Abstract
Integrin-linked kinase (ILK) is a serine-threonine kinase and scaffold protein with well defined roles in focal adhesions in integrin-mediated cell adhesion, spreading, migration, and signaling. Using mass spectrometry–based proteomic approaches, we identify centrosomal and mitotic spindle proteins as interactors of ILK. α- and β-tubulin, ch-TOG (XMAP215), and RUVBL1 associate with ILK and colocalize with it to mitotic centrosomes. Inhibition of ILK activity or expression induces profound apoptosis-independent defects in the organization of the mitotic spindle and DNA segregation. ILK fails to localize to the centrosomes of abnormal spindles in RUVBL1-depleted cells. Additionally, depletion of ILK expression or inhibition of its activity inhibits Aurora A–TACC3/ch-TOG interactions, which are essential for spindle pole organization and mitosis. These data demonstrate a critical and unexpected function for ILK in the organization of centrosomal protein complexes during mitotic spindle assembly and DNA segregation.
Introduction
Integrin-linked kinase (ILK) is a signaling and scaffold protein localized to focal and fibrillar adhesions (Hannigan et al., 2005; Legate et al., 2006). Identified as an interactor of integrin β1 and 3 cytoplasmic domains (Hannigan et al., 1996), ILK also regulates cell survival, proliferation, migration, and angiogenesis and Pi3 kinase–dependent signal transduction (Hannigan et al., 2005). By interacting with the focal adhesion proteins PINCH, paxillin, and α- and β-parvin, ILK regulates integrin-mediated cell adhesion and cytoskeletal dynamics within focal adhesions to regulate cell adhesion, spreading, and migration (Legate et al., 2006). Tissue-specific gene knockout studies have revealed several essential roles of ILK in embryonic development, tissue homeostasis, and organ function (Bendig et al., 2006; White et al., 2006; Lorenz et al., 2007). In addition, ILK appears to be differentially required for cell survival and growth in normal versus cancer cells (Troussard et al., 2006). The diversity of phenotypes observed in these studies suggests complex regulation of ILK activity and adaptor functions.
To identify novel ILK protein–protein interactions that will provide further insights into the diverse functions of ILK, we analyzed ILK complexes by stable isotope labeling with amino acids in cell culture (SILAC)–based mass spectrometry (Dobreva et al., 2008). In addition to identifying known interactors, such as PINCH and α-parvin, we also identified, with equal robustness, tubulin, and tubulin-interacting proteins, especially those known to localize to centrosomes, such as ch-TOG (XMAP215 and CKAP5; Gergely et al., 2003) and RUVBLl (Pontin 52; Gartner et al., 2003). Ch-TOG has been shown to be essential for organizing spindle poles as well as stabilizing spindle microtubules (Gergely et al., 2003). RUVBL1 is an ATP helicase and has several established nuclear functions (Weiske and Huber, 2005). However, this protein also binds to tubulin and has been shown to localize to centrosomes within mitotic spindles (Gartner et al., 2003).
In this paper we show that in addition to its focal adhesion functions, ILK localizes to centrosomes with several newly identified binding partners and plays an essential role in mitotic spindle assembly and mitosis.
Results and discussion
Proteomic analysis of ILK interactors within the cytoskeleton identifies α- and β-tubulin, ch-TOG, and RUVBL1
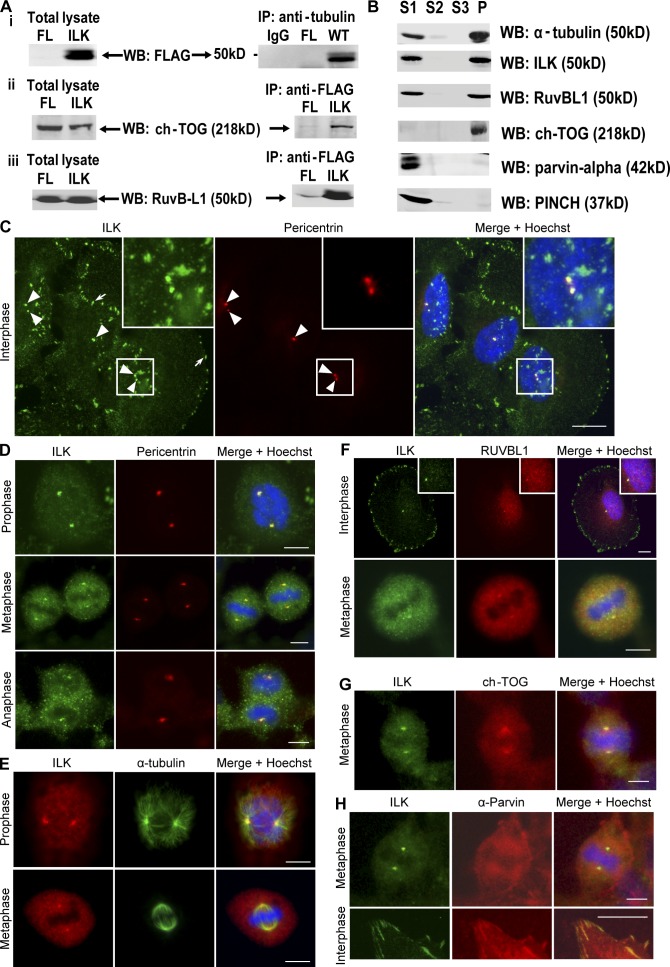
To identify novel ILK-interacting proteins in the cytoskeleton, ILK was immunoprecipitated from cytoskeletal HEK293 cell extracts and immune complexes were resolved by SDS-PAGE and analyzed by SILAC-based gel-enhanced liquid chromatography/tandem mass spectrometry (GelC-MS/MS). Details of isotope labeling and analysis are described in Dobreva et al. (2008). Together with cytoskeletal proteins already known to bind ILK, e.g., PINCH and α-parvin (Hannigan et al., 2005; Legate et al., 2006), several novel interactors were identified. A high proportion of these proteins are known to associate with the mitotic spindle and/or centrosomes. Our attention was drawn to α- and β-tubulin as well as to the tubulin binding proteins ch-TOG and RUVBL1. To confirm these interactions, anti-FLAG immunoprecipitates from cytoskeletal extracts of FLAG-ILK–expressing cells were Western blotted with antibodies to α- and β-tubulin, ch-TOG, and RUVBL1. As shown in Fig. 1 A, these proteins could be readily detected in FLAG-ILK, but not FLAG, immunoprecipitations. In addition, endogenous interactions were also confirmed (Dobreva et al., 2008). Yeast two-hybrid analysis indicated that the interaction of ILK with β-tubulin and RUVBL1 is not direct (unpublished data).
Figure 1.
ILK interacts with tubulin, RUVBL1, and ch-TOG and localizes to centrosomes. (A) FLAG-ILK was immunoprecipitated from the cytoskeleton of HEK293 cells, and the presence of α- and β-tubulin, ch-TOG, and RUVBL1 was determined by Western blot. (B) Detergent-soluble fractions (S1 and S2), isolation buffer–soluble fraction (S3), and purified mitotic spindles (P) from HeLa cells Western blotted for indicated proteins are shown. (C–H) HeLa cells costained for ILK and indicated proteins as well as DNA. Insets are close-ups of small squares (C) or of centrosomes (F). Arrowheads, centrosomes; arrow, focal adhesions. Bars, ∼10 μm.
As ILK associated with proteins that localize to mitotic spindles and/or centrosomes, we next purified mitotic spindles (including centrosomes), as previously described (Sauer et al., 2005), and assessed whether ILK was present there. Enrichment of mitotic spindles and centrosomes was confirmed by immunofluorescence microscopy (not depicted), and Western blotting revealed that in addition to known mitotic spindle proteins, such as α-tubulin and ch-TOG, a proportion of ILK also associated with the purified spindles together with RUVBL1, whereas PINCH and α-parvin (known focal adhesion ILK interactors) were not present on spindles (Fig. 1 B). These data show that ILK can partition between actin and tubulin cytoskeletal networks by associating with distinct protein components. We therefore examined the subcellular localization of ILK to determine whether it localizes to mitotic spindles and/or centrosomes.
ILK localizes to interphase and mitotic centrosomes
Immunofluorescence localization of ILK with a monoclonal anti-ILK antibody shows a largely focal and fibrillar adhesion pattern of ILK (Fig. 1 C) as previously reported (Sepulveda and Wu, 2006). However, costaining with pericentrin, a resident centrosomal protein, reveals that ILK is also present in centrosomes in interphase cells (Fig. 1 C). Cells at various stages of mitosis also show clear ILK-pericentrin colocalization (Fig. 1 D). The centrosome localization of ILK was confirmed by costaining with an alternative polyclonal anti-ILK antibody and an anti–α-tubulin antibody to stain microtubules and the mitotic spindle (Fig. 1 E). ILK staining was clearly concentrated at the centrosomes/microtubule organizing centers (MTOCs) from which the mitotic spindle radiates. ILK also localized to the centrosomes in HEK293 and IMR-90 cells (Fig. S1, A–C, available at http://www.jcb.org/cgi/content/full/jcb.200710074/DC1). Staining cells with IgG controls and secondary antibodies confirmed the specificity of the ILK centrosomal staining (Fig. S1, D and E). As an additional control, it can be seen that upon ILK knockdown with siRNA, ILK staining is no longer observed in interphase (Fig. S1 F) or mitotic (see Fig. 4 B) centrosomes.
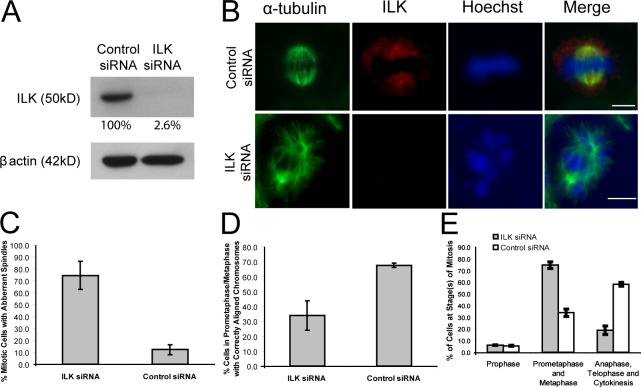
Figure 4.
ILK siRNA results in aberrant mitotic spindles, misaligned chromosomes, and the prevention of progress through mitosis. (A) Western blot of HeLa cell extracts after ILK siRNA treatment. The amount of ILK remaining after knockdown is shown as a percentage of ILK in control siRNA and is adjusted according to β-actin levels. (B) HeLa cells treated with control or ILK siRNA show similar aberrant spindle and misaligned chromosome phenotypes to the QLT-0267–treated cells. Bars, ∼10 μm. (C–E) Histograms showing the percentage of mitotic cells with aberrant spindles (C), with misaligned chromosomes (D), and in the various stages of cell division (E) in ILK and control siRNA-treated cells. Data are mean ± SD from three independent experiments in which >50 mitotic cells were analyzed.
Fig. 1 (F and G) shows colocalization in centrosomes of ILK with two of its newly identified associating proteins, RUVBL1 and ch-TOG. Ch-TOG shows partial colocalization with ILK as, in addition to being present in centrosomes, it is also found on the mitotic spindle as previously reported (Gergely et al., 2003). We also assessed whether α-parvin, a well-established ILK binding partner, localizes to centrosomes. Fig. 1 H shows cells costained for ILK and α-parvin in both interphase and mitosis. Although the α-parvin antibody clearly stains focal adhesions where it colocalizes with ILK (Fig. 1 H, bottom), it does not show colocalization with the prominent ILK staining at the centrosomes (Fig. 1 H, top), confirming the differential partitioning of ILK and α-parvin that is shown in Fig. 1 B.
RUVBL1 depletion causes a mitotic spindle defect and prevents localization of ILK to the centrosomes
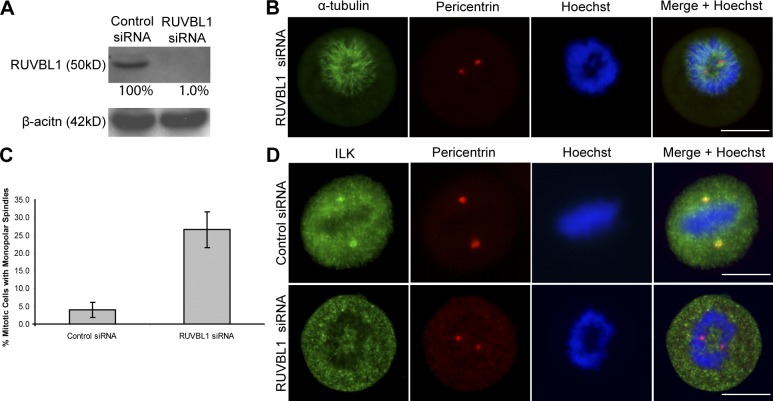
Because RUVBL1 was specifically enriched with ILK in the SILAC-based proteomic analysis, we wanted to determine whether it might be involved in recruiting ILK to the centrosomes. We therefore depleted RUVBL1 expression with siRNA (Fig. 2 A). This resulted in the disruption of the mitotic spindle and DNA segregation as shown in Fig. 2 B. Quantification of this effect showed that 27% of mitotic cells showed this phenotype in RUVBL1 siRNA cells compared with only 4% in control cells (Fig. 2 C). Although centrosomes were intact, as shown by pericentrin localization, ILK failed to localize to the centrosomes in the RUVBL1-depleted cells (Fig. 2 D) in 100% of cells that showed the spindle defect. These data suggest that RUVBL1 is involved in recruiting ILK to the centrosomes, although it is possible that the severity of the spindle phenotype observed may indirectly prevent the localization of ILK to the centrosome.
Figure 2.
RUVBL1 is required for ILK localization to the centrosome and its absence leads to mitotic spindle defects. (A) RUVBL1 protein was depleted by RUVBL1 siRNA as shown by Western blot. (B) RUVBL1 siRNA–treated cells. (C) Histogram showing the percentage of disrupted mitotic cells in control and RUVBL1 siRNA–treated cells. Data are mean ± SD of three independent experiments in which >100 mitotic cells were counted for each condition. (D) Cells stained with ILK and pericentrin show localization of ILK to centrosomes in control but not RUVBL1 siRNA–treated cells. Bars, ∼10 μm.
Collectively, these results demonstrate that ILK can localize simultaneously to two distinct cellular regions, focal adhesions, and the centrosome by interacting with different protein partners and cytoskeletal components.
Inhibition of ILK activity and expression disrupts mitotic spindle organization
The striking localization of ILK to centrosomes suggests that it may play an important role in centrosome integrity, mitotic spindle organization, or microtubule polymerization. To address the role of the kinase activity of ILK in these functions, we initially used an ILK inhibitor, QLT-0267, which is a highly selective inhibitor of ILK activity (Troussard et al., 2006).
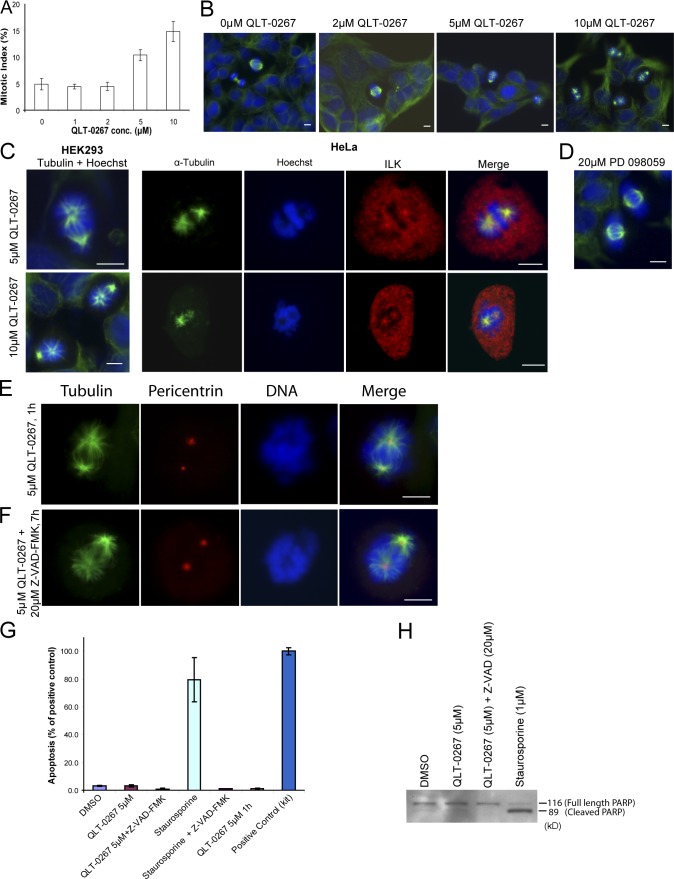
HEK293 or HeLa cells were exposed to increasing concentrations of QLT-0267 for 7 h and then fixed and immunostained for α-tubulin and DNA to identify mitotic cells. Examination of these cells revealed the appearance of abnormal mitotic spindles at 5 μM QLT-0267, whereas at 10 μM, 100% of mitotic cells showed spindle defects (Fig. 3 B). Specifically, there was asymmetrical localization of microtubules to spindle poles. It has previously been shown that QLT-0267 inhibits ILK kinase activity with an IC50 of between 2 and 5 μM, depending on the cell type (Troussard et al., 2006). This correlates very well with the effect on the mitotic spindle that is presented in this paper, suggesting that ILK kinase activity is likely to be the driving force behind the phenotypes observed. Quantification of the proportion of cells in mitosis showed that QLT-0267 had a dose-dependent effect on the mitotic index (Fig. 3 A), suggesting that the observed spindle defects were leading to an arrest in mitosis. Exposure over the same time period to other similar small molecule kinase inhibitors, such as the Mek inhibitor PD098059, did not have any effects on spindle assembly (Fig. 3 D). To determine whether the ILK inhibitor affected ILK localization or centrosomal integrity, we immunostained cells that were exposed to 5 μM QLT-0267 for ILK, ch-TOG, and pericentrin. ILK still localized to the centrosomes of the abnormal mitotic spindles (Fig. 3 C), as did ch-TOG and pericentrin (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200710074/DC1), indicating that the ILK inhibitor does not alter the localization of these proteins.
Figure 3.
Treatment with the ILK inhibitor QLT-0267 causes apoptotic-independent effects on mitotic spindles and an accumulation of cells in mitosis. (A) HEK293 cells were treated with increasing concentrations of QLT-0267 for 7 h and the mitotic index was calculated. Data are mean ± SD of three independent experiments in which >400 cells were counted for each condition. (B and C) Cells were treated with QLT-0267 for 7 h and stained as indicated. (D) Cells were treated with the MEK inhibitor PD098059. (E and F) HeLa cells were treated with QLT-0267 for 1 h (E) or QLT-0267 + Z-VAD-FMK for 7 h (F). Bars, ∼10 μm. (G) Quantification of apoptosis in indicated samples by Cell Death Detection ELISA PLUS. All samples, except QLT-0267 5 μm 1 h, are from the 7-h time point. Data are mean ± SD from two experiments. (H) Indicated cell lysates (all 7-h treatments) were Western blotted for PARP.
Inhibition of ILK expression (Fukuda et al., 2003) or kinase activity (Troussard et al., 2006) can induce apoptosis, with QLT-0267 causing apoptosis after 18 h of exposure (Troussard et al., 2006). The mitotic spindle defects seen here, however, are observed after just 7 h. Nevertheless, to verify that the observed spindle phenotype was not an indirect result of the cells undergoing apoptosis, two approaches were taken. First, cells were treated with 5 μM QLT-0267 for just 1 h. Fig. 3 E shows that these cells showed the same spindle phenotype as those treated for 7 h. Second, cells were treated with 5 μM QLT-0267 together with the antiapoptotic pan-caspase inhibitor Z-VAD-FMK for 7 h. These cells also showed the same spindle phenotype, suggesting that this effect occurs in the absence of apoptosis (Fig. 3 F). Fig. 3 (G and H) verifies that little or no apoptosis is seen at 1 or 7 h of treatment with 5 μM QLT-0267 and that what little may be occurring can be successfully blocked with Z-VAD-FMK. These data demonstrate that ILK activity is required locally for the maintenance of mitotic spindle integrity during mitosis.
Although QLT-0267 is highly selective for ILK and the effects of the inhibitor on the activation of downstream targets of ILK, such as Akt, can be rescued by overexpression of active, but not inactive, ILK (Troussard et al., 2006), it is still possible that the observed defects were caused by off-target effects. We therefore used siRNA to silence ILK expression to determine whether ILK is essential for mitotic spindle assembly and mitosis. As shown in Fig. 4 A, ILK siRNA resulted in >95% inhibition of ILK expression in HeLa cells. Examination of mitotic spindles in control and ILK siRNA–treated cells confirmed depletion of ILK expression in centrosomes and clearly showed a large increase in the accumulation of abnormal mitotic figures in the ILK siRNA–treated cells (Fig. 4 C). Cells showed similar disrupted phenotypes of mitotic spindles (Fig. 4 B) to those observed for QLT-0267 treatment, with asymmetrical and unorganized spindles resulting in misaligned chromosomes. The percentage of cells in a prometaphase/metaphase-like state with correctly aligned chromosomes was much greater in the control than in the ILK siRNA–treated cells (Fig. 4 D). The percentage of cells in later phases of mitosis declined dramatically in the ILK siRNA–treated cells, suggesting that cells were arrested in early stages of mitosis (Fig. 4 E). This is consistent with the increased mitotic index of cells treated with the ILK inhibitor, suggesting that perturbation of ILK function causes a block in mitosis.
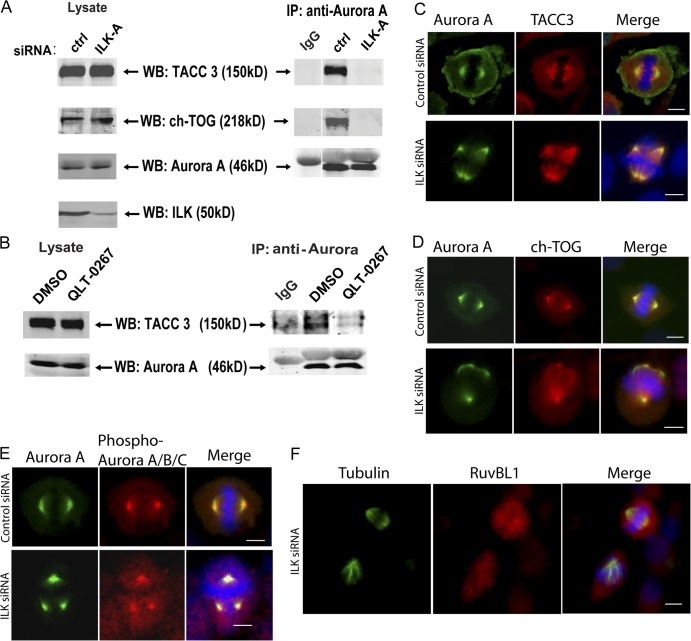
ILK is required for Aurora A–TACC3/ch-TOG interactions
The current paradigm for the regulation of mitotic spindle organization is that Aurora A kinase phosphorylates TACC3, which recruits ch-TOG, a protein required for microtubule polymerization and spindle organization (Gergely et al., 2003; Barros et al., 2005; Kinoshita et al., 2005). To determine the role of ILK in mitotic spindle organization, we analyzed whether the interaction between Aurora A and TACC3/ch-TOG is affected in the absence of ILK. As shown in Fig. 5 A, although immunoprecipitation of Aurora A from control siRNA-treated cells results in the copurification of both TACC3 and ch-TOG, these proteins are not associated with Aurora A in ILK siRNA–treated cells. The ILK inhibitor QLT-0267 also resulted in the inhibition of interaction of TACC3 with Aurora A (Fig. 5 B), indicating that the kinase activity of ILK may be required for its proper function in the centrosome. Examination of the localization of Aurora A, TACC3, and ch-TOG demonstrated that although the spindles are disrupted in these cells, these proteins still colocalize to centrosomes at sites of microtubule nucleation (Fig. 5, C and D). Therefore, although the interactions between these proteins are impaired in ILK-depleted cells, as seen by coimmunoprecipitation, ILK depletion does not cause a severe mislocalization of these proteins. To test whether ILK may have an effect on the activation of Aurora A, we also costained siRNA-treated cells for total and phospho–T288–Aurora A. As can be seen in Fig. 5 E, however, phospho–T288–Aurora A is still present at centrosomes treated with ILK siRNA, suggesting that ILK does not affect the activation of Aurora A in this manner. ILK siRNA–treated cells were also stained for RUVBL1 to examine the potential effect of ILK depletion on RUVBL1 localization. Fig. 5 F shows two mitotic cells in ILK siRNA–treated cells. One cell appears normal and RUVBL1 can be seen to localize to one of the centrosomes. In the cell showing an aberrant spindle, however, no RUVBL1 can be seen at the centrosome. As Fig. 2 shows that RUVBL1 disruption leads to loss of ILK localization at the centrosome, these data suggest that ILK and RUVBL1 may localize to the centrosome as a co-complex and are dependent on each other for their centrosomal localization.
Figure 5.
ILK siRNA causes a disruption of Aurora A–TACC3/chTOG interaction. (A) HeLa cells were transfected with control or ILK siRNA and synchronized with nocodazole. Aurora A kinase was immunoprecipitated from cytoskeletal extracts with a monoclonal anti–Aurora A antibody and then Western blotting was performed with polyclonal antibodies against Aurora A, TACC3, and ch-TOG. (B) A similar experiment to A was performed on cells treated with QLT-0267 and a DMSO control. (C–F) Effect of ILK siRNA on localization of ILK-interacting proteins. Hela cells were transfected and stained as indicated. Phospho–Aurora A/B/C staining is presumably phospho–Aurora A rather than Aurora B or C, as it colocalizes with total Aurora A; Aurora B is not found on centrosomes, and Aurora C is only expressed in testis. Bars, ∼10 μm.
In this paper, we provide two novel and unexpected results. First, we have shown that ILK localizes to focal adhesions and centrosomes simultaneously in interphase cells. ILK also colocalizes with the tubulin-interacting proteins ch-TOG and RUVBL1 through all phases of mitosis. Surprisingly, the focal adhesion partner of ILK, α-parvin (Legate et al., 2006), did not localize in centrosomes, whereas RUVBL1 appears to be required for localization of ILK to centrosomes. These results suggest that ILK associates with distinct protein partners to localize to distinct subcellular regions. Although unexpected for ILK, the localization of focal adhesion proteins to centrosomes is not without precedent, as several focal adhesion proteins, such as HEF1 (Pugacheva and Golemis, 2005), Ajuba (Hirota et al., 2003), zyxin (Hirota et al., 2000), and paxillin (Herreros et al., 2000), have also been shown to localize and function in centrosomes and MTOCs.
Second, ILK plays an essential role in mitotic spindle organization because disruption of its kinase activity or inhibition of its expression has profound effects on this structure. The similar phenotype of ch-TOG- and ILK-depleted spindles (Gergely et al., 2003) suggests that ILK may play an important role in ch-TOG function, such as in the regulation or maintenance of ch-TOG–TACC3 interaction (Gergely et al., 2003; LeRoy et al., 2007). Disruption of the interaction between Aurora A and TACC3/ch-TOG in ILK-depleted cells suggests that ILK is required in the centrosomes for the maintenance of these interactions. It remains to be determined whether ILK regulates Aurora A kinase activity or TACC3 phosphorylation.
Supporting a functional role for ILK at the centrosome, a siRNA screen to identify protein kinases involved in regulating the cell cycle in Drosophila melanogaster cells identified ILK as being required for mitosis (Bettencourt-Dias et al., 2004). ILK silencing resulted in abnormal spindle assembly and altered chromosomal segregation. In addition, integrin function can regulate mitotic spindle assembly and cytokinesis (Reverte et al., 2006). This interesting finding suggests a complex interplay between adhesion complexes and the MTOC and requires further investigation.
Our findings raise some interesting and important questions. First, are the focal adhesion and centrosomal functions of ILK connected or are they independent? The simultaneous localization of ILK to focal adhesions and centrosomes would suggest independent roles, although it will be interesting to determine whether ILK function in centrosomes alters in cells in which focal adhesions are disrupted or integrin activity is modulated. Second, what are the precise functions and targets of ILK in the centrosome and mitotic spindle? Based on our results, ILK appears to be required for the formation of the Aurora A–TACC3/ch-TOG complex. In addition, some of the known signaling effectors of ILK, namely Akt, GSK-3, and β-catenin, have all been shown to localize in centrosomes and mitotic spindles (Wakefield et al., 2003; Huang et al., 2007).
In this paper, we have identified a novel role of ILK in mitosis as a centrosomal protein required for mitotic spindle organization by regulating the interplay between Aurora A and TACC3/ch-TOG. The precise functions of ILK in the centrosomes will be the focus of future studies.
Materials and methods
Cell culture and SILAC
HEK293 cells expressing FLAG were cultured in 42 mg/liter of normal isotopic abundance arginine and 73 mg/liter of normal isotopic abundance lysine, whereas cells expressing FLAG-ILK were in 43.5 mg/liter 13C6-arginine and 75 mg/liter 2H4-lysine (Cambridge Isotope Laboratories, Inc.). All SILAC media was based on arginine- and lysine-free DME (Caisson Laboratories, Inc.) supplemented with 10% dialyzed FBS (Invitrogen), 1% l-Glu, 1% Pen/Str (Thermo Fisher Scientific), and 200 μg/ml geneticin.
Isolation of the cytoskeleton
The cytoskeleton was extracted as previously described (Arregui et al., 1994; Dobreva et al., 2008). In brief, cells were rinsed with CSB (10 mM Pipes, pH 6.2, 50 mM KCl, 10 mM EGTA, 3 mM MgCl2, and 2 M glycerol) and soluble proteins were removed with CSB containing 1% Triton X-100, 1 mM PMSF, 50 μg/ml leupeptin, 50 μg/ml aprotinin, 2 mM NaF, and 1 mM Na3VO4 for 2 min at 37°C. The remaining cytoskeleton was collected in extraction buffer (20 mM Tris-HCl, pH 7.4, 80 mM KCl, 30 mM MgCl2, 1 mM EGTA, 0.25 M NaCl, 1 mM DTT, 50 μg/ml leupeptin, 1 mM PMSF, and 1% Triton X-100), sonicated, and dialyzed in TBS, pH 8.0 (0.05 M Tris, 0.138 M NaCl, and 0.0027 M KCl), containing 1 mM EDTA. Precipitate was removed by centrifugation and protein concentration was measured by the BCA method (Thermo Fisher Scientific).
Isolation of mitotic spindles
Mitotic spindles were purified as described previously (Sauer et al., 2005) with modification. In brief, HeLa cells were grown in p15 plates until confluence and synchronized with 1 μM nocodazole for 16 h. The mitotic cells were then collected by shake off and centrifugation at 300 g, followed by a PBS wash, and released into normal medium for 40 min until most cells had reached metaphase (verified by immunofluorescence staining with DAPI for metaphase plate formation). Microtubules were then stabilized by the addition of 5 μg/ml taxol for 3 min. The cells were then collected, washed with PBS containing 2 μg/ml latrunculin B, 1 mM PMSF, and 5 μg/ml taxol, and incubated in lysis buffer (100 mM Pipes, 1 mM Mg2SO4, 2 mM EGTA, 0.5% NP-40, 5 μg/ml taxol, 2 μg/ml latrunculin B, 200 μg/ml DNase I, 10 μg/ml RNase A, 1 U/ml micrococcal nuclease, 20 U/ml benzonase, Complete protease inhibitors [Roche], 1 mM PMSF, and 20 mM β-glycerophosphate) for 15 min at 37°C (fraction S1 collected). The cells were harvested by centrifugation at 700 g for 2 min, resuspended in the same buffer, incubated for 5 min at 37°C, and harvested again by centrifugation (fraction S2 collected). The mitotic spindles were isolated by incubating the lysed cell ghosts in isolation buffer (1 mM Pipes, 5 μg/ml taxol, and Complete protease inhibitors) for 5 min and collected by centrifugation at 1,500 g for 3 min (fraction S3 collected). The pellet was resuspended in isolation buffer (fraction P and mitotic spindles) and the presence of mitotic spindles and spindle-associated proteins was confirmed by immunofluorescence staining for tubulin, pericentrin, and ILK. The spindles were then disrupted by sonication and proteins were resolved on gradient SDS-PAGE gels. Presence of known spindle proteins, such as tubulin and ch-TOG, were confirmed by Western blotting.
Immunoprecipitation, GeLC-MS/MS, and data analysis
Equal amounts of cytoskeleton from FLAG and FLAG-ILK–expressing cells were combined and precleared with protein A beads (Sigma-Aldrich) for 1 h at 4°C. The remaining supernatant was immunoprecipitated overnight at 4°C with 50 μl ANTI-FLAG M2 affinity gel (Sigma-Aldrich). The beads were collected by centrifugation, washed three times with washing buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1% Triton X-100, 2 mM NaF, 1 mM Na3VO4, and one tablet of protease inhibitor [Roche]) and boiled in sample buffer. Proteins were resolved on 5–15% SDS-PAGE gels and stained with blue-silver (Candiano et al., 2004) before in-gel digestion (Shevchenko et al., 1996). Extracted peptides were concentrated/desalted/filtered on Stop and Go Extraction Tips (Rappsilber et al., 2003) and analyzed by GeLC-MS/MS (Thermo Fisher Scientific) as previously described (Chan et al., 2006). Fragment spectra were extracted and the resulting peak list was searched against the human International Protein Index database (v3.29; 68,161 sequences) using Mascot (v2.1; Matrix Science). SILAC ratios were extracted as previously described (Foster et al., 2006).
Coimmunoprecipitation and Western blot analysis
Immunoprecipitation was performed as described in the previous section and proteins were resolved on SDS-PAGE gels. Transfer membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences) and incubated overnight with primary antibodies diluted in TBS, 5% BSA, or 0.05% Tween. The primary antibodies were detected by Alexa Fluor 680 (Invitrogen) or IRDye 800–conjugated secondary antibodies (Rockland Immunochemicals, Inc.) diluted at 1:5,000 in 5% nonfat dry milk, TBS buffer, and 0.2% NP-40 and visualized with the Odyssey IR imaging system (LI-COR Biosciences).
Immunofluorescence microscopy
HeLa and HEK293 cells were cultured under standard conditions and fixed in −20°C methanol for 10 min. PBS containing 1% BSA was added to cells for 30 min to block nonspecific interactions. Primary antibodies were then added for 16 h at 4°C in goat serum/gelatin blocking buffer, cells were washed, and secondary antibodies were added for 1 h at room temperature also in blocking buffer. HOECHST was added to stain DNA and coverslips were mounted in mounting medium (Vector Laboratories). Cells were viewed under a fluorescence microscope (Axioplan 2; Carl Zeiss, Inc.) using a 40×/0.75 objective (Plan-Neofluar; Carl Zeiss, Inc.) at room temperature. Images were acquired using a camera (AxioCam HRc; Carl Zeiss, Inc.) and AxioVision3.1 software (Carl Zeiss, Inc.), saved as tif files, and processed using Photoshop CS2 (Adobe). Only brightness/contrast adjustments were made to images.
Antibodies, ILK inhibitor, and siRNA
The following primary antibodies were used: rabbit anti-RUVBL1 (Proteintech); rabbit anti–ch-TOG, anti-ILK, and anti-pericentrin (AbCam); rabbit anti–α-parvin and mouse DM1A α-tubulin, anti–Aurora A, and anti–β-actin (Sigma-Aldrich); mouse anti-ILK (Millipore); rabbit anti-TACC3 (Santa Cruz Biotechnology, Inc.); and rabbit anti-PARP and anti–phospho–Aurora A (Thr288)/B (Thr232)/C (Thr198; Cell Signaling Technology). Secondary antibodies used for immunofluorescence labeling were Alexa Fluor goat anti–mouse (488 nm) and goat anti–rabbit (594 nm; Invitrogen). QLT-0267 (Quadra Logic Technology, Inc.) was diluted in DMSO and added to cells for 7 h unless otherwise stated. ILK (AAG ACG CTC AGC AGA CAT GTG GA), RUVBL1, or control nonsilencing (AAT TCT CCG AAC GTG TCA CGT) siRNAs (QIAGEN) were introduced into cells by using siLentFect reagent (Bio-Rad Laboratories), according to the manufacturer's instructions. To achieve sufficient ILK knockdown in centrosomes, cells were transfected twice (at day 0 and 2) with 100 nM ILK siRNA and then fixed/harvested on day 5. RUVBL1 siRNA was transfected once on day 0 and cells were fixed/harvested on day 3. Apoptosis was measured using Cell Death Detection ELISA PLUS (Roche). The assay and data analysis were performed according to the manufacturer's instructions.
Online supplemental material
In Fig. S1, ILK localizes to centrosomes in metaphase in three human cell lines and can be detected there using three different anti-ILK antibodies. Cells were also transfected with either ILK or control siRNA, stained for ILK, pericentrin, and HOECHST and imaged in interphase. In Fig. S2, pericentrin and ch-TOG still localize to the spindle poles in cells treated with QLT-0267, a specific small molecule inhibitor of ILK. Although the tubulin staining shows an aberrant mitotic spindle, pericentrin and ch-TOG still localize to the sites of spindle nucleation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200710074/DC1.
Supplementary Material
Acknowledgments
We thank Quadra Logic Technology, Inc. for supplying QLT-0267. We also thank Professor Michel Roberge (University of British Columbia, Vancouver, Canada) for valuable discussions during the course of this work.
This work was supported by grants to S. Dedhar from the National Cancer Institute of Canada, with funds raised by the Canadian Cancer Society and the Terry Fox Foundation, and by a grant from the Canadian Institutes of Health Research (MOP-77688) to L.J. Foster. L.J. Foster is the Canada Research Chair in Organelle Proteomics, a Michael Smith Foundation Scholar, and a Peter Wall Institute Early Career Scholar.
A.B. Fielding and I. Dobreva contributed equally to this paper.
Abbreviations used in this paper: ILK, integrin-linked kinase; MTOC, microtubule organizing center; SILAC, stable isotope labeling with amino acids in cell culture.
References
- Arregui, C.O., S. Carbonetto, and L. McKerracher. 1994. Characterization of neural cell adhesion sites: point contacts are the sites of interaction between integrins and the cytoskeleton in PC12 cells. J. Neurosci. 14:6967–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros, T.P., K. Kinoshita, A.A. Hyman, and J.W. Raff. 2005. Aurora A activates D-TACC–Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 170:1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig, G., M. Grimmler, I.G. Huttner, G. Wessels, T. Dahme, S. Just, N. Trano, H.A. Katus, M.C. Fishman, and W. Rottbauer. 2006. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 20:2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., R. Giet, R. Sinka, A. Mazumdar, W.G. Lock, F. Balloux, P.J. Zafiropoulos, S. Yamaguchi, S. Winter, R.W. Carthew, et al. 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 432:980–987. [DOI] [PubMed] [Google Scholar]
- Candiano, G., M. Bruschi, L. Musante, L. Santucci, G.M. Ghiggeri, B. Carnemolla, P. Orecchia, L. Zardi, and P.G. Righetti. 2004. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 25:1327–1333. [DOI] [PubMed] [Google Scholar]
- Chan, Q.W., C.G. Howes, and L.J. Foster. 2006. Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics. 5:2252–2262. [DOI] [PubMed] [Google Scholar]
- Dobreva, I., A.B. Fielding, L.J. Foster, and S. Dedhar. 2008. Mapping the integrin-linked kinase interactome using SILAC. J. Proteome Res. In press. [DOI] [PubMed]
- Foster, L.J., A. Rudich, I. Talior, N. Patel, X. Huang, L.M. Furtado, P.J. Bilan, M. Mann, and A. Klip. 2006. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC)*. J. Proteome Res. 5:64–75. [DOI] [PubMed] [Google Scholar]
- Fukuda, T., K. Chen, X. Shi, and C. Wu. 2003. PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J. Biol. Chem. 278:51324–51333. [DOI] [PubMed] [Google Scholar]
- Gartner, W., J. Rossbacher, B. Zierhut, T. Daneva, W. Base, M. Weissel, W. Waldhäusl, M.S. Pasternack, and L. Wagner. 2003. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil. Cytoskeleton. 56:79–93. [DOI] [PubMed] [Google Scholar]
- Gergely, F., V.M. Draviam, and J.W. Raff. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan, G., A.A. Troussard, and S. Dedhar. 2005. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat. Rev. Cancer. 5:51–63. [DOI] [PubMed] [Google Scholar]
- Hannigan, G.E., C. Leung-Hagesteijn, L. Fitz-Gibbon, M.G. Coppolino, G. Radeva, J. Filmus, J.C. Bell, and S. Dedhar. 1996. Regulation of cell adhesion and anchorage-dependent growth by a new [beta]1-integrin-linked protein kinase. Nature. 379:91–96. [DOI] [PubMed] [Google Scholar]
- Herreros, L., J.L. Rodriguez-Fernandez, M.C. Brown, J.L. Alonso-Lebrero, C. Cabanas, F. Sanchez-Madrid, N. Longo, C.E. Turner, and P. Sanchez-Mateos. 2000. Paxillin localizes to the lymphocyte microtubule organizing center and associates with the microtubule cytoskeleton. J. Biol. Chem. 275:26436–26440. [DOI] [PubMed] [Google Scholar]
- Hirota, T., T. Morisaki, Y. Nishiyama, T. Marumoto, K. Tada, T. Hara, N. Masuko, M. Inagaki, K. Hatakeyama, and H. Saya. 2000. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J. Cell Biol. 149:1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598. [DOI] [PubMed] [Google Scholar]
- Huang, P., T. Senga, and M. Hamaguchi. 2007. A novel role of phospho-[beta]-catenin in microtubule regrowth at centrosome. Oncogene. 26:4357–4371. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., T.L. Noetzel, L. Pelletier, K. Mechtler, D.N. Drechsel, A. Schwager, M. Lee, J.W. Raff, and A.A. Hyman. 2005. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170:1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate, K.R., E. Montanez, O. Kudlacek, and R. Fassler. 2006. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 7:20–31. [DOI] [PubMed] [Google Scholar]
- LeRoy, P.J., J.J. Hunter, K.M. Hoar, K.E. Burke, V. Shinde, J. Ruan, D. Bowman, K. Galvin, and J.A. Ecsedy. 2007. Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 67:5362–5370. [DOI] [PubMed] [Google Scholar]
- Lorenz, K., C. Grashoff, R. Torka, T. Sakai, L. Langbein, W. Bloch, M. Aumailley, and R. Fassler. 2007. Integrin-linked kinase is required for epidermal and hair follicle morphogenesis. J. Cell Biol. 177:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva, E.N., and E.A. Golemis. 2005. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat. Cell Biol. 7:937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J., Y. Ishihama, and M. Mann. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670. [DOI] [PubMed] [Google Scholar]
- Reverte, C.G., A. Benware, C.W. Jones, and S.E. LaFlamme. 2006. Perturbing integrin function inhibits microtubule growth from centrosomes, spindle assembly, and cytokinesis. J. Cell Biol. 174:491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, G., R. Korner, A. Hanisch, A. Ries, E.A. Nigg, and H.H.W. Sillje. 2005. Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics. 4:35–43. [DOI] [PubMed] [Google Scholar]
- Sepulveda, J.L., and C. Wu. 2006. The parvins. Cell. Mol. Life Sci. 63:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- Troussard, A.A., P.C. McDonald, E.D. Wederell, N.M. Mawji, N.R. Filipenko, K.A. Gelmon, J.E. Kucab, S.E. Dunn, J.T. Emerman, M.B. Bally, and S. Dedhar. 2006. Preferential dependence of breast cancer cells versus normal cells on integrin-linked kinase for protein kinase B/Akt activation and cell survival. Cancer Res. 66:393–403. [DOI] [PubMed] [Google Scholar]
- Wakefield, J.G., D.J. Stephens, and J.M. Tavare. 2003. A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J. Cell Sci. 116:637–646. [DOI] [PubMed] [Google Scholar]
- Weiske, J., and O. Huber. 2005. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-{beta }-catenin-mediated transcription. J. Cell Sci. 118:3117–3129. [DOI] [PubMed] [Google Scholar]
- White, D.E., P. Coutu, Y.-F. Shi, J.-C. Tardif, S. Nattel, R. St. Arnaud, S. Dedhar, and W.J. Muller. 2006. Targeted ablation of ILK from the murine heart results in dilated cardiomyopathy and spontaneous heart failure. Genes Dev. 20:2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.