Abstract
Height has been used for more than a century as a model by which to understand quantitative genetic variation in humans. We report that the entire genome appears to contribute to its additive genetic variance. We used genotypes and phenotypes of 11,214 sibling pairs from three countries to partition additive genetic variance across the genome. Using genome scans to estimate the proportion of the genomes of each chromosome from siblings that were identical by descent, we estimated the heritability of height contributed by each of the 22 autosomes and the X chromosome. We show that additive genetic variance is spread across multiple chromosomes and that at least six chromosomes (i.e., 3, 4, 8, 15, 17, and 18) are responsible for the observed variation. Indeed, the data are not inconsistent with a uniform spread of trait loci throughout the genome. Our estimate of the variance explained by a chromosome is correlated with the number of times suggestive or significant linkage with height has been reported for that chromosome. Variance due to dominance was not significant but was difficult to assess because of the high sampling correlation between additive and dominance components. Results were consistent with the absence of any large between-chromosome epistatic effects. Notwithstanding the proposed architecture of complex traits that involves widespread gene-gene and gene-environment interactions, our results suggest that variation in height in humans can be explained by many loci distributed over all autosomes, with an additive mode of gene action.
Research into the genetics of complex traits has moved from the estimation of genetic variance in populations to the detection and identification of variants that are associated with or directly cause variation. The standard paradigm has been to perform linkage studies in pedigrees, followed by fine-mapping or candidate-gene studies with the use of association. Recently, genomewide association (GWA) studies, which rely on linkage disequilibrium between observed and causal variants, have become a reality—in particular, for the study of common disease in human populations.1–4 The success of both linkage and association studies depends on the frequency and distribution of individual gene effects in the population. Rare variants with large effects can most readily be mapped in pedigrees, whereas common variants with moderate effects can be mapped using an association study. Multiple rare variants in the same gene, each with a moderate effect on the phenotype, can be detected using linkage studies but would be hard to find in an association study.
Despite the large research effort in the past decade or so, the nature of complex-trait variation—in terms of the number of causal variants, their frequency in the population, and the size of their effects—is still largely unknown.5 Emerging evidence suggests that there are common variants with effects large enough to be detected for a range of phenotypes across a number of species, but the number of identified causal variants remains small.6,7 Other evidence suggests that multiple rare variants in the same gene may segregate in the population, each with an effect large enough to increase susceptibility to disease.8 In human populations, there has frequently been inconsistency of linkage to disease and quantitative phenotypes across multiple samples and populations, with few examples of clear-cut replication. One possible explanation is that, for most phenotypes, the effects of causal variants are too small to be detected by linkage—that is, that most studies have been underpowered. Association analyses are much more powerful for detecting small effects but, again, are dependent on the actual distribution of effect sizes. Recent reports of associated and replicated SNPs from GWA studies show that the effect sizes of individual common variants are typically small.1–4 Both linkage and association studies suffer from a multiple-testing problem, because they are generally hypothesis generating. There is also a conceptual problem with the null hypothesis in nearly all gene-mapping studies. The null hypothesis for a test at a given location in the genome is that there is no genetic variation associated with that location, despite the fact that we know that there is heritability, often considerable, for the phenotype in question. Hence, a priori, the null hypothesis cannot be true for all test locations. This assumption is particularly worrisome for linkage studies because of the strong linkage disequilibrium within families, such that many linked genes of small effects would result in false evidence of a major gene of large effect.9 We recently showed that a number of these problems disappear if the emphasis is on estimation of variance rather than on hypothesis testing.10
The actual genomewide relationship between pairs of relatives varies because of segregation and can be estimated using dense genetic markers for each pair. For full siblings, for example, the average proportion of the genome shared identical by descent (IBD) is 50%, with a range of ∼38% to ∼62%.10 By calculating the covariance between the proportion of the genome shared and the similarity of siblings for the phenotype, we were able to estimate the genetic variance free from assumptions about nongenetic sources of resemblance between relatives.10 In this study, we apply the principle of chromosome- and genomewide-realized relationships10,11 to partition genetic variance across the genome. We use a very large sample of sibling pairs with genomewide marker genotypes, a well-studied12 quantitative phenotype in humans (i.e., height), and the independent segregation of chromosomes, to partition genetic variation across chromosomes. We show that at least six chromosomes are responsible for genetic variation but that the hypothesis that all chromosomes contribute variation cannot be rejected. We find no evidence of dominance or epistatic variation. Thus, our data are consistent with a large number of underlying variants acting additively across all chromosomes to affect height in humans.
The data comprised quasi-independent sibling pairs (QISPs) from three studies in Australia (AU), the United States (US), and the Netherlands (NL). All individuals were of European descent. Pairs of MZ twins were excluded, but QISPs, including a single MZ individual, were maintained. Descriptions of the pedigrees, phenotypes, and genotypes have all been given elsewhere.10,13–15 In brief, for each of the three samples, QISPs were created from pairs of siblings within a nuclear family. Pairs were included if they had both phenotypic and genomewide genotypic information, with a minimum of 210 microsatellite markers per individual and an average of >400 markers for each of the studies.10,13–15 Height measurements were adjusted for sex and for age at measurement, and standardized residuals (Z scores) were calculated for each individual for each sample separately, to avoid the influence of heterogeneous variances across populations. There were 5,952, 3,996, and 1,266 QISPs for the AU, US, and NL samples, respectively, with a total sample size of 11,214. There were 1,936 brother-brother pairs, 4,011 sister-sister pairs, and 5,267 brother-sister pairs. After adjustment for sex and age, the sibling correlations for the AU, US, and NL samples were 0.432, 0.502, and 0.451, respectively, and the sibling correlation in the entire sample was 0.461. The brother-brother, sister-sister, and brother-sister correlations in the entire sample, after adjustments for age (and for the mean difference in sex in the brother-sister pairs), were 0.494, 0.479, and 0.435, respectively.
Additive coefficients of relationship were calculated using Merlin16 for each chromosome and genomewide for all three samples, as described elsewhere.10 For the X chromosome, IBD probabilities were estimated using MINX. The genetic length of the chromosomes was taken from independent pedigree data.17 We estimated from the marker data the proportions of individual chromosomes and of the genome as a whole that are shared IBD between all 11,214 pairs of siblings.10,13,14 These proportions are coefficients of additive relationship, which are, on average, 0.5 for full siblings but which vary considerably around their expectation, both between chromosomes for the same full-sib pair and between full-sib pairs for the same chromosome.10,18 The mean±SD of genomewide additive relationships in our sample of 11,214 sibling pairs was 0.4994±0.036 and the range was 0.309 to 0.644, consistent with previous results and with theory.10
Variance components were estimated by maximum likelihood, as implemented in the statistical package Mx19 and described elsewhere.10 Mixed linear models were fitted, including nongenetic family effects, chromosome and genomewide additive genetic effects, and residual effects. We first estimated genetic variance associated with the entire genome, by fitting a model that estimated the covariation between phenotypic similarity and the coefficient of additive relationship. For this whole-genome analysis, we confirmed our previous results,10 which were based on a smaller data set of 4,919 pairs from only one source of data. The estimate of heritability for stature from genomewide IBD from the sample of 11,214 sibling pairs was 0.86 (95% CI 0.49–0.95; P<.00001). The estimate of the proportion of phenotypic variation due to nongenetic family effects was 0.03 (P=.38), which is statistically nonsignificant. In addition to the genomewide additive effect, we fitted a genomewide dominance effect, using the probability of sharing two alleles IBD, averaged across the genome.10 The estimated proportions of variance due to additive and dominance effects were 0.699 and 0.160, respectively, but the dominance component was not significantly different from zero (P=.35). However, statistical power to separate these effects is low in our sibling-pair design, since the genomewide additive and dominance coefficients are highly correlated (r=0.911; n=11,214), as predicted by theory.10
After the genomewide analyses, we estimated genetic variance associated with individual chromosomes, using chromosomewide coefficients of additive relationship. The proportion of additive genetic variance explained by a particular chromosome was estimated in two ways—first, by fitting a full model that included effects due to a single chromosome and a reduced model in which no chromosomal effects were fitted, and, second, by fitting a full model containing effects for all 22 autosomes and a reduced model that fitted 21 autosomes only. Table 1 shows that, for the individual-chromosome analyses, 6 of 22 estimates of chromosomal heritability were significantly different from zero at P<.05 and of these 3 at P<.01. The six most significant chromosomes were, in order of the size of the test statistic, 17, 4, 3, 18, 15, and 8. This order is not the same as that of estimated chromosomal heritability, because SEs of estimates are larger for longer chromosomes.10 The estimate of the proportion of variance due to nongenetic family effects (f2) in the single-chromosome analyses captures the variation due to the other autosomes not fitted—for all 22 estimates, the sum of the estimates of f2 and (1/2)h2 was ∼0.459, the observed overall sibling correlation. Very similar estimates and test statistics were obtained from a full model with 22 additive genetic-variance components, from which chromosomal heritabilities were dropped one by one (table 1), consistent with the absence of any large between-chromosome epistatic effects.
Table 1. .
Estimates of Variance Proportions from Single-Chromosome Analyses and a Joint Analysis of All 22 Autosomes
| Single-Chromosome Analyses |
Combined-Chromosome Analysis |
|||||||
| Chromosome | f2a | h2ib | e2c | LRTd | P | h2i | LRTe | P |
| 1 | .4285 | .0607 | .5108 | 1.201 | .137 | .0633 | 1.418 | .117 |
| 2 | .4525 | .0131 | .5344 | .065 | .399 | .0097 | .037 | .424 |
| 3 | .4023 | .1134 | .4843 | 5.704 | .008 | .1160 | 6.269 | .006 |
| 4 | .4036 | .1124 | .4840 | 5.938 | .007 | .1082 | 5.705 | .008 |
| 5 | .4458 | .0264 | .5278 | .319 | .286 | .0196 | .191 | .500 |
| 6 | .4336 | .0506 | .5158 | 1.294 | .128 | .0508 | 1.370 | .500 |
| 7 | .4284 | .0616 | .5100 | 2.019 | .078 | .0630 | 2.230 | .068 |
| 8 | .4234 | .0708 | .5058 | 2.778 | .048 | .0856 | 4.172 | .021 |
| 9 | .4482 | .0216 | .5302 | .277 | .299 | .0325 | .663 | .500 |
| 10 | .4590 | .0000 | .5410 | .000 | .500 | .0000 | .000 | .500 |
| 11 | .4590 | .0000 | .5410 | .000 | .500 | .0000 | .000 | .500 |
| 12 | .4365 | .0451 | .5184 | 1.121 | .145 | .0489 | 1.434 | .500 |
| 13 | .4545 | .0089 | .5366 | .056 | .406 | .0006 | .000 | .500 |
| 14 | .4427 | .0323 | .5250 | .728 | .197 | .0185 | .246 | .500 |
| 15 | .4241 | .0703 | .5056 | 3.353 | .034 | .0760 | 4.028 | .022 |
| 16 | .4556 | .0069 | .5375 | .035 | .426 | .0180 | .251 | .308 |
| 17 | .4023 | .1142 | .4834 | 9.019 | .001 | .1124 | 8.967 | .001 |
| 18 | .4237 | .0703 | .5060 | 3.753 | .026 | .0622 | 3.013 | .041 |
| 19 | .4437 | .0309 | .5253 | .759 | .192 | .0317 | .840 | .500 |
| 20 | .4575 | .0031 | .5395 | .008 | .464 | .0037 | .012 | .456 |
| 21 | .4590 | .0000 | .5410 | .000 | .500 | .0000 | .000 | .500 |
| 22 | .4590 | .0000 | .5410 | .000 | .500 | .0000 | .000 | .500 |
| Total | … | .9126 | … | 38.427 | … | .9205 | 40.846 | … |
Proportion of variance due to sibling resemblance not accounted for by single-chromosomal genetic effects.
Proportion of variance due to additive genetic effects on the chromosome.
Proportion of variance due to individual environmental effects.
The likelihood-ratio test (LRT) statistic from comparing a full model fitting three variance components with a reduced model fitting two variance components. The P value was calculated assuming that the LRT statistic is distributed as 0 or χ21, each with a probability of 1/2.
The LRT statistic from comparing a full model fitting 24 variance components (22 additive genetic, 1 common environmental, and 1 residual) with a reduced model fitting 23 variance components, by dropping the ith additive genetic-variance component. The P value was calculated in the same way as that in the single-chromosome analyses.
We estimated variance on the X chromosome separately from the autosomes and separately for brother-brother, sister-sister, and brother-sister pairs, because the expected additive genetic covariance between siblings depends on their sex and on assumptions regarding dosage compensation.20 There was no evidence of additive genetic variance for height on the X chromosome for all three groups. The estimates of the proportion of variance due to additive effects on the X chromosome was 0.007 in brother-brother pairs (P=.47), 0.081 in sister-sister pairs (P=.39), and 0.00 in brother-sister pairs (P=.50).
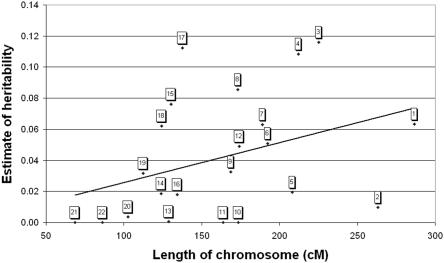
Figure 1 shows the relationship between the genetic length of the chromosome and the estimate of the proportion of additive genetic variance attributed to it in the single-chromosome analyses. In general, the longer the chromosome, the more variation it explains. A weighted least-squares regression was performed, with use of the empirical variance of chromosomal additive coefficients as weights, because this variance is inversely proportional to the sampling variance of the estimate of heritability.10 The relationship was highly statistically significant in a no-intercept model (F test, P<.0001), and adding an intercept after the regression was not statistically significant (P=.683). The slope of the regression line (0.03 heritability per 100 cM) is consistent with what would be expected if variance were apportioned according to genetic length, with the assumption of 0.86 for the overall heritability and a total sex-averaged map length of 2,864 cM for the autosomes.17 The correlation (r=0.23) between our estimates of chromosomal heritability and the number of genes per chromosome, obtained from Ensembl build 36, was smaller than the correlation with chromosomal length. The estimates and log-likelihoods of models in which all 22 additive genetic components were fitted were compared with those for the model in which a single genomewide additive genetic component was fitted. The drop in the log-likelihood of the data was 19.2 (P=.57, χ2 test with 21 df), which means that the more parsimonious model of variance contributed by chromosomes proportional to their length was not rejected.
Figure 1. .
Estimate of chromosomal heritability from a joint analysis of all 22 autosomes (labeled on graph) as a function of the genetic length of the chromosome. The figure shows that there is a relationship between the genetic length of a chromosome and the amount of variance it explains. P<.0001 for the regression coefficient from a weighted least-squares analysis if no intercept is fitted. P=.683 if an intercept is fitted after the regression.
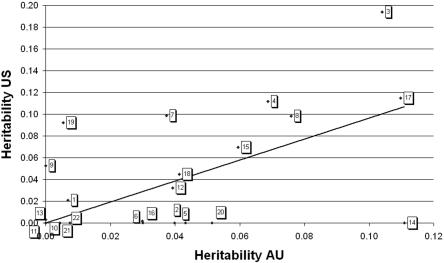
What is the minimum number of chromosomes needed to explain genetic variance for height? To address this question, we ordered chromosomes according to the amount of genetic variance they explained from the joint analysis (table 1) and, in a stepwise procedure, added one chromosome at a time. We compared the log-likelihoods, stopping when the addition of an extra chromosome did not improve the fit after accounting for the number of parameters in the model. To compare models, we used Akaike’s information criterion (AIC), calculated as −2(difference in log-likelihood) + 2(number of additive genetic-variance components), between the model and the null model of no additive genetic-variance components. In all models, a nongenetic family effect (f2) was fitted. The best and most parsimonious model, based on AIC, includes six chromosomes: 17, 4, 3, 18, 15, and 8, in order of statistical significance. The scaled AIC values for fitting 1–8 chromosomes were −7.02, −10.41, −13.73, −15.35, −16.36, −19.09, −19.03, and −18.51, with a minimum of six fitted genetic-variance components. Hence, our data indicate that at least 6 but as many as 22 chromosomes may contribute to additive genetic variation for height. For internal validation, we estimated the proportion of additive genetic variance attributable to each chromosome separately for the two largest data sets (AU and US). Figure 2 shows that the estimates are positively correlated. The slope of the regression line for the 22 pairs of heritability estimates in a no-intercept model is close to unity (0.96) and is highly significantly different from zero (P<.0001), which indicates that the estimates are generally similar across the two data sets. The partial correlation between the heritability estimates after conditioning on the length of the chromosome remained positive and significant (partial r=0.54; P=.011, two-sided), supporting the consistency of heritability estimates across data sets. Variable heritability estimates between chromosomes in excess of sampling variation imply that chromosomes do not contribute equally to additive genetic variance, even after an adjustment for their length.
Figure 2. .
Relationship between heritability estimates from the AU and US data sets. There is a highly significant correlation between the estimates of chromosomal heritability from the two data sets (no-intercept model, P<.0001; model with intercept, P=.009). The relationship remains significant when conditioning on the length of the chromosome (partial correlation 0.540; P=.011, two-sided).
We next investigated whether the proportion of variance explained by an autosome in our data is correlated with the number of times that suggestive or significant linkage21 has been reported for that chromosome in the literature, excluding reports that are partially or wholly based on the data in this study. For each chromosome, the number of LOD scores >1.9, defined as being “suggestive” of linkage,21 were counted from reported whole-genome linkage scans for height.22–30 From these scans, there were 2, 7, 5, 2, and 6 chromosomes for which 0, 1, 2, 3, and 4 linkages were reported, respectively, for an average of 2.1 linkages per autosome. We found a positive and significant correlation between our estimate of chromosomal heritability and the number of reported linkages from independent studies (Spearman’s nonparametric rank correlation 0.586; P=.002, one-tailed). This relationship remained significant after a linear adjustment of the latter for the length of the chromosome (P=.013). For three of the six most significant chromosomes in our data—namely, 3, 4, and 15—there have been at least three independent reports of suggestive or significant linkage, although, for our most significant chromosome (17), there has been only a single report of suggestive linkage in the literature. Counting the number of peak LOD scores from the literature may be an inaccurate quantification of assessing the importance of individual chromosomes in explaining variation because of reporting bias and because more false-positive results are likely to be reported for longer chromosomes. Nevertheless, even after an adjustment for chromosome length and despite the large sampling errors of estimates, we found a significant (but not perfect) relationship between our estimates of chromosomal heritability and the number of times suggestive or significant linkage had been reported.
To our knowledge, this is the first attempt to attribute additive genetic variation for a complex trait in humans to specific chromosomes by partitioning the total variance. We estimate that additive genetic variation for height in humans is contributed by all autosomes, with a minimum of six that are responsible, and that there is no significant evidence against the hypothesis that all chromosomes contribute to genetic variance in proportion to their length. For five of these (chromosomes 3, 4, 8, 15, and 18), there have been multiple independent reports of linkage in the literature.
Estimates of the effects of individual chromosomes on variation in a quantitative trait have been reported for Drosophila,31,32 and a method to estimate variance associated with whole chromosomes was proposed for experimental line crosses.33 For human pedigrees, a method to estimate the contribution of chromosome regions or whole chromosomes to variation in a quantitative trait, by the estimation of IBD sharing of sibling pairs from (sparse) marker data, was described elsewhere.34 The principle of genome partitioning and whole-genome analysis, as a multistage approach toward individual QTL mapping, was proposed by Schork.11 We have performed the first large-scale application of such methods to the quantitative trait height and have shown how additive genetic variance in three populations is distributed over chromosomes.
If indeed all chromosomes contribute variance in proportion to their length, then the best-case scenario for gene mapping is that all of them harbor a single QTL. For such an “average” QTL that explains, say, 3.9% (∼0.90 of 23) of the phenotypic variance, a linkage study with 57,830 sib pairs would be needed to detect it with a probability of 0.80 at a type I error rate of 0.0001.35 This is much larger than the sample sizes in all reported genomewide linkage studies for height in humans. The largest study comprises data from ∼4,000 equivalent full-sib pairs plus >10,000 more-distantly related (and therefore less informative) relative pairs, with approximately sufficient power to detect QTLs explaining 10% of the phenotypic variance in a genome linkage scan.13 A number of genome linkage scans have been reported with smaller sample size (typically <1,000 sibling pairs), and a number of loci, notably those on chromosome 9 and on the X chromosome, appear to be “replicated.”14,24–27 There is no evidence of genetic variation associated with chromosome 9 or with the X chromosome in our study. The study by Liu et al.,13 who reported significant linkage results on chromosomes 9 and X, was based on large extended pedigrees, whereas, in our study, we have extracted the linkage information from only the full-sibling pairs from that sample. Large pedigrees contain many more contrasts between relatives and therefore have more power to detect a QTL than does an analysis based on the full-sib pairs only. Statistical replication in linkage studies for complex traits is problematic because of the imprecision with which loci are mapped.36 Most of the reported studies, apart from that by Liu et al.,13 are characterized by small sample size and by analysis of data for males and females separately, thereby effectively creating even smaller samples. There is some evidence of a sex-by-genotype interaction for height in humans,37,38 but the additive genetic correlation across the sexes is ∼0.8–0.9,38 so we would expect that most trait loci have similar effects in males and females. From the brother-brother, sister-sister, and brother-sister correlations in our data—0.494, 0.479, and 0.435, respectively—and with the assumption of a pure additive model of family resemblance, we estimate a genetic correlation coefficient across the sexes of 0.435/√(0.494×0.479)=0.894.
Our results have implications for GWA studies for height and other complex traits, including disease. GWA studies are typically powered to detect loci that explain at least 0.5%–1% of the phenotypic variation. If the total genetic variance explained per chromosome is ∼5%, this puts an upper bound to the effect sizes that can be detected. Many complex traits, including most diseases, have lower heritabilities than that of height, typically 30%–50%; so, if the genetic variance for these complex traits is distributed over all chromosomes, individual chromosomes will explain only of the order of 1%–3% of the phenotypic variation. Further, if the partitioning of variation across chromosomes, implying many trait loci, can be extrapolated to the partitioning of variation on an individual chromosome (for which we currently have no evidence), then the effect sizes at individual loci or over small intervals may become too small to be detected. In the near future, we will be able to test these hypotheses for quantitative traits, using GWA studies that are currently being conducted. From our study, we predict that, if GWA studies of height are successful in locating significant loci, then the associated variants are likely to be on chromosomes 3, 4, 8, 15, 17, and 18.
The main limitation of our study, as in most studies of the genetics of complex traits in humans, is sample size. Despite having a sample of >11,000 sibling pairs with genomewide marker data and a highly heritable phenotype, we have insufficient power to estimate precisely the contribution of each chromosome to genetic variance and to estimate nonadditive genetic variance. However, since height is a phenotype that is easy to measure and is collected in many cohort studies, and since researchers are willing to collaborate to increase sample size, it should be feasible in the near future to further dissect quantitative genetic variation for height by linkage and/or by association with the use of sample sizes of tens of thousands of individuals.
In conclusion, with a large sample size of 11,214 sibling pairs, we estimated how genetic variance is apportioned in the genome. The hypothesis that chromosomes explain additive genetic variance in proportion to their length could not be rejected in our data. Despite the recent suggestion that variation due to epistasis is too-often neglected in complex-trait studies,39 we found no evidence of any nonadditive genetic variance for height, the complex trait we studied. Our results imply, at least for the quantitative trait height in humans, that genetic variation can be explained by many loci distributed over all the autosomes with an additive mode of gene action.
Acknowledgments
Australian-based authors P.M.V., S. Macgregor, B.B., G.Z., S.G., S. Medland, G.W.M., and N.G.M. thank the twins and their families, for their participation; Marlene Grace, Ann Eldridge, and Dixie Statham, for collection of data; Anjali Henders and Megan Campbell, for managing sample processing; and David Smyth and Harry Beeby, for information technology support. Genome scans of Australian data were supported by Australian National Health and Medical Research Council (NHMRC) Program in Medical Genomics grant 219178, by Center for Inherited Disease Research at Johns Hopkins University grant N01-HG-65403 (to Dr. Jeff Trent), and by Mammalian Genotyping Service (Marshfield, WI; director Dr. James Weber) grants (to Drs. Daniel T. O’Connor, David Duffy, Patrick Sullivan, and Dale Nyholt; Drs. Eline Slagboom and Bas Heijmans in Leiden, The Netherlands; and Drs. Peter Reed and Jeff Hall). This research was supported in part by National Institute on Alcohol Abuse and Alcoholism grants AA007535, AA013320, AA013326, AA014041, AA07728, AA10249, and AA11998 and by NHMRC grants 941177, 951023, 950998, 981339, 241916, 941944, 389892, and 443036.
U.S.-based authors Y.-Z.L. and H.-W.D. were partially supported by National Institutes of Health grants K01 AR02170-01, R01 AR45349-01, and R01 GM60402-01A1 and by State of Nebraska grant LB595. The study also benefited from grants from National Science Foundation of China, Huo Ying Dong Education Foundation, HuNan Province, Xi’an Jiaotong University, and the Ministry of Education of China. The genotyping experiments were performed by Marshfield Center for Medical Genetics and were supported by National Heart, Lung, and Blood Institute Mammalian Genotyping Service contract number HV48141.
Netherlands-based authors J.-J.H., G.W., and D.I.B. acknowledge the following grants: Genetic basis of anxiety and depression (Netherlands Organization for Scientific Research [NWO] 904-61-090), Database Twin register (NWO 575-25-006), Genetics of individual differences in smoking (NWO 985-10-002), Resolving cause and effect in the association between regular exercise and psychological well-being (NWO-MW 904-61-193), Spinozapremie (NWO/Spinoza 56-464-14192), Centre Neurogenomics and Cognition Research–VU, Center Medical Systems Biology (NWO Genomics), Twin-family database for behavior genetics and genomics (NWO 480-04-004), and Genome-wide analyses of European twin and population cohorts to identify genes predisposing to common diseases (European Union/QLRT-2001-01254).
Web Resource
The URL for data presented herein is as follows:
References
- 1.The Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al (2006) A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314:1461–1463 10.1126/science.1135245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al (2007) A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 39:207–211 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- 4.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, et al (2006) A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38:617–619 10.1038/ng1800 [DOI] [PubMed] [Google Scholar]
- 5.Barton NH, Keightley PD (2002) Understanding quantitative genetic variation. Nat Rev Genet 3:11–21 10.1038/nrg700 [DOI] [PubMed] [Google Scholar]
- 6.Glazier AM, Nadeau JH, Aitman TJ (2002) Finding genes that underlie complex traits. Science 298:2345–2349 10.1126/science.1076641 [DOI] [PubMed] [Google Scholar]
- 7.Korstanje R, Paigen B (2002) From QTL to gene: the harvest begins. Nat Genet 31:235–236 10.1038/ng0702-235 [DOI] [PubMed] [Google Scholar]
- 8.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH (2004) Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305:869–872 10.1126/science.1099870 [DOI] [PubMed] [Google Scholar]
- 9.Visscher PM, Haley CS (1996) Detection of putative quantitative trait loci in line crosses under infinitesimal genetic models. Theor Appl Genet 93:691–702 10.1007/BF00224064 [DOI] [PubMed] [Google Scholar]
- 10.Visscher PM, Medland SE, Ferreira MA, Morley KI, Zhu G, Cornes BK, Montgomery GW, Martin NG (2006) Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet 2:e41 10.1371/journal.pgen.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schork NJ (2001) Genome partitioning and whole-genome analysis. Adv Genet 42:299–322 [DOI] [PubMed] [Google Scholar]
- 12.Galton F (1886) Hereditary stature. Nature 33:295–298 [Google Scholar]
- 13.Liu YZ, Xiao P, Guo YF, Xiong DH, Zhao LJ, Shen H, Liu YJ, Dvornyk V, Long JR, Deng HY, et al (2006) Genetic linkage of human height is confirmed to 9q22 and Xq24. Hum Genet 119:295–304 10.1007/s00439-006-0136-y [DOI] [PubMed] [Google Scholar]
- 14.Willemsen G, Boomsma DI, Beem AL, Vink JM, Slagboom PE, Posthuma D (2004) QTLs for height: results of a full genome scan in Dutch sibling pairs. Eur J Hum Genet 12:820–828 10.1038/sj.ejhg.5201229 [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Evans DM, Duffy DL, Montgomery GW, Medland SE, Gillespie NA, Ewen KR, Jewell M, Liew YW, Hayward NK, et al (2004) A genome scan for eye color in 502 twin families: most variation is due to a QTL on chromosome 15q. Twin Res 7:197–210 10.1375/136905204323016186 [DOI] [PubMed] [Google Scholar]
- 16.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 17.Kong X, Murphy K, Raj T, He C, White PS, Matise TC (2004) A combined linkage-physical map of the human genome. Am J Hum Genet 75:1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagnon A, Beise J, Vaupel JW (2005) Genome-wide identity-by-descent sharing among CEPH siblings. Genet Epidemiol 29:215–224 10.1002/gepi.20090 [DOI] [PubMed] [Google Scholar]
- 19.Neale MC, Boker SM, Xie G, Maes HH (2003) Mx: statistical modeling. Virginia Institute for Psychiatric and Behavioral Genetics, Richmond [Google Scholar]
- 20.Kent JW Jr, Dyer TD, Blangero J (2005) Estimating the additive genetic effect of the X chromosome. Genet Epidemiol 29:377–388 10.1002/gepi.20093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 22.Dempfle A, Wudy SA, Saar K, Hagemann S, Friedel S, Scherag A, Berthold LD, Alzen G, Gortner L, Blum WF, et al (2006) Evidence for involvement of the vitamin D receptor gene in idiopathic short stature via a genome-wide linkage study and subsequent association studies. Hum Mol Genet 15:2772–2783 10.1093/hmg/ddl218 [DOI] [PubMed] [Google Scholar]
- 23.Hirschhorn JN, Lindgren CM, Daly MJ, Kirby A, Schaffner SF, Burtt NP, Altshuler D, Parker A, Rioux JD, Platko J, et al (2001) Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am J Hum Genet 69:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay N, Finegold DN, Larson MG, Cupples LA, Myers RH, Weeks DE (2003) A genome-wide scan for loci affecting normal adult height in the Framingham Heart Study. Hum Hered 55:191–201 10.1159/000073203 [DOI] [PubMed] [Google Scholar]
- 25.Perola M, Ohman M, Hiekkalinna T, Leppavuori J, Pajukanta P, Wessman M, Koskenvuo M, Palotie A, Lange K, Kaprio J, et al (2001) Quantitative-trait-locus analysis of body-mass index and of stature, by combined analysis of genome scans of five Finnish study groups. Am J Hum Genet 69:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sale MM, Freedman BI, Hicks PJ, Williams AH, Langefeld CD, Gallagher CJ, Bowden DW, Rich SS (2005) Loci contributing to adult height and body mass index in African American families ascertained for type 2 diabetes. Ann Hum Genet 69:517–527 10.1046/j.1529-8817.2005.00176.x [DOI] [PubMed] [Google Scholar]
- 27.Sammalisto S, Hiekkalinna T, Suviolahti E, Sood K, Metzidis A, Pajukanta P, Lilja HE, Soro-Paavonen A, Taskinen MR, Tuomi T, et al (2005) A male-specific quantitative trait locus on 1p21 controlling human stature. J Med Genet 42:932–939 10.1136/jmg.2005.031278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shmulewitz D, Heath SC, Blundell ML, Han Z, Sharma R, Salit J, Auerbach SB, Signorini S, Breslow JL, Stoffel M, et al (2006) Linkage analysis of quantitative traits for obesity, diabetes, hypertension, and dyslipidemia on the island of Kosrae, Federated States of Micronesia. Proc Natl Acad Sci USA 103:3502–3509 10.1073/pnas.0510156103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiltshire S, Frayling TM, Hattersley AT, Hitman GA, Walker M, Levy JC, O’Rahilly S, Groves CJ, Menzel S, Cardon LR, et al (2002) Evidence for linkage of stature to chromosome 3p26 in a large U.K. family data set ascertained for type 2 diabetes. Am J Hum Genet 70:543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Bleecker ER, Jongepier H, Howard TD, Koppelman GH, Postma DS, Meyers DA (2002) Major recessive gene(s) with considerable residual polygenic effect regulating adult height: confirmation of genomewide scan results for chromosomes 6, 9, and 12. Am J Hum Genet 71:646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies RW, Workman PL (1971) The genetic relationship to two quantitative characters in Drosophila melanogaster. I. Responses to selection and whole chromosome analysis. Genetics 69:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison BJ, Mather K (1950) Polygenic variability in chromosomes of Drosophila melanogaster obtained from the wild. Heredity 4:295–312 [DOI] [PubMed] [Google Scholar]
- 33.Visscher PM, Haley CS (1998) Power of a chromosomal test to detect genetic variation using genetic markers. Heredity 81:317–326 10.1046/j.1365-2540.1998.00398.x [DOI] [Google Scholar]
- 34.Goldgar DE (1990) Multipoint analysis of human quantitative genetic variation. Am J Hum Genet 47:957–967 [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149–150 10.1093/bioinformatics/19.1.149 [DOI] [PubMed] [Google Scholar]
- 36.Visscher PM, Goddard ME (2004) Prediction of the confidence interval of quantitative trait loci location. Behav Genet 34:477–482. 10.1023/B:BEGE.0000023652.93162.e8 [DOI] [PubMed] [Google Scholar]
- 37.Silventoinen K (2003) Determinants of variation in adult body height. J Biosoc Sci 35:263–285 10.1017/S0021932003002633 [DOI] [PubMed] [Google Scholar]
- 38.Silventoinen K, Sammalisto S, Perola M, Boomsma DI, Cornes BK, Davis C, Dunkel L, De Lange M, Harris JR, Hjelmborg JV, et al (2003) Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res 6:399–408 10.1375/136905203770326402 [DOI] [PubMed] [Google Scholar]
- 39.Carlborg O, Haley CS (2004) Epistasis: too often neglected in complex trait studies? Nat Rev Genet 5:618–625 10.1038/nrg1407 [DOI] [PubMed] [Google Scholar]