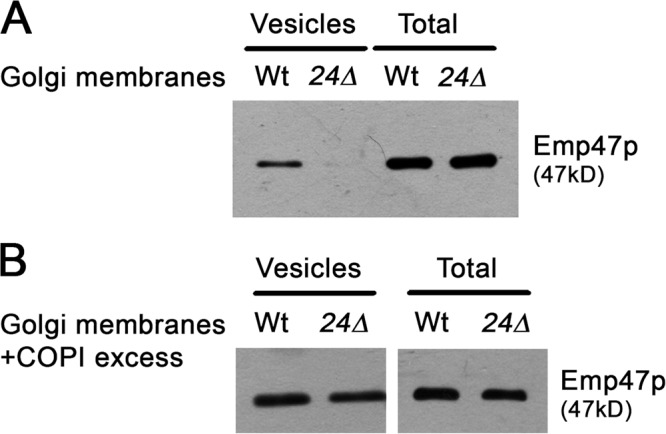
Golgi membranes derived from emp24Δ mutant cells are defective for the generation of COPI vesicles in vitro. Vesicles were generated from wild-type and emp24Δ (RH4443) Golgi membranes, which were incubated with GTP in the absence (A) or the presence of an excess of COPI components (B). The vesicles were purified over a velocity gradient, and subsequently floated on a Nycodenz gradient. Vesicle-containing fractions were collected and pooled, TCA precipitated, resolved on SDS-PAGE, and analyzed by immunoblot using antibody against Emp47p.
The p24 complex requirement for budding was observed only under coat-limiting conditions. Increasing the amount of coatomer and Arf1p in the budding assays restored COPI vesicle generation from emp24Δ Golgi membranes (Fig. 3 B). This COPI dosage dependence of budding strongly suggests that p24 proteins might facilitate the budding process by stabilizing coatomer on the Golgi membrane. This possibility is supported by a previous report showing that overexpression of EMP24 suppresses sec21-3 temperature sensitivity (Sandmann et al., 2003). Thus, the phenotype enhancement in the emp24Δ sec21-1 mutant might be due to the failure of cargo proteins to create enough priming complexes that would support budding in the absence of p24 proteins. Increasing the stability of coatomer on Golgi membranes might rescue the emp24Δ sec21-1 mutant phenotype. Because ArfGAPs have been shown to potentiate the binding of coatomer to cargo proteins and consequently promote the stabilization of COPI priming complexes (Rein et al., 2002; Lee et al., 2005), we tested whether the temperature sensitivity of emp24Δ sec21-1 and erv25Δ sec21-1 is suppressed by overexpression of Glo3p and Gcs1p. Indeed, overexpression of GLO3 was able to restore growth of emp24Δ sec21-1 and erv25Δ sec21-1 at the restrictive temperature, whereas overexpression of GCS1 did not rescue the lethal phenotypes (Fig. 4 A). This result is consistent with the role of Glo3p as a specific component of the COPI vesicle coat through association with the coatomer subunit Sec21p (Lewis et al., 2004), and provides further evidence that p24 family proteins are important to stabilize priming complexes on Golgi membranes, which allows efficient polymerization of the COPI coat.
Figure 4.
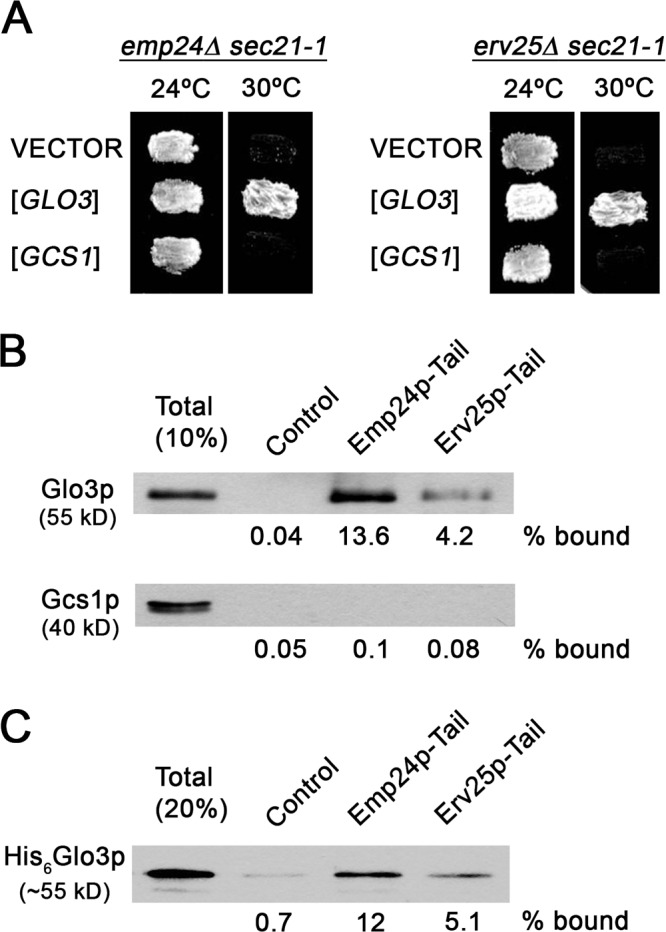
Specific genetic and physical interactions between p24 members and COPI coat components. (A) The emp24Δ sec21-1 and erv25Δ sec21-1 cells were transformed with empty plasmid (YEp352) or a high-copy plasmid carrying either the GLO3 (pPPL43) or the GCS1 gene (pPP421). Transformed cells were tested for growth at 24 and 30°C. (B and C) Synthetic peptides corresponding to cytoplasmic domains of Emp24p (RRFFEVTSLV) and Erv25p (KNYFKTKHII) were coupled to thiopropyl-Sepharose beads and incubated with cytosol (B) or recombinant Glo3p (C). Bound material was resolved by SDS-PAGE and analyzed by immunoblot using antibodies against Glo3p and Gcs1p.
If our hypothesis is correct that p24 proteins are relevant for COPI priming complex formation, they should efficiently recruit COPI coat components such as coatomer and Glo3p. It has already been demonstrated that Emp24p and Erv25p bind coatomer (Belden and Barlowe, 2001b). Next, we tested whether Glo3p is also able to interact specifically with p24 proteins. Peptides corresponding to the C-terminal 10 amino acids of Emp24p or Erv25p were coupled to Sepharose beads and incubated with cytosol. We detected binding of Glo3p to both tail sequences (Fig. 4 B). In contrast, Gcs1p was not recruited to either the Emp24p or the Erv25p tail, supporting the idea that Gcs1p is not part of the COPI coat complex. Furthermore, recombinant Glo3p bound directly to the p24 tails (Fig. 4 C). Our data demonstrate that p24 proteins recruit COPI coat components. Together, our results show that p24 proteins can form COPI priming complexes and that these complexes are required to form retrograde COPI-coated vesicles efficiently from Golgi membranes.
UPR compensates the loss of retrograde transport function in the absence of p24 proteins
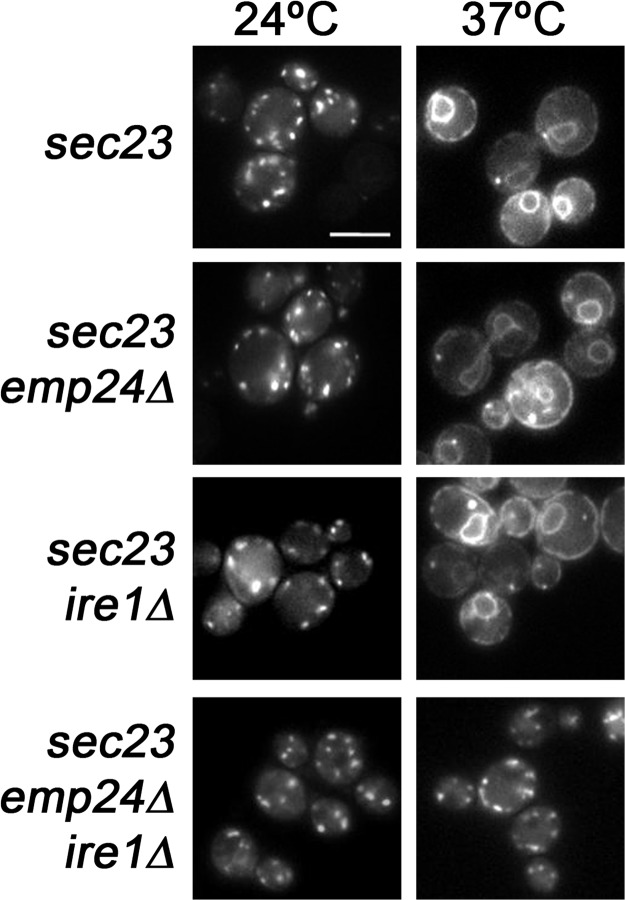
We have established in the previous paragraph that p24 complex form priming complexes. However, p24 genes are not essential in yeast. Indeed, deletion of all eight p24 family members does not produce severe transport phenotypes (Springer et al., 2000). Yet, subclasses of secretory proteins accumulate in the ER. How can this apparent discrepancy between the importance of p24 proteins for vesicle generation and the mild phenotypes of their deletion be reconciled? Perhaps a compensatory mechanism exists that helps the cell to cope with the loss of p24 proteins. Indeed, deletion of ERV25 leads to UPR activation (Belden and Barlowe, 2001a). Moreover, the concomitant loss of ERV25 (or EMP24) and IRE1, an ER-localized kinase that activates the UPR, results in slow growth and more severe transport defects (Belden and Barlowe, 2001a). Accordingly, a genetic interaction between emp24Δ and hac1Δ, a transcriptional activator for a set of UPR-regulated genes, was observed during the genetic screen (unpublished data). We wanted to test directly whether UPR activation is responsible for the lack of an obvious retrograde trafficking defect in the emp24Δ strain. We assessed this possibility by determining whether the Golgi protein Rer1p, which cycles through the ER (Sato et al., 1997), is efficiently recycled back to the ER in an emp24Δ ire1Δ strain. We blocked ER exit with the temperature-sensitive sec23-1 (COPII) allele to trap cycling proteins in the ER (Sato et al., 1997). If retrograde transport of Rer1p-GFP depends on both the p24 complex and on UPR, an emp24Δ ire1Δ sec23-1 triple mutant strain should not accumulate Rer1p in the ER upon shift to the restrictive temperature (37°C). As shown in Fig. 5, in sec23-1, emp24Δ sec23-1, and ire1Δ sec23-1 mutant cells at 24°C, Rer1p-GFP showed punctuate staining typical for Golgi in yeast, whereas at 37°C, the ER-characteristic nuclear ring staining was observed. In contrast, when Rer1p-GFP was expressed in emp24Δ ire1Δ sec23-1 triple mutant cells at 37°C, no ER staining was observed and Rer1-GFP remained in the Golgi (Fig. 5). This result demonstrates that retrograde traffic is affected in the emp24Δ ire1Δ sec23-1 mutant cells at 37°C, indicating that UPR can compensate for the loss of p24 function in retrograde transport from Golgi to the ER. This compensatory effect could rely on the control that the UPR exerts over many aspects of secretory function, including retrograde transport. Indeed, genes involved in Golgi retrieval, such as genes encoding for several coatomer subunits (i.e., SEC27) are up-regulated by the UPR (Travers et al., 2000). Because the p24 complex requirement for budding is overcome by increasing the amount of coatomer, it remains possible that the up-regulation of the COPI genes helps the cell to cope with the loss of the p24 proteins. In addition, it is also possible that activity of COPI is up-regulated post-translationally by the UPR, for instance by phosphorylation. Indeed, the phosphorylation of several coatomer subunits has been reported (Sheff et al., 1996).
Figure 5.
UPR compensates the loss of retrograde transport function in the absence of p24 proteins. Cells expressing Rer1p-GFP were observed by fluorescence microscopy at 24°C, or after the shift to 37°C for 20 min. Bar, 5 μm.
This study further supports the notion that certain transmembrane cargoes, such as the p24 protein family members, might be essential components in the regulation of vesicle formation instead of simply being a passive traveler. p24 proteins could recruit initially the deactivated form of Arf1p, (Gommel et al., 2001) facilitating the formation of a COPI priming complex and subsequently enabling efficient budding. In addition, the ability of p24 proteins to oligomerize and to present multiple coatomer-binding motifs might promote COPI budding by docking coatomer more firmly to the membrane (Bethune et al., 2006). Overexpression of coatomer-binding tails in oligomeric complexes has been shown to overcome the lethality of sec21-3 at the nonpermissive temperature presumably by stabilizing coatomer on the Golgi (Sandmann et al., 2003). Moreover, p24 complex might play a key role in the stabilization of priming complexes by modulating the ArfGAP activity (Goldberg, 2000; Lanoix et al., 2001). Thus, other cargo proteins with lower coatomer affinity, like the single tail exposing Emp47p, could be captured by stabilized priming complexes and then be sorted into COPI vesicles.
Materials and methods
Yeast strains and growth conditions
Strains of Saccharomyces cerevisiae used for this work are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200710025/DC1). Standard genetic manipulations were used throughout (Sherman, 1991). Strain RH6148 was made by replacement of EMP24 ORF by natMX (Goldstein and McCusker, 1999) disruption cassette in the Y2922 background. Strain MMY104 was obtained by replacing the entire IRE1 coding sequence of RH1433 with a HphMX disruption cassette (Goldstein and McCusker, 1999). Strains MMY103 and MMY91 were constructed by crossing MMY104 with RH4443. Strain MMY369 was constructed by crossing RH441 with PC238. Strains MMY409, MMY410, MMY419, and MMY518 were constructed by crossing MMY369 and RH1469 with BGY418. Cells were grown in either rich medium YPUAD (1% yeast extract, 2% peptone, 2% glucose, and 40 mg/ml each of adenine and uracil) or minimal medium SD (0.67% yeast nitrogen base without amino acids, 2% glucose, and the required nutrients).
Synthetic lethal screen
Synthetic lethal screen was performed as described by Tong et al. (2001), using an emp24Δ mutant (RH6148) as a bait strain. The yeast knockout collection (YKO) and wild-type strain Y2922 were provided by C. Boone (University of Toronto, Ontario, Canada; Tong et al., 2001). In total, 72 double-deletion combinations were found that resulted in a potential synthetic growth defect.
Plasmids
To generate pMM63 and pMM67, the SacI–SmaI fragment from pNY24-3HA-GLO3 (Yahara et al., 2006) containing the GLO3 gene was subcloned in the vectors pRS416 and pRS315, respectively. Other plasmids used in this study are pSKY5/RER1-0 (a CEN-based URA3 plasmid carrying Rer1-GFP; Sato et al., 1997), pPPL43 and pPP421 (a 2μ-based URA3 plasmid with GLO3 and GCS1, respectively; Poon et al., 1999), pJS209 (a 2μ-based URA3 plasmid with ERD2; Semenza et al., 1990), and pJC31 (a CEN-based TRP1 plasmid with UPRE1,2-CYC-lacZ; (Cox and Walter, 1996).
Pulse-chase analysis of CPY and invertase
Radiolabeling and immunoprecipitations were performed as described by Sutterlin et al. (1998). For invertase secretion, cells were induced as described by Kübler et al. (1994) and internal and external invertase was assayed as described by Gaynor and Emr (1997).
Analysis of Kar2 secretion
Extracellular Kar2p secretion was analyzed as described by Belden and Barlowe (2001a).
β-Galactosidase assay
Assays of β-galactosidase activity in extracts of yeast cells containing the UPRE-LacZ fusion construct, pJC31, were performed as described by Cox and Walter (1996).
In vitro Golgi budding assay
Golgi membranes and COPI components were purified as described by Spang and Schekman (1998). The Golgi budding assay was performed as described by Lewis et al. (2004) with several modifications. For the Golgi budding reactions, membranes were incubated in the presence of 0.1 mM GTP without (coat-limited conditions) or with an excess of coatomer (250 μg/ml), and Arf1 protein (80 μg/ml) at 30°C for 30 min in a total volume of 180 μl. Samples were loaded on a Ficoll-sucrose gradient consisting of 135 μl of 60% (wt/vol) sucrose, 360 μl of 7.5%, 450 μl of 5, 4, and 3%, and 360 μl of 2% (wt/wt) Ficoll in 15% (wt/vol) sucrose in 20 mM Hepes, pH 6.8, 5 mM Mg(OAc)2, and 150 mM KOAc (B88*). The vesicles were separated from the Golgi apparatus by centrifugation for 90 min at 42,000 rpm (TLS55 rotor; Beckman Coulter). 540 μl from the top was discarded and the next 540 μl were mixed with an equal volume of 80% Nycodenz in B88*, and overlaid with 276 μl of 35, 25, 20, and 15% and 184 μl of 10% Nycodenz in B88*. The gradients were centrifuged for 16 h at 50,000 rpm (TLS55 rotor; Beckman Coulter). Fractions (150 μl) were collected from the top. Vesicle-containing fractions (5, 6, 7) were collected and pooled, TCA precipitated, resolved on SDS-PAGE, and analyzed by immunoblot.
Pull-down assay
Synthetic peptides corresponding to the 10 C-terminal amino acids of Emp24p (RRFFEVTSLV) and Erv25p (KNYFKTKHII) with an N-terminal cysteine residue were generated (JPT Peptide Technologies) and linked to thiopropyl-Sepharose 6B (GE Healthcare) as described by Belden and Barlowe (2001b). Cytosol from a pep4Δ strain (RH732) was obtained as described by Muñiz et al. (2000). Recombinant Glo3p was purified as described by Rein et al., (2002). In vitro binding reactions were performed as described by Belden and Barlowe (2001b). The immunoblot was quantified by densitometry.
Light microscopy
For fluorescence microscopy of GFP-tagged strains, log-phase cells grown in minimal media were observed directly. Acquisition was performed at 24°C using a Leica DMR microscope equipped with an objective lens (HCX PL APO 63×/1.32 OIL Ph3), a DC 350F camera, and Image Manager 50 v1.20 following the instructions of the manufacturer.
Online supplemental material
Table S1 provides information about the yeast strains used during this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200710025/DC1.
Supplementary Material
Acknowledgments
We thank F. Letourneur, A. Nakano, P. Poon, B. Glick, C. Barlowe, C. Boone, P. Walter, and C. de Virgilio for materials and advice; and A. Velasco for critical reading of the manuscript.
This work was supported by Ministerio de Educación y Ciencia grant (BFU2005-01642) and an EMBO short-term fellowship (to M. Muñiz), grants from the Swiss National Science Foundation (to H. Riezman and A. Spang) and the University of Geneva (to H. Riezman) and the University of Basel (to A. Sprang), grant N303 101 32/3456 (to J. Kaminska), and a University of Seville fellowship (to A. Aguilera).
Abbreviations used in this paper: CPY, carboxypeptidase Y; UPR, unfolded protein response.
References
- Belden, W.J., and C. Barlowe. 2001. a. Deletion of yeast p24 genes activates the unfolded protein response pathway. Mol. Biol. Cell. 12:957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden, W.J., and C. Barlowe. 2001. b. Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. J. Biol. Chem. 276:43040–43048. [DOI] [PubMed] [Google Scholar]
- Bethune, J., M. Kol, J. Hoffmann, I. Reckmann, B. Brugger, and F. Wieland. 2006. Coatomer, the coat protein of COPI transport vesicles, discriminates endoplasmic reticulum residents from p24 proteins. Mol. Cell. Biol. 26:8011–8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremser, M., W. Nickel, M. Schweikert, M. Ravazzola, M. Amherdt, C.A. Hughes, T.H. Sollner, J.E. Rothman, and F.T. Wieland. 1999. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 96:495–506. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 87:391–404. [DOI] [PubMed] [Google Scholar]
- Dogic, D., B. de Chassey, E. Pick, D. Cassey, Y. Lefkir, S. Hennecke, P. Cosson, and F. Letourneur. 1999. The ADP-ribosylation factor GTPase-activating protein Glo3p is involved in ER retrieval. Eur. J. Cell Biol. 78:305–310. [DOI] [PubMed] [Google Scholar]
- Duden, R., M. Hosobuchi, S. Hamamoto, M. Winey, B. Byers, and R. Schekman. 1994. Yeast beta- and beta'-coat proteins (COP). Two coatomer subunits essential for endoplasmic reticulum-to-Golgi protein traffic. J. Biol. Chem. 269:24486–24495. [PubMed] [Google Scholar]
- Elrod-Erickson, M.J., and C.A. Kaiser. 1996. Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol. Biol. Cell. 7:1043–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, G., R.G. Parton, M. Rojo, and J. Gruenberg. 2003. The trans-membrane protein p25 forms highly specialized domains that regulate membrane composition and dynamics. J. Cell Sci. 116:4821–4832. [DOI] [PubMed] [Google Scholar]
- Esmon, P.C., B.E. Esmon, I.E. Schauer, A. Taylor, and R. Schekman. 1987. Structure, assembly, and secretion of octameric invertase. J. Biol. Chem. 262:4387–4394. [PubMed] [Google Scholar]
- Eugster, A., G. Frigerio, M. Dale, and R. Duden. 2000. COPI domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 19:3905–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullekrug, J., T. Suganuma, B.L. Tang, W.J. Hong, B. Storrie, and T. Nilsson. 1999. Localization and recycling of gp27 (hp24 gamma(3)): complex formation with other p24 family members. Mol. Biol. Cell. 10:1939–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, E.C., and S.D. Emr. 1997. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J. Cell Biol. 136:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, J. 2000. Decoding of sorting signals by coatomer through a GTPase switch in the COPI coat complex. Cell. 100:671–679. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.L., and J.H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- Gommel, D.U., A.R. Memon, A. Heiss, F. Lottspeich, J. Pfannstiel, J. Lechner, C. Reinhard, J.B. Helms, W. Nickel, and F.T. Wieland. 2001. Recruitment to Golgi membranes of ADP-ribosylation factor 1 is mediated by the cytoplasmic domain of p23. EMBO J. 20:6751–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler, E., F. Schimmöler, and H. Riezman. 1994. Calcium-independent calmodulin requirement for endocytosis in yeast. EMBO J. 13:5539–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix, J., J. Ouwendijk, A. Stark, S. Szafer, D. Cassel, K. Dejgaard, M. Weiss, and T. Nilsson. 2001. Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J. Cell Biol. 155:1199–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, C., J. Paiement, M. Dominguez, L. Roy, S. Dahan, J.N. Gushue, and J.J.M. Bergeron. 1999. Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. J. Cell Biol. 146:285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.Y., J.S. Yang, W.J. Hong, R.T. Premont, and V.W. Hsu. 2005. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J. Cell Biol. 168:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S.M., P.P. Poon, R.A. Singer, G.C. Johnston, and A. Spang. 2004. The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol. Biol. Cell. 15:4064–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch, M., D.C. Henthorn, J.M. Herrmann, R. Wilson, D.Y. Thomas, J.J.M. Bergeron, R.C.E. Solari, and A. Rowley. 1999. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell. 10:1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz, M., C. Nuoffer, H.P. Hauri, and H. Riezman. 2000. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148:925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz, M., P. Morsomme, and H. Riezman. 2001. Protein sorting upon exit from the endoplasmic reticulum. Cell. 104:313–320. [DOI] [PubMed] [Google Scholar]
- Poon, P.P., D. Cassel, A. Spang, M. Rotman, E. Pick, R.A. Singer, and G.C. Johnston. 1999. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18:555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, W.A., L. Grzyb, M. Veenhuis, J.A. Kahana, P.A. Silver, and T.A. Rapoport. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein, U., U. Andag, R. Duden, H.D. Schmitt, and A. Spang. 2002. ARF-GAP-mediated interaction between the ER-Golgi v-SNAREs and the COPI coat. J. Cell Biol. 157:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, M., R. Pepperkok, G. Emery, R. Kellner, E. Stang, R.G. Parton, and J. Gruenberg. 1997. Involvement of the transmembrane protein p23 in biosynthetic protein transport. J. Cell Biol. 139:1119–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese, O.W., C.A. Reinke, B.J. Bevis, A.T. Hammond, I.B. Sears, J. O'Connor, and B.S. Glick. 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153:47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann, T., J.M. Herrmann, J. Dengjel, H. Schwarz, and A. Sprang. 2003. Suppression of coatomer mutants by a new protein family with COPI and COPII binding motifs in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:3097–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., M. Sato, and A. Nakano. 1997. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc. Natl. Acad. Sci. USA. 94:9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, J.C., K.G. Hardwick, N. Dean, and H.R.B. Pelham. 1990. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 61:1349–1357. [DOI] [PubMed] [Google Scholar]
- Sheff, D., M. Lowe, T.E. Kreis, and I. Mellman. 1996. Biochemical heterogeneity and phosphorylation of coatomer subunits. J. Biol. Chem. 271:7230–7236. [DOI] [PubMed] [Google Scholar]
- Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21. [DOI] [PubMed] [Google Scholar]
- Sohn, K., L. Orci, M. Ravazzola, M. Amherdt, M. Bremser, F. Lottspeich, K. Fiedler, J.B. Helms, and F.T. Wieland. 1996. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J. Cell Biol. 135:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A. 2002. ARF1 regulatory factors and COPI vesicle formation. Curr. Opin. Cell Biol. 14:423–427. [DOI] [PubMed] [Google Scholar]
- Spang, A., and R. Schekman. 1998. Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 143:589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, S., A. Spang, and R. Schekman. 1999. A primer on vesicle budding. Cell. 97:145–148. [DOI] [PubMed] [Google Scholar]
- Springer, S., E. Chen, R. Duden, M. Marzioch, A. Rowley, S. Hamamoto, S. Merchant, and R. Schekman. 2000. The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 97:4034–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes, M.A., M.W. Craighead, M.H. Hoe, N. Lampen, S. Geromanos, P. Tempst, and J.E. Rothman. 1995. An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc. Natl. Acad. Sci. USA. 92:8011–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, T., B. Esmon, and R. Schekman. 1982. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase-Y to the vacuole. Cell. 30:439–448. [DOI] [PubMed] [Google Scholar]
- Sutterlin, C., M.V. Escribano, P. Gerold, Y. Maeda, M.J. Mazon, T. Kinoshita, R.T. Schwarz, and H. Riezman. 1998. Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem. J. 332:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H.Y., M. Evangelista, A.B. Parsons, H. Xu, G.D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C.W.V. Hogue, H. Bussey, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368. [DOI] [PubMed] [Google Scholar]
- Travers, K.J., C.K. Patil, L. Wodicka, D.J. Lockhart, J.S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. [DOI] [PubMed] [Google Scholar]
- Yahara, N., K. Sato, and A. Nakano. 2006. The Arf1p GTPase-activating protein Glo3p executes its regulatory function through a conserved repeat motif at its C-terminus. J. Cell Sci. 119:2604–2612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.