Abstract
Protein aggregates and nuclear inclusions (NIs) containing components of the ubiquitin–proteasome system (UPS), expanded polyglutamine (polyQ) proteins, and transcriptional coactivators characterize cellular responses to stress and are hallmarks of neurodegenerative diseases. The biological function of polyQ-containing aggregates is unknown. To analyze proteasomal activity within such aggregates, we present a nanoparticle (NP)-based method that enables controlled induction of sodium dodecyl sulfate–resistant inclusions of endogenous nuclear proteins while normal regulatory mechanisms remain in place. Consistent with the idea that the UPS maintains quality control, inhibition of proteasomal proteolysis promotes extra large protein aggregates (1.4–2 μm), whereas formation of NP-induced NIs is found to be inversely correlated to proteasome activation. We show that global proteasomal proteolysis increases in NP-treated nuclei and, on the local level, a subpopulation of NIs overlaps with focal domains of proteasome-dependent protein degradation. These results suggest that inclusions in the nucleus constitute active proteolysis modules that may serve to concentrate and decompose damaged, malfolded, or misplaced proteins.
Introduction
Proteasomal proteolysis enables nuclear processes of gene expression and regulation of the cell cycle. The proteasome machinery's involvement in protein degradation is spatially regulated through self-compartmentalization at the molecular level (Voges et al., 1999) and segregation to distinct subcellular loci (Pines and Lindon, 2005). Consistent with their functional interactions, nuclear components of the ubiquitin–proteasome system (UPS) are predominately localized in euchromatic regions as well as in the periphery or within subnuclear compartments, such as splicing factor–containing speckles and promyelocytic leukemia (PML) nuclear bodies (NBs; Wojcik and DeMartino, 2003; von Mikecz, 2006). PML NBs may serve as proteolysis centers because they accumulate malfolded forms of mutated virus nucleoprotein (Anton et al., 1999), recruit proteasomal regulator subunit 11S and PML under conditions of proteasome inhibition (Lallemand-Breitenbach et al., 2001), and host proteasomal proteolysis of ectopic substrate DQ-ovalbumin (Rockel et al., 2005).
Although 20–30% of newly synthesized proteins undergo rapid degradation as defective ribosomal products, quality control by the UPS is also important for protection of cells against aggregation of damaged mature proteins caused by harsh environmental and disease conditions. Formation of nuclear inclusions (NIs) that contain the general transcription factor TATA binding protein, the transcriptional coactivator CREB binding protein (CBP), ubiquitin (Ub), and proteasomes is associated with expansion of polyglutamine (polyQ) repeats in inherited neurodegenerative disorders, e.g., Huntington's disease and spinocerebellar ataxia (Ross, 2002). Research on mechanisms of protein aggregation and their role in disease pathology focuses on formation of insoluble fibrillar deposits called amyloids (Ross et al., 2003), large amorphous protein assemblies, and proteolysis.
Global impairment of the UPS in polyQ diseases is implied by studies that show inefficient degradation of polyQ proteins and inhibition of proteasomal activity by irreversible sequestration of substrates within proteasomes (Bence et al., 2001; Holmberg et al., 2004; Venkatraman et al., 2004; Bennett et al., 2005). Subcellular topology is put forward as an important factor in protein aggregation by other studies (Janer et al., 2006; Qin et al., 2006). Expression of the PML isoform IV induces the formation of distinct NBs that recruit mutant ataxin-7 and host its degradation by proteasomes (Janer et al., 2006). A subpopulation of endogenous PML NBs seems to locally increase the capacity to degrade polyQ proteins. The open question concerning the biological function of protein aggregation is whether NIs embody permanent storage sites of damaged and misplaced proteins or active proteolytic centers.
We show in this paper that nanoparticles (NPs) induce insoluble SDS-resistant NIs. These NIs copy physiologically relevant processes because in contrast to other cell-based protein aggregation models, they contain aberrant assemblies of endogenous nuclear proteins with an intact UPS in place. Three lines of evidence suggest that NIs constitute sites of proteasomal protein degradation. First, global proteasomal activity is increased in nuclear fractions of silica-NP–treated cells. Second, formation of silica-NP–induced NIs can be reduced by activation of proteasomes and increased by inhibition of proteasome-dependent proteolysis. Third, a significant subset (30%) of silica-NP–induced NIs overlaps with proteasomal degradation of a model substrate.
Results and discussion
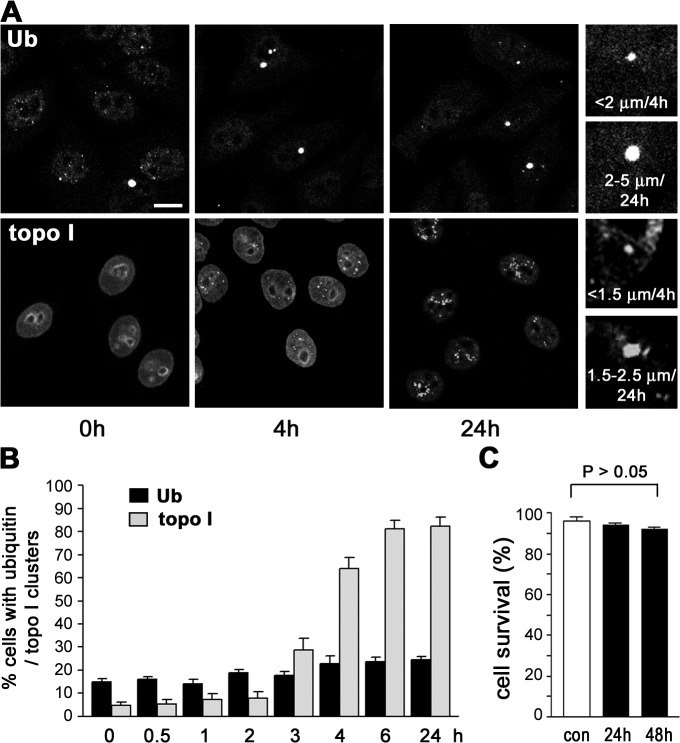
To obtain standardized experimental conditions for aggregation of endogenous proteins in the nucleus, we treated cells with nanosized silicium dioxide particles (silica-NPs or nanosilica). Silica-NPs seed inclusions of Ub (Fig. 1 A, top) or topoisomerase I (topo I; Fig. 1 A, bottom) in the nucleoplasm. Ub-NIs of up to 2 μm are detectable in untreated control cells and grow in number and size after the addition of silica-NPs (2–5 μm; Fig. 1, A [closeups] and B). In contrast, nuclear clustering of topo I is rare (5%; Fig. 1 B) in untreated controls where topo I typically localizes at the nucleolar rim and diffusely in the nucleoplasm (Fig. 1 A, bottom, 0 h). After nanosilica incubation, bright irregularly shaped topo I–NIs appear, which grow over time (Fig. 1 A, bottom, closeup) and occur in 81% of nuclei after 6 h (Fig. 1 B). Hence, analysis of topo I–NIs represents a sensitive readout for intranuclear protein aggregation that exceeds the conventionally used detection of aggregated Ub in sensitivity and specificity. Because topo I is critically involved in DNA replication, recombination, and transcription, topo I–NIs may represent deposits of the protein at sites of stalled nuclear processes resulting from harsh environmental conditions.
Figure 1.
Silica-NPs induce Ub- and topo I–containing inclusions in the nucleus. (A) Confocal IF of Ub (top) and topo I (bottom) shows time-dependent (0, 4, and 24 h) formation of NIs after treatment of cells with silica-NPs. Closeups show representative NIs after 4 or 24 h of silica-NP treatment. Bar, 10 μm. (B) Quantitation of cells with Ub- or topo I–NIs after silica-NP incubation for the indicated times. Values represent means ± SD from three experiments. (C) Viability remains unchanged in cells treated with silica-NPs for the indicated times versus untreated controls (con). Values represent means ± SD from three experiments.
It has been suggested that formation of NIs is promoted by the nuclear environment (Perez et al. 1998). Accordingly, we show that although silica-NPs penetrate the cytoplasm and the nucleus, protein aggregates exclusively form in the nucleoplasm (Figs. S1 and S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200708131/DC1; Chen and von Mikecz, 2005). Increase of silica-NP concentration does not result in alteration of the NI number nor in larger size, which further underlines the specificity of the experiments (Fig. S2 B and not depicted). Because soluble silicium dioxide is cytotoxic (Nash et al., 1966), cell viability was assessed in untreated versus silica-NP–treated cells. Cell survival ranged insignificantly between 96 (untreated) and 92% (Fig. 1 C, 48-h nanosilica), suggesting that silica-NPs do not dissolve in cell culture and that Ub- and topo I–NIs are not caused by cytotoxic effects or cell death. Generation of NIs by nanosilica clearly differs from that by proteasome inhibitor MG132, which induces severe alterations of cell morphology and reduction of cell survival (Ding et al., 2002; unpublished data).
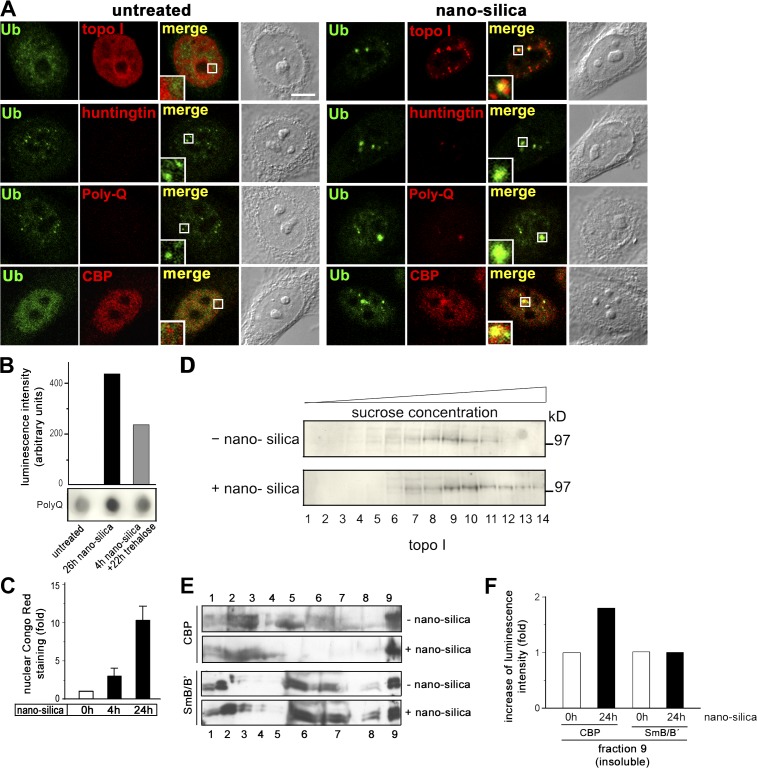
NIs feature a specific composition of aggregated proteins (Rajan et al., 2001). To determine which proteins segregate into silica-NP–induced NIs, double-labeling immunofluorescence (IF) was performed, which showed that Ub (Fig. 2 A, green) and topo I (Fig. 2 A, red) partially colocalize in nuclear clusters (Fig. 2 A, top, right, inset). Colocalization also occurs between Ub and the NI markers huntingtin, polyQ, and CBP (Fig. 2 A, right, bottom, insets, red). Notably, Ub forms a shell around huntingtin and polyQ (Fig. 2 A, insets). A comprehensive summary of results demonstrates that the protein composition of silica-NP–induced Ub- and topo I–NIs is identical to polyQ-induced nuclear protein aggregates (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200708131/DC1). To further validate nanosilica-induced NIs, aggregates and constituent proteins were analyzed for their solubility. Filter retardation assays reveal that silica-NPs induce a SDS-resistant polyQ-containing protein fraction (Fig. 2 B) that is sensitive to the protein aggregation inhibitor trehalose. Trehalose reduces polyQ retardation to ∼50%. Because positive signals in membrane filter assays indicate an amyloid-like structure (Wanker et al., 1999), the azo dye Congo red, which preferentially binds to β-sheet–containing amyloid fibrils (Cooper, 1969), was used to test silica-NP–induced NIs. Nuclear Congo red staining increases 4.3-fold after 4 h and 17.3-fold after 24 h of treatment with silica-NPs (Fig. 2 C), which suggests amyloid formation. Consistent with the idea that components of NIs are less soluble, sucrose step gradients show that topo I, which participates in distinct fractions of untreated cells (Fig. 2 D, lanes 5–12), shifts to fractions with higher sucrose concentration/density (Fig. 2 D, lanes 6–14) and that the population of CBP that copurifies with the nuclear matrix increases 1.8-fold in silica-NP–treated cells (Fig. 2, E [lane 9] and F). In summary, the localization, composition, and biochemical characterization of Ub- and topo I–NIs defines them as genuine SDS-resistant protein inclusions in the nucleoplasm.
Figure 2.
Silica-NP–induced NIs contain insoluble polyQ proteins and amyloid. (A) Confocal IF of Ub (green) and topo I (top, red) or signature proteins of NIs (bottom, huntingtin, PolyQ, and CBP, red) defines partial colocalization after 4 h of silica-NP treatment (nanosilica, merge, insets). Bar, 5 μm. (B) Filter retardation assay, followed by a dot blot of polyQ proteins, shows induction of SDS-resistant NIs by nanosilica that is mitigated by coincubation with the protein aggregation inhibitor trehalose. (C) Cells were treated with silica-NPs for the indicated times and analyzed cytochemically with Congo red. Quantitation of fluorescence intensity indicates an induction of nuclear β-amyloid staining by silica-NPs. Values represent means ± SD from three independent experiments (n = 120). (D) Nuclear extracts of untreated and silica-NP–treated (24 h) cells were separated according to their density on step sucrose gradients and subjected to Western blot with an anti–topo I antibody. (E) Nuclear extracts from untreated or silica-NP–treated cells were further fractioned and analyzed by Western blot to determine the levels of CBP (250 kD) and spliceosomal component SmB/B' (28/29 kD; control) that are soluble (lane 1), DNA bound (lanes 2–5), high salt extractible (lanes 6–8), and associated with the nuclear matrix (lane 9). (F) Quantitation of luminescent signals of E (lane 9).
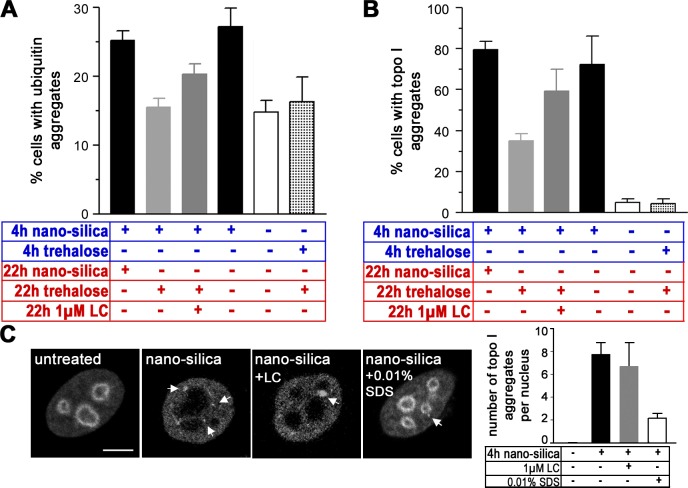
To characterize the role of the UPS in the assembly of Ub- and topo I–NIs, proteolytic activity was modulated with the proteasome inhibitor lactacystin (Fig. 3, A and B). Treatment with silica-NPs induces Ub-NIs in 25% and topo I–NIs in 80% of nuclei. Addition of trehalose after 4 h of silica-NP treatment reduces formation of Ub-NIs to 15% and topo I–NIs to 35%. Coincubation with lactacystin mitigates the effect of trehalose. Under conditions of proteasome inhibition, 20% of nuclei contain Ub-NIs and 60% contain topo I–NIs, suggesting that proteasomal activity is involved in formation of nuclear protein aggregates. Cotreatment with nanosilica and lactacystin induces extra large topo I–NIs (Fig. 3 C), whereas their number per nucleus remains unchanged.
Figure 3.
Involvement of proteasomal activity in the formation of Ub- and topo I–NIs. Cells were left untreated (white bars) or treated with silica-NPs or trehalose for 4 h (blue), followed by treatment with silica-NPs, trehalose, or a combination of trehalose and the proteasome inhibitor lactacystin for an additional 22 h (red). Quantitation of cells with Ub (A) and topo I aggregates (B) shows that trehalose significantly reduces formation of silica-NP–induced NIs, whereas addition of lactacystin partly attenuates the effect of trehalose. Values represent means ± SD from three experiments. (C) Micrographs show confocal IF of topo I in representative cells that were left untreated or treated with silica-NPs, a combination of silica-NPs and 1 μM lactacystin, or a combination of silica-NPs and 0.01% SDS. Note that proteasomal inhibition by lactacystin does not alter the number of NIs but induces larger aggregates (micrograph, nanosilica + LC, arrow). Arrows designate topo I–NIs. Values represent means ± SD from three experiments (n = 120). LC, lactacystin. Bar, 5 μm.
We next investigated the consequences of proteasomal activation. Active centers of 20S proteasomes are secluded from the exterior cellular space within a hollow cavity that is closed by a gating structure (Voges et al., 1999). Destabilization of the gate by low concentrations of SDS promotes the entry of substrates and their degradation to peptides (Dahlmann et al., 1993). In cells that are coincubated with silica-NPs and 0.01% SDS, topo I localizes at the nucleolar rim, throughout the nucleoplasm, and in a few small clusters (Fig. 3 C, right, micrograph). Size and number of silica-NP–induced topo I–NIs is reduced, confirming that proteasomal activity correlates with protein aggregation.
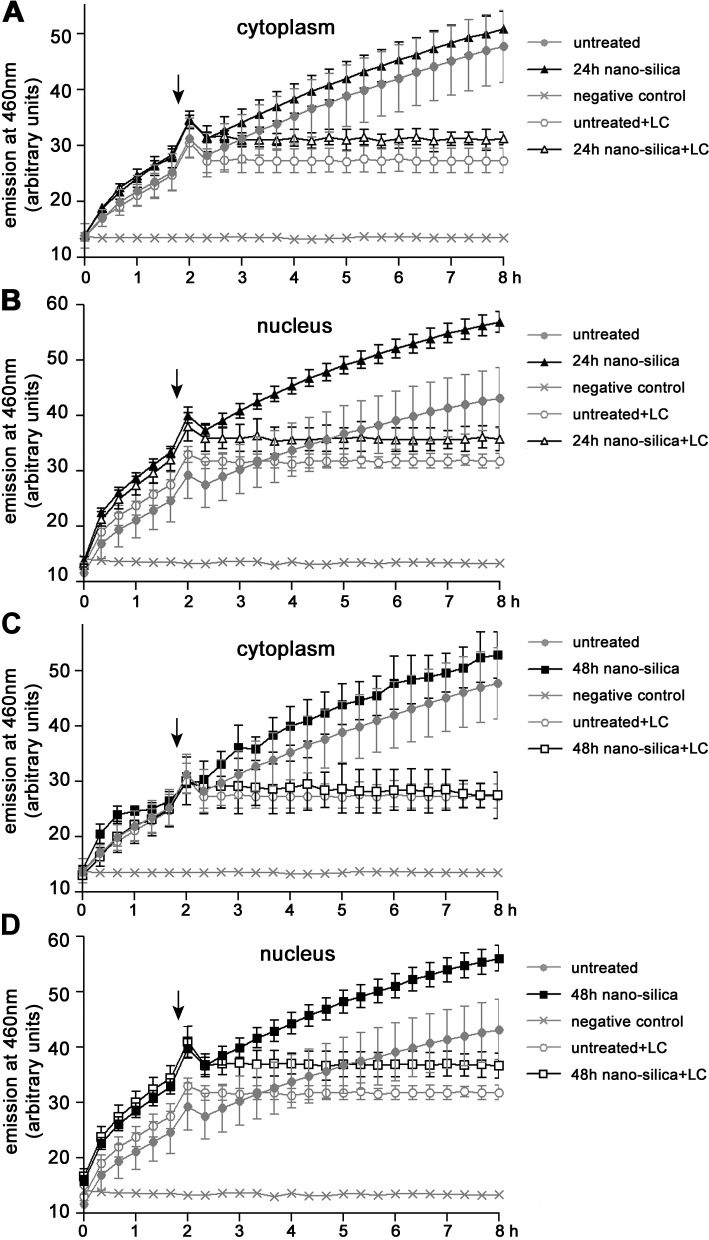
To strengthen this idea, global proteasomal activity was measured in vitro by incubation of the fluorogenic precursor substrate succinyl (Suc)-LLVY-aminomethylcoumarin (AMC) with cytoplasmic or nuclear protein fractions. Although proteasomal activity is similar in the cytoplasm of untreated and silica-NP–treated cells (Fig. 4, A and C), accumulation of cleaved substrate increases in nanosilica-treated nuclei (Fig. 4 B). The induction of fluorescent Suc-LLVY-AMC is slight but not significant after 4 h (not depicted; Chen and von Mikecz, 2005) and significant after 24 (Fig. 4 B) and 48 h (Fig. 4 D) of treatment with nanosilica, suggesting a sustainable induction of global proteasomal activity in nuclei over time that correlates with NI formation. These results contradict the view that the UPS is generally impaired under conditions of protein aggregation (Bence et al., 2001; Bennett et al., 2005) but confirm observations that in cells that develop protein aggregates because of knockdown of the endosomal transport factor ESCRT-III, proteasome activity is increased (Filimonenko et al., 2007).
Figure 4.
Induction of global proteasomal activity in the nucleus by silica-NPs. Cells were either untreated or treated with silica-NPs for 24 or 48 h, followed by preparation of cytoplasmic and nuclear extracts. Proteasomal activity of extracts was analyzed by incubation with the fluorogenic substrate Suc-LLVY-AMC and measurement of fluorescence intensity for 8 h (closed symbols). Specificity of proteasomal degradation was tested by addition of lactacystin after 2 h (arrows, open symbols). The graphs show proteasomal activity of cytoplasmic (A and C) and nuclear (B and D) extracts. Values of fluorescence intensity represent means ± SD from three experiments. LC, lactacystin.
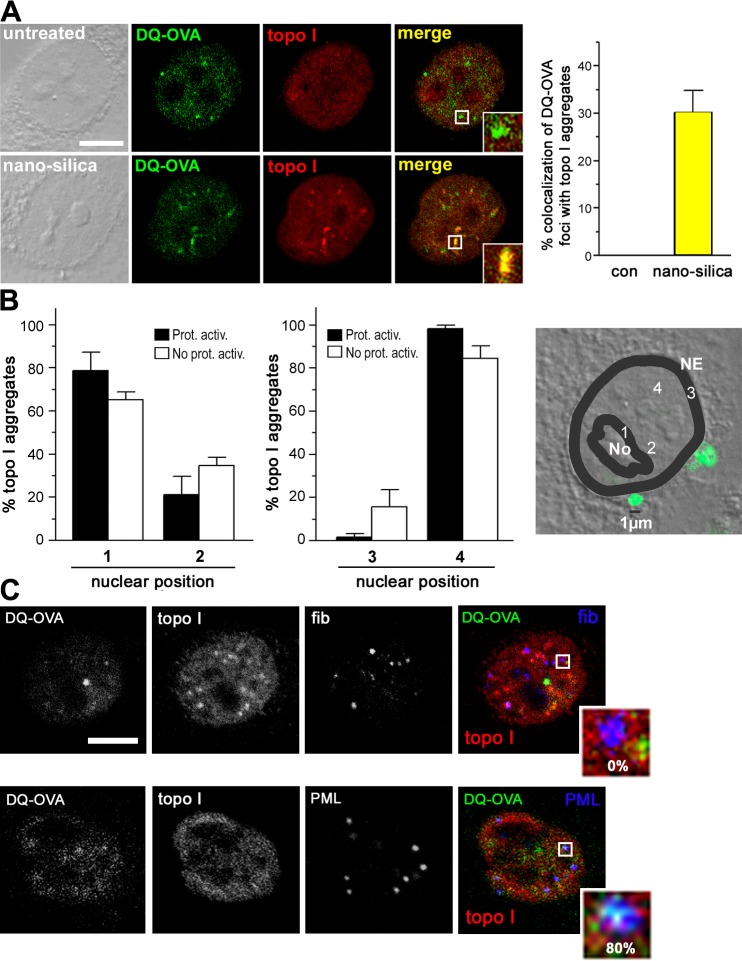
The rate of proteolysis is markedly affected by poly-Ub chain length. Thus, the mere presence of Ub in silica-induced NIs does not characterize them as proteolytic centers. To analyze proteasomal activity at the local level, we developed a microinjection-based method that pinpoints degradation of the ectopic fluorogenic substrate DQ-ovalbumin (DQ-OVA; Rockel et al., 2005). Confocal IF of cells that were microinjected into the nucleus with DQ-OVA (Fig. 5 A, green) and colabeled with antibodies against topo I (Fig. 5 A, red) shows no overlap in untreated cells (Fig. 5 A, top, inset) but shows partial colocalization of topo I–NIs with DQ-OVA proteolysis after silica-NP treatment (Fig. 5 A, bottom, inset). The quantitation reveals that 30% of topo I–NIs overlap with DQ-OVA foci, suggesting that a significant subpopulation of silica-NP–induced NIs are proteolytically active and, thus, represent proteolysis centers. Given the transient nature of DQ-OVA foci (Rockel et al., 2005), it is tempting to speculate that the subset of proteolytically active NIs might increase when viewed over time. Another, yet not excluding, possibility is that this subpopulation has distinct characteristics.
Figure 5.
A subpopulation of silica-NP–induced NIs hosts proteasomal degradation of DQ-OVA. In situ localization of proteasomal protein degradation was performed by microinjection of the fluorogenic substrate DQ-OVA into nuclei. (A) Double labeling of DQ-OVA and antibodies against topo I in untreated (top) or silica-NP–treated (4 h; bottom) cells shows that a subpopulation of NIs contains proteasomal activity (bottom, merge and inset). Quantitation of colocalization between green and red fluorescence signals indicates that 30% of topo I–NIs overlaps with focal domains of DQ-OVA degradation. Values represent means ± SD from three experiments. Bar, 5 μm. (B) Analysis of the nuclear positioning of topo I aggregates reveals no significant difference between proteolytically active versus inactive NIs. The micrograph describes the positioning method. Cells were incubated with 1 μm silica particles as size standards and areas were defined as follows: juxtaposed to nucleoli (1); random to nucleoli (2); juxtaposed to the nuclear envelope (3); and random to the nuclear envelope (4). Values represent data ± SD from four independent experiments (n = 30). con, control; NE, nuclear envelope; No, nucleolus; No prot. activ., topo I–NIs without proteasomal activity; Prot. Activ., topo I–NIs with proteasomal activity. (C) Micrographs of representative cells treated with silica-NPs for 4 h. Triple labeling of microinjected DQ-OVA (green) with topo I (red) and fibrillarin (fib; top, blue) or PML (bottom, blue) shows that 80% of NIs with proteolytic activity partially colocalize with PML (bottom, inset), whereas no overlap could be detected with fibrillarin (top, inset).
To investigate their distinctive features, the location (Fig. 5 B) and composition (Fig. 5 C) of proteolytically active and inactive NIs was compared. A nuclear positioning analysis reveals that proteolytically active NIs are juxtaposed (79%) or randomly positioned (21%) to nucleoli, and juxtaposed (2%) or randomly positioned (98%) to the nuclear envelope (Fig. 5 B, closed bars). Similar results are observed for NIs without proteolytic activity (Fig. 5 B, open bars), ruling out that proteolysis in protein aggregates depends on their localization. Next, the protein composition was defined. PML and fibrillarin participate in NBs and silica-NP–induced NIs (Chen and von Mikecz, 2005). Triple labeling experiments show that 80% of proteolytically active topo I–NIs contain PML, whereas none comprise fibrillarin located to NBs, probably Cajal NBs (Fig. 5 C). PML (Fig. 5 C, blue) forms a spherical wrap around topo I (Fig. 5 C, red) and DQ-OVA (Fig. 5 C, green), thus colocalizing with proteolytically active NIs in subdomains of the NB (Fig. 5 C, bottom, inset, white). By codetection of PML, NIs, and proteolysis at the same nuclear locations, we confirm PML NBs as proteolytic centers and offer an explanation for the prevention of accumulation of polyQ proteins by NBs that contain the PML isoform IV and UPS components (Janer et al., 2006). Consequently, the role of PML IV in the formation of proteolytically active NIs should be investigated. Consistent with the concept that all proteins with RING finger domains may act as E3 Ub ligases (Freemont, 2000), it is possible that PML, or isoforms thereof, mediates ubiquitination and proteasomal proteolysis of proteins concentrated in NIs.
The results presented here suggest that the UPS is triggered as a cellular defense mechanism in response to the formation of aberrant protein aggregates in the nucleus. Like other nuclear processes, this proteolytic response is spatially organized, namely in proteolytically active NIs that may represent key structures in the clearance of protein depositions. However, proteolysis within NIs may also contribute to toxicity by degradation of essential nuclear proteins and impairment of functions such as transcription (Ross and Pickart, 2004). Proteolytically active NIs potentially act as sink holes for components of the transcription machinery, thereby blocking expression of gene subsets that ultimately alter cellular fate. Consistent with this idea, we showed previously that nanosilica induce an irreversible arrest of cell proliferation with distinctive features of cellular senescence (Chen and von Mikecz, 2005). Further analyses are required to comprehend protein degradation in NIs. Are certain substrates degraded preferentially (i.e., substrate specificity) and does proteolysis of distinct proteins impair nuclear functions? It is essential to learn more about the biological function and proteasomal regulation of wild-type polyQ proteins versus the polyQ tract mutants. Particular polyQ proteins may be incompletely degraded, block the proteasomes involved (Holmberg et al., 2004; Venkatraman et al., 2004), and transform the respective NIs into areas of permanent segregation. An alternative nonexclusive possibility is that protein aggregates compete for proteasome-dependent proteolysis, thus leaving certain aggregates doomed for permanent persistence. A picture is emerging where polyQ toxicity does not depend on a structural transition occurring above a specific threshold but, rather, that polyQ tracts are inherently toxic sequences whose deleterious effects grow along a length-dependent toxicity gradient (Klein et al., 2007). In this case, balanced proteolysis in the nucleus would be constantly required to regulate formation of small soluble oligomers that are seeded by proteins with wild-type polyQ tracts and to avoid large amorphous assemblies or highly ordered fibrillar amyloids. In perspective, it will be important to define physiological versus pathological thresholds of UPS activity and substrate specificity in the context of their nuclear location.
Materials and methods
Particles
Unlabeled plain silica particles sized 50 or 70 nm (silica-NPs or nanosilica) and FITC-labeled 70-nm or 1-μm silica particles were purchased from G. Kisker and Postnova Analytics. Silica particles were added directly to the culture medium at concentrations of 25 or 50 μg/ml where indicated.
Chemicals
DNase was purchased from Roche. Suc-LLVY-AMC and lactacystin were obtained from Affinity BioReagents and Qbiogene, respectively. All other chemicals were obtained from Sigma-Aldrich.
Cell culture and treatments
HEp-2 cells (American Tissue Culture Collection) were routinely cultured in RPMI 1640 medium supplemented with 10% FBS, nonessential amino acids, l-glutamine, and penicillin and streptomycin (pen/strep) in a humidified incubator at 37°C with 5% CO2. Cells were either untreated or incubated with nanosilica particles for 4, 24, or 48 h, respectively. For analysis of the interaction between protein aggregation and proteasomal proteolysis, cells were initially incubated with silica-NPs for 4 h and then further cultivated in silica-NP–free medium containing 100 μM trehalose in the presence or absence of 1 μM lactacystin. Note that none of the treatments increased DNA damage (Chen and von Mikecz, 2005).
Antibodies
The following antibodies were used for IF or Western blot where indicated: rabbit anti-Ub (FL-76; Santa Cruz Biotechnology, Inc.) at 1:25; human serum against topo I (Chen and von Mikecz, 2005) at 1:200 and 1:50 (Western blot); mouse anti-huntingtin (Millipore) at 1:400; mouse anti-PML (PG-M3; Santa Cruz Biotechnology, Inc.) at 1:10; mouse anti-fibrillarin (72B9; provided by E. Tan, Scripps Research Institute, La Jolla, CA) at 1:50; rabbit anti-TATA binding protein (N-12; Santa Cruz Biotechnology, Inc.) at 1:100; mouse anti-ataxin 3 (Millipore) at 1:10; mouse anti-Ub (P4D1; Santa Cruz Biotechnology, Inc.) at 1:10; rabbit anti-Ub (Sigma-Aldrich) at 1:100 (Western blot); rabbit anti-CBP (C20; Santa Cruz Biotechnology, Inc.) at 1:25 and 1:200 (Western blot); and mouse anti-polyglutamine (1C2; Chemicon) at 1:400 and 1:1,000 (Western blot and filter assay). In addition to proteins with expanded polyglutamine (CAG) tracts, anti-polyQ antibodies (clone 1C2; Millipore) detect the respective wild-type proteins. Secondary antibodies (Jackson ImmunoResearch Laboratories) conjugated to FITC, rhodamine, or Cy5 were used at dilutions of 1:100 in IF. Secondary antibodies were used at 1:10,000 (anti–rabbit or anti–human IgG) and 1:5,000 (anti–mouse IgG) in Western blots.
Filter retardation assay
Dot-blot filter retardation assay was applied to detect SDS-resistant polyglutamine-containing protein aggregates as previously described (Wanker et al., 1999). In brief, cells were washed and harvested in PBS, pelleted, and lysed on ice for 30 min in lysis buffer containing 50 mM Tris-HCl, pH 8.8, 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, and 1 mM EDTA supplemented with a protease inhibitor cocktail (Roche). Insoluble material was collected by centrifugation for 5 min at 14,000 rpm in a microfuge (Eppendorf) at 4°C. Pellets were resolved in DNase buffer containing 20 mM Tris-HCl, pH 8.0, 15 mM MgCl2, and 0.5 mg/ml DNase I and incubated for 1 h at 37°C. Incubation was terminated by adjusting the mixture to 20 mM EDTA, 2% SDS, and 50 mM DTT, followed by heating at 98°C for 5 min. Protein samples were further diluted in 2% SDS and filtered through a cellulose acetate membrane (Whatman). Filters were blocked in PBS containing 0.5% Tween 20 with 5% nonfat dried milk, followed by Western blot detection of polyQ proteins.
Sucrose gradients
Adherent cells were detached by trypsinization, washed twice with PBS, and lysed with a lysis buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 15 mM MgCl2, 1% Triton X-100, and protease inhibitor) at 4°C for 10 min. After centrifugation at 2,500 g for 20 min, the pellet was resuspended in 20 mM Tris-HCl buffer, pH 8, and sonicated. Nuclear extracts from either untreated or nanosilica-treated HEp-2 cells were loaded onto 13 × 64-mm ultracentrifuge tubes (Beckman Coulter) that were prepared with a step gradient formed by layers of 20 mM Tris-HCl buffer, pH 8, containing 0–65% (5% steps) sucrose, respectively. The tubes were centrifuged for 15 h at 180,000 g at 4°C using a rotor (50.2Ti; Beckman Coulter) and an ultracentrifuge (Beckman Coulter). Fractions were collected and prepared for Western blot detection.
Biochemical fractionation and solubility assay
To determine the solubility of cellular proteins, biochemical fractionation was performed according to the protocol for the enrichment of nuclear matrix proteins (He et al., 1990). To detect CBP (as representative polyQ protein) and SmB/B' (as nuclear control protein), fractions were subjected to Western blotting.
Cell viability assay
Cells were seeded simultaneously in culture flasks with the same density and left untreated or treated with silica-NPs. At the indicated times, cells were trypsinized and counted by hemocytometer (n = 100–200). Cell viability was assessed by Trypan blue exclusion.
Proteasome activity assays
Analysis of proteasomal activity of nuclear and cytoplasmic protein fractions and of subnuclear localization of proteasome-dependent proteolysis has been previously described (Chen and von Mikecz, 2005; Rockel, et al. 2005).
IF and microscopy
Subconfluent cells on glass coverslips were either fixed with 3.7% formaldehyde in PBS for 10 min and permeabilized with 1% Triton X-100 for 3 min at room temperature or fixed with methanol for 5 min followed by permeabilization in acetone for 2 min at −20°C. Incubations with primary antibodies against various proteins were performed as previously described (Chen and von Mikecz, 2005). Samples were observed with a confocal laser microscope (Fluoview IX70; Olympus) using a 60×/1.25 UPlanFl objective. Channels were scanned sequentially (488 nm for FITC, 568 nm for rhodamine, and 647 nm for Cy5). Controls established the specificity of fluorochrome-conjugated antibodies for their respective immunoglobulins and that signals in green, red, and far red channels were derived from the respective fluorochrome. No cross talk was observed in multiple staining experiments.
Congo red staining
Cells on coverslips were untreated or treated with silica-NPs for 4 and 24 h, fixed with 4% formaldehyde, and permeabilized with 1% Triton X-100 for 3 min at room temperature. After washing with PBS, cells were incubated in PBS with 10 mg/ml BSA for 20 min and then stained with 0.2% Congo red solution for 10–20 min. Cells were washed with PBS and covered with mounting medium. Images were acquired at 488-nm excitation and at 568-nm emission.
Quantification of fluorescence and luminescence intensities
Quantitative analysis of fluorescence was performed with the Metamorph image analysis software package (MDS Analytical Technologies). For measurement of fluorescence intensities within nuclear domains (e.g., nuclear envelope, nucleolus, and nucleoplasm), regions of interest were positioned manually based on corresponding differential interference contrast images. Images were background corrected by reference regions outside the cells but within the field of view, which corresponded to identical-sized regions of interest within the nucleus. In double-labeling experiments, signals were defined as colocalizing in the range of hue (31–54), intensity (0–255), and saturation (106–251; HIS color model; Metamorph software). For each experiment, the fluorescence intensities of 100–200 nuclei were determined. Luminescence intensities of Western blot and filter assays were quantified with the AlphaEaseFC software (Alpha Innotech). Figures were assembled in Photoshop (Adobe).
Spatial positioning
1-μm labeled silica particles were coincubated with cells and used as size standards. The nuclear position of topo I–containing protein aggregates was defined as juxtaposed to the nucleolus (<1 μm distance) or random to nucleoli (>1 μm distance) and juxtaposed to the nuclear envelope (<1 μm distance) or random to the nuclear envelope (>1 μm distance). The nucleoli and nuclear envelope were located based on differential interference contrast.
Statistical analysis
Analysis of variance was used to analyze the statistical significance between control and nanosilica particle–treated groups. P-values <0.05 were considered to be statistically significant.
Online supplemental material
Fig. S1 contains confocal microscopy showing that silica-NPs translocate to the cytoplasm and nucleoplasm. Fig. S2 contains confocal microscopy showing that protein aggregation is independent of the concentration of silica-NPs. Table S1 shows a comprehensive summary of protein composition and biochemical properties of polyQ- versus silica-NP–induced NIs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708131/DC1.
Supplementary Material
Acknowledgments
We thank Eng Tan for donation of antibodies.
This work was supported by Deutsche Forschungsgemeinschaft, through grant SFB 728, and by Bundesministerium für Umwelt, Naturschutz und Reaktorsicherheit.
Abbreviations used in this paper: AMC, aminomethylcoumarin; CBP, CREB binding protein; IF, immunofluorescence; NB, nuclear body; NI, nuclear inclusion; NP, nanoparticle; PML, promyelocytic leukemia; UPS, ubiquitin–proteasome system.
References
- Anton, L.C., U. Schubert, I. Bacik, M.F. Princiotta, P.A. Wearsch, J. Gibbs, P.M. Day, C. Realini, M.C. Rechsteiner, J.R. Bennink, and J.W. Yewdell. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J. Cell Biol. 146:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence, N.F., R.M. Sampat, and R.R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555. [DOI] [PubMed] [Google Scholar]
- Bennett, E.J., N.F. Bence, R. Jayakumar, and R.R. Kopito. 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell. 17:351–365. [DOI] [PubMed] [Google Scholar]
- Chen, M., and A. von Mikecz. 2005. Formation of nucleoplasmic protein aggregates impairs nuclear function in response to SiO2 nanoparticles. Exp. Cell Res. 305:51–62. [DOI] [PubMed] [Google Scholar]
- Cooper, J.H. 1969. An evaluation of current methods for the diagnostic histochemistry of amyloid. J. Clin. Pathol. 22:410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann, B., B. Becher, A. Sobek, C. Ehlers, F. Kopp, and L. Kuehn. 1993. In vitro activation of the 20S proteasome. Enzyme Protein. 47:274–284. [DOI] [PubMed] [Google Scholar]
- Ding, Q., J.J. Lewis, K.M. Strum, E. Dimayuga, A.J. Bruce-Keller, J.C. Dunn, and J.N. Keller. 2002. Polyglutamine expansion, protein aggregation, proteasomal activity, and neuronal survival. J. Biol. Chem. 277:13935–13942. [DOI] [PubMed] [Google Scholar]
- Filimonenko, M., S. Stuffers, C. Raiborg, A. Yamamoto, L. Malerod, E.M.C. Fisher, A. Isaacs, A. Brech, H. Stenmark, and A. Simonsen. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont, P.S. 2000. Ring for destruction. Curr. Biol. 10:R84–R87. [DOI] [PubMed] [Google Scholar]
- He, D.C., J.A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg, C.I., K.E. Staniszewski, K.N. Mensah, A. Matouschek, and R.I. Morimoto. 2004. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 23:4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janer, A., E. Martin, M.-P. Muriel, M. Latouche, H. Fujigasaki, M. Ruberg, A. Brice, Y. Trottier, and A. Sittler. 2006. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J. Cell Biol. 174:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F.A.C., A. Pastore, L. Masino, G. Zeder-Lutz, H. Nierengarten, M. Oulad-Abdelghani, D. Altschuh, J.-L. Mandel, and Y. Trottier. 2007. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: towards a length-dependent toxicity gradient. J. Mol. Biol. 371:235–244. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P.P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor α degradation. J. Exp. Med. 193:1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, T., A.C. Allison, and J.S. Harington. 1966. Physico-chemical properties of silica in relation to its toxicity. Nature. 210:259–261. [DOI] [PubMed] [Google Scholar]
- Perez, M.K., H.L. Paulson, S.J. Pendse, S.J. Saionz, N.M. Bonini, and R.N. Pittman. 1998. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 143:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines, J., and C. Lindon. 2005. Proteolysis: anytime, any place, anywhere? Nat. Cell Biol. 7:731–735. [DOI] [PubMed] [Google Scholar]
- Qin, Q., R. Inatome, A. Hotta, M. Kojima, H. Yamamura, H. Hirai, T. Yoshizawa, H. Tanaka, K. Fukami, and S. Yanagi. 2006. A novel GTPase, CRAG, mediates promyelocytic leukemia protein–associated nuclear body formation and degradation of expanded polyglutamine protein. J. Cell Biol. 172:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan, R.S., M.E. Illing, N.F. Bence, and R.R. Kopito. 2001. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. USA. 98:13060–13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel, T.D., D. Stuhlmann, and A. von Mikecz. 2005. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 118:5231–5242. [DOI] [PubMed] [Google Scholar]
- Ross, C.A. 2002. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and relating disorders. Neuron. 35:819–822. [DOI] [PubMed] [Google Scholar]
- Ross, C.A., and C.M. Pickart. 2004. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 14:703–711. [DOI] [PubMed] [Google Scholar]
- Ross, C.A., M.A. Poirier, E.E. Wanker, and M. Amzel. 2003. Polyglutamine fibrillogenesis: the pathway unfolds. Proc. Natl. Acad. Sci. USA. 100:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman, P., R. Wetzel, M. Tanaka, N. Nukina, and A.L. Goldberg. 2004. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 14:95–104. [DOI] [PubMed] [Google Scholar]
- Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015–1067. [DOI] [PubMed] [Google Scholar]
- von Mikecz, A. 2006. The nuclear ubiquitin-proteasome system. J. Cell Sci. 119:1977–1984. [DOI] [PubMed] [Google Scholar]
- Wanker, E.E., E. Scherzinger, V. Heiser, A. Sittler, H. Eickhoff, and H. Lehrach. 1999. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 309:375–386. [DOI] [PubMed] [Google Scholar]
- Wojcik, C., and G.N. DeMartino. 2003. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35:579–589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.