Abstract
Mechanisms by which tumor cells evade detection by the host’s immune system are thought to play a role in progression to malignancy, but this has not been investigated in the context of neurofibromatosis type 1 (NF1). NF1 is an autosomal dominant disorder, in which aggressive peripheral nerve tumors, known as malignant peripheral nerve sheath tumors (MPNSTs), develop in 5–10% of patients. Large scale gene expression profiling of a MPNST-derived cell line, T265, and normal human Schwann cells (hSCs) identified a large group of immune function genes down-regulated in T265 cells. Here we report that the aberrant expression of immune system related genes extends beyond MHC class I and II genes in T265 cells to include a transcription factor (MHC2TA) and other critical components of the antigen processing and presentation apparatus. TAP1, the transporter-activator protein that loads peptide antigens onto MHC class I molecules, is down-regulated, and CD74, a chaperone protein whose function is in processing and transport of MHC class II molecules, is down-regulated and alternatively spliced to produce an RNA transcript not evident in normal human Schwann cells. These findings reveal multiple molecular pathways and at least two cellular mechanisms acting to reduce the normal immune system molecules involved in antigen processing and presentation in cells derived from a peripheral nerve sheath tumor. Acquiring a “silent” immune signature may be a critical step in the progress towards malignancy in MPNSTs.
Keywords: NF1, T265, Schwann, Splicing, MHC2TA, TAP1, CD74, Cancer, MHC
Cellular progression to malignancy is often accompanied by reduced expression of immune system genes in tumor cells [10]. This is thought to allow tumor cells to evade detection and destruction by the host’s immune system [10]. This process has not been fully investigated in peripheral nerve sheath tumors, but preliminary evidence from a microarray screen supports this hypothesis [4]. A large scale gene expression profile comparison between normal human Schwann cells and T265 cells, a cell line derived from a malignant peripheral nerve sheath tumor [1], suggests wide-spread down-regulated expression of most major histocompatibility complex (MHC) genes in the tumor-derived cells [4]. Schwann cells are most likely the primary tumorigenic cell type in these tumors [3,5,12], which are derived from patients with neurofibromatosis type 1 (NF1), an autosomal dominant condition that can develop into an MPNST in 5–10% of patients. In this study we confirm and extend these findings to include a transcription factor, accessory proteins involved in antigen processing and presentation, and posttranscriptional processing of CD74 mRNA.
T265 cells and normal human Schwann cells were prepared and grown as previously described [4]; total RNA was isolated using the TRIzol extraction method with modifications. Three to five micrograms of total RNA from T265 cells and normal human Schwann cells (hSCs) was used in a labeling reaction and hybridized to cDNA microarrays; raw hybridization values and statistical analyses are as previously described [2,4]. Semi-quantitative (LightCycler) RT-PCR was carried out as previously described [4], with modifications for specific primer sets; GAPDH was used as a reference housekeeping gene.
A selection of immune function transcripts that exhibit down-regulation in T265 cells from a microarray screen were validated by semi-quantitative RT-PCR (Table 1). Multiple cDNA clones of the same genes are present on the microarray and are listed in order to show consistency between clones. In addition to validating the down-regulation of immune function genes identified from the microarray data, we were interested in determining potential mechanisms by which such a broad down-regulation of MHC class I and class II expression was produced in T265 cells.
Table 1.
Microarray data and LightCycler validation of down-regulation of immune function genes
| Gene name | Gene symbol | Z-test | LightCycler PCRa,b |
|---|---|---|---|
| Major histocompatability complex, class I, A | HLA-A | −22.88 | −5712 |
| 1 Major histocompatability complex, class I, C | HLA-C | −14.58 | −62.5 |
| 2 Major histocompatability complex, class I, C | HLA-C | −20.92 | – |
| 1 Major histocompatability complex, class II, DR beta | HLA-DRB | −24.64 | −1893 |
| 2 Major histocompatability complex, class II, DR beta | HLA-DRB | −9.51 | – |
| 1 Major histocompatability complex, class II, DO alpha | HLA-DOA | −8.01 | – |
| 2 Major histocompatability complex, class II, DO alpha | HLA-DOA | −12.67 | – |
| 1 Major histocompatability complex, class II, DQ beta | HLA-DQB | −10.74 | −27.3 |
| 2 Major histocompatability complex, class II, DQ beta | HLA-DQB | −16.27 | – |
| 3 Major histocompatability complex, class II, DQ beta | HLA-DQB | −27.61 | – |
Selection of deregulated immune function genes in MPNST-derived Schwann cell line T265, meeting the criteria p ≤ 0.01 (±2.58 Z-test) from three independent microarray experiments. Significance of this difference in mRNA abundance is given as a Z-test value. Multiple copies of the same cDNA on the array are numbered consecutively. Semi-quantitative RT-PCR (LightCycler) validation of the abundance of four mRNAs indicated from microarray analysis.
Fold change is with respect to the mRNA abundance in human Schwann cells.
mRNA abundance in hSC and T265 cells was compared to a control housekeeping gene, glyceraldehyde phosphate dehydrogenase (GAPDH), for relative quantification.
We tested the hypothesis that expression of the transcription factor MHC2TA, which is critical to the regulation of genes required for MHC class II antigen presentation [6], was suppressed in T265 cells, and observed a significant decrease in the transcript levels of this key transcription factor (−4.4-fold by semi-quantitative RT-PCR). These data suggest that down-regulation of class II mRNAs (Table 1) may be the result of reduced transcription from CTIIA-responsive promoters. However, destabilization of class II mRNAs, deletion of class II loci, loss of epigenetic regulation of these loci, or genomic instability by other mechanisms may also contribute to the marked suppression of immune system genes.
CD74 has a key role in antigen expression by MHC class II [11], therefore we analyzed the transcript levels of CD74 in normal human Schwann cells and T265 cells. We observed a large decrease (−1212-fold) in transcript levels of this key molecule in T265 cells compared to hSCs.
We also found by Western blot analysis that expression of the transporter-activator protein (TAP1) was decreased in T265 cells (Fig. 1). Reduced TAP1 expression results in a loss of surface expression of MHC class I molecules and is also associated with an increase in malignancy of tumors [9].
Fig. 1.
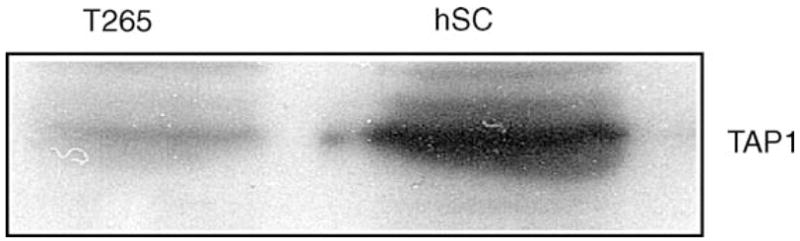
Western analysis of TAP1 levels in T265 cells and normal human Schwann cells. Western analysis of TAP1, a transporter required for correct MHC class I antigen presentation, in hSCs and T265 cells. TAP1 is down-regulated in T265 cells. Total cell lysates were prepared and 35 μg added to each lane, equal protein loading was confirmed by Coomassie staining of the gel.
We were especially interested in looking at, not only the down-regulation of transcripts, but the expression pattern of alternatively spliced products. To accomplish this goal we performed a more detailed transcript analysis of CD74 in hSC and T265 cells, revealing not only down-regulation of this transcript in T265 cells, but expression of alternative splice products between these cell types. The human CD74 gene spans 11.1 kb and is comprised of 8 potential exons which undergo alternative splicing. In Fig. 2A, alternate usage of Exons 5 through 8 generate several alternatively spliced transcripts [7,8]. Using semi-quantitative LightCycler RT-PCR we observed a shift in the melting curve of amplification of transcripts for CD74 in hSCs and T265 cells (Fig. 2B) that may result from the presence of different gene products. We therefore analyzed this region using the same primers as used in the LightCycler PCR (Table 2). In both hSC and T265 cells, there was a ~200-bp product corresponding to sequences in the NCBI database; this product was also present in the T265 cells at reduced levels. However, in T265 cells there was also an additional ~500-bp product, which may correspond to transcripts where intron 7 is utilized as part of exon 7, these products were sequence-verified (In Fig. 2A, referred to as exon 7B).
Fig. 2.
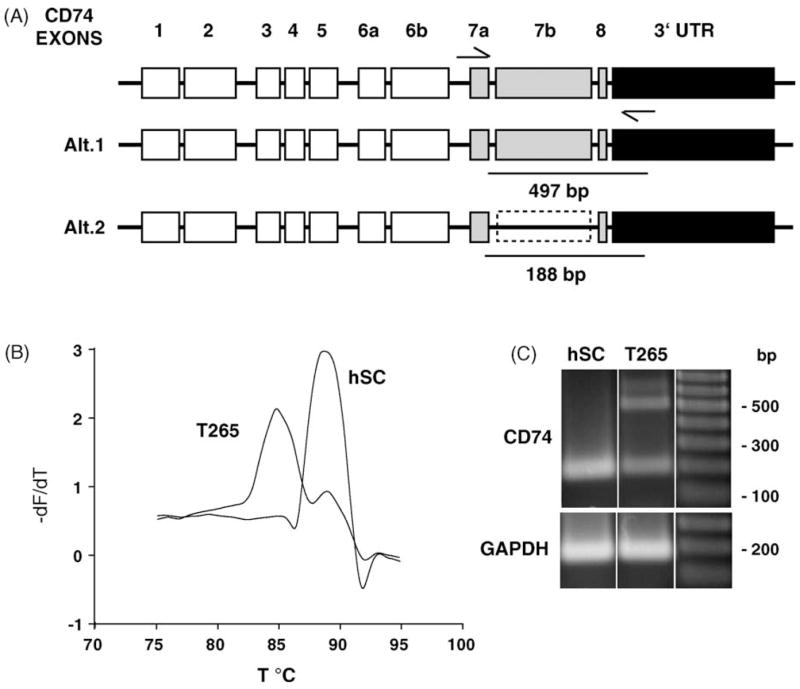
CD74 is differentially regulated in hSC and T265. (A) CD74, a MHC class II chaperone, exon structure and potential alternative splice forms. CD74 spans 11 kb on chromosome 5 and is comprised of 8 exons. Exon 7b may arise from alternate usage of splice donor and acceptor sites that utilize intron sequence. (B) Melting curve analysis of LightCycler PCR products demonstrates an additional product present in T265 cell PCR sample. Melting curve analysis was performed following LightCycler PCR. Plotted is the dF/dT as a function of temperature (°C) to define the melting point of amplified PCR products. The peaks correspond to different products, at 85 and 89 °C. There is an additional product that is present in the T265 cells, evident by the presence of the second peak at 85 °C. (C) RT-PCR of hSC and T265 cells demonstrate the presence of an alternatively spliced transcript. Primers corresponding to exon 7a and within the 3′UTR, amplify a single product of 188 bp in hSC, and an additional 497 bp product in the cancer cell line. This larger product may arise from utilization of intronic sequence between exons 7a and 8.
Table 2.
LightCycler PCR primers used in this study
| Gene symbol | 5′-Primer sequence-3′ | Exon/Intron | Product size |
|---|---|---|---|
| CD74 | AAGAGTCACTGGAACTGG
TCTCATGGGATGAGGTACAGG |
Exon 7a-3′-UTR | 188, 497 bp |
| MHC2TA | CTGGTCCACTCAGTCCATAGC
GAACTGTGTCCCAGAACATCC |
3′-UTR | 143 bp |
| HLA-DQB1 | TACTGGAACAGCCAGAAGGAC
GAAATCTGTCACCGAGCAGAC |
– | 189 bp |
| HLA-DRB1 | TACTGGAACAGCCAGAAGGAC
GGCTGGGTCTTTGAAGGATAC |
– | 146 bp |
| HLA-C | TGTGTCTGCGTTCCTGTTAGC
AGACATCCAGCCCACTTCTCT |
- | 141 bp |
| HLA-A | GACCAGGAGACACGGAATGTG
AATCCTTGCCGTCGTAGGCGT |
– | 171, 428 bp |
| GAPDH | TGCACCACCAACTGCTTA
GGATGCAGGGATGATGTTC |
– | 177 bp |
There is considerable interest in elucidating the molecular mechanisms responsible for the progression to malignancy in NF1. In a small fraction of cases the disease can develop into a malignant and potentially fatal disease. It is becoming clear that the immune system has a well-developed surveillance mechanism for the monitoring of, and ultimate destruction of tumors, however this has not been investigated in NF1. It is also apparent that most tumors appear to utilize multiple strategies to avoid killing by the host’s immune system. These may include down-regulation of the apparatus processing MHC class I antigen presentation, and down-regulation of transcription factors vital for MHC class II transcription. All may result in non-functional or dysfunctional protein products and therefore an attenuated immune response to the tumor.
The present study provides evidence for all of these mechanisms in T265 cells. Aberrant expression of components central to the functioning of the immune system would clearly result in an attenuated immune response to any tumor, but this data may also have therapeutic implications. Treatment of MPNSTs by immune therapy approaches, such as T-cell based immunotherapy, could be complicated by severely impaired antigen presentation in these tumor-derived Schwann cells (T265). Aberrant expression of MHC class I and subsequent lack of antigen processing would hamper recognition by cytotoxic T cells or natural killer cells and therefore severely impede destruction of tumors by this type of therapeutic approach.
Taken as a whole we have identified multiple mechanisms by which T265 cells globally perturb the expression and presentation of class I and class II antigens. In addition, we have identified specific transcripts that are subject to down-regulation and alternative splicing in T265 cells. Wide deregulation of genes involved in immune function may represent a set of mutations that are acquired early in the progression to malignancy. This may in turn contribute to evasion of the host’s immune system in the tumor micro-environment thus significantly enhancing the progression to malignancy in this tumor type.
Acknowledgments
We would like to thank George DeVries and Robert Farrer for normal human Schwann cells and the T265 cell line. We would also like to thank Kevin Becker and Bill Wood, NIA Gene Expression and Genomics unit for providing the cDNA microarrays, and Nancy Ratner for helpful discussions. This work was supported by the National Institute of Child Health and Human Development.
References
- 1.Badache A, Muja N, DeVries G. Expression of kit in neurofibromin-deficient human Schwann cells: role in Schwann cell hyperplasia associated with type 1 neurofibromatosis. Oncogene. 1998;17:795–800. doi: 10.1038/sj.onc.1201978. [DOI] [PubMed] [Google Scholar]
- 2.Cheadle C, Cho-Chung YS, Becker KG, Vawter MP. Application of z-score transformation to Affymetrix data. Appl Bioinform. 2003;2:209–217. [PubMed] [Google Scholar]
- 3.Kluwe L, Friedrich R, Mautner V. Loss of NF1 allele in Schwann cells but not fibroblasts derived from an NF1-associated neurofibroma. Genes Chromosomes Cancer. 1999;24:283–285. doi: 10.1002/(sici)1098-2264(199903)24:3<283::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 4.Lee PR, Cohen JE, Tendi EA, Farrer R, DeVries GH, Becker KG, Fields RD. Transcriptional profiling in an MPNST-derived cell line and normal human Schwann cells. Neuron Glia Biol. 2004;1:135–147. doi: 10.1017/s1740925x04000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legius E, Marchuk DA, Collins FS, Glover TW. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumor suppressor gene hypothesis. Nat Genet. 1993;3:122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- 6.LeibundGut-Landman S, Waldburger J-M, Krawczyk M, Otten LA, Suter T, Fontana A, Acha-Orbea H, Reith W. Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 7.O’Sullivan DM, Larhammar D, Wilson MC, Peterson PA, Quaranta V. Structure of the human Ia-associated invariant (gamma)-chain gene: identification of 5′ sequences shared with major histocompatibility complex class II genes. Proc Natl Acad Sci USA. 1986;83:4484–4488. doi: 10.1073/pnas.83.12.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Sullivan DM, Noonan D, Quaranta V. Four Ia invariant chain forms derive from a single gene by alternate splicing and alternate initiation of transcription/translation. J Exp Med. 1987;166:444–460. doi: 10.1084/jem.166.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seliger B, Maeurer MJ, Ferrone S. TAP off–tumors on. Immunol Today. 1997;18:292–299. doi: 10.1016/s0167-5699(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 10.Seliger B, Maeurer MJ, Ferrone S. Antigen-processing machinery breakdown and tumor growth. Immunol Today. 2000;21:455–464. doi: 10.1016/s0167-5699(00)01692-3. [DOI] [PubMed] [Google Scholar]
- 11.Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta. 2002;1542:1–13. doi: 10.1016/s0167-4889(01)00166-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]


