Abstract
We report mutations in the gene for topoisomerase I–binding RS protein (TOPORS) in patients with autosomal dominant retinitis pigmentosa (adRP) linked to chromosome 9p21.1 (locus RP31). A positional-cloning approach, together with the use of bioinformatics, identified TOPORS (comprising three exons and encoding a protein of 1,045 aa) as the gene responsible for adRP. Mutations that include an insertion and a deletion have been identified in two adRP-affected families—one French Canadian and one German family, respectively. Interestingly, a distinct phenotype is noted at the earlier stages of the disease, with an unusual perivascular cuff of retinal pigment epithelium atrophy, which was found surrounding the superior and inferior arcades in the retina. TOPORS is a RING domain–containing E3 ubiquitin ligase and localizes in the nucleus in speckled loci that are associated with promyelocytic leukemia bodies. The ubiquitous nature of TOPORS expression and a lack of mutant protein in patients are highly suggestive of haploinsufficiency, rather than a dominant negative effect, as the molecular mechanism of the disease and make rescue of the clinical phenotype amenable to somatic gene therapy.
Retinitis pigmentosa (RP) is a clinically and genetically heterogeneous disorder with an incidence of 1 in 3,500, or a total of 1.8 million people affected worldwide. Affected individuals experience a progressive degeneration of the photoreceptors, which eventually results in severe visual impairment. The mode of inheritance of RP can be autosomal dominant (adRP), autosomal recessive, X linked, or digenic.1 To date, at least 16 causative genes have been identified for adRP (RetNet) (MIM numbers 114760, 602225, 607643, 602275, 146690, 604485, 162080, 607301, 607300, 606419, 179605, 180380, 180721, 603937, 607331, and 607292), and the products of these genes are associated with photoreceptor structure; cellular function, including the phototransduction cascade; or gene expression, including transcription and mRNA splicing.2
Recently, we reported a new locus for adRP (RP31) in a three-generation family based in the province of Quebec, Canada.3 After exclusion of all known loci for adRP, a genomewide search established positive linkage to a marker from the short arm of chromosome 9 (LOD score of 6.3 at recombination fraction q=0). The linked region is flanked by markers D9S157 and AFMe153td9 on 9p22-p21.1, corresponding to a physical distance of 15 Mb.3
Since the previous report,3 17 members with RP from this family, ranging in age from 8 to 64 years, were clinically reexamined at the Montreal Children’s Hospital Research Institute. Age at onset of symptoms ranged from 10 to 50 years and differed between the generations, and three affected patients were found to be asymptomatic. Visual acuities (range 20/20 to finger counting) were maintained in most patients, since 16 of 17 had better than 20/40 acuity, and 11 of 17 had 20/20 acuity at the most recent visit. Visual-field sizes ranged from 10° to 80°, and electroretinography (ERG) abnormalities were highly variable as well, with early rod dysfunction followed by cone defects. The earliest sign of disease (found in four children) was an unusual perivascular cuff of retinal pigment epithelium (RPE) atrophy, which was found surrounding the superior and inferior arcades (fig. 1). This progressed to a diffuse pigmentary retinopathy with choroidal sclerosis. Three patients with the disease haplotype were asymptomatic and had completely normal retinal appearance; nonetheless, their ERG abnormalities were similar to those in the symptomatic patients. This phenotype differs significantly from the clinical diagnosis associated with the other 16 published adRP gene defects. A detailed description of the phenotype will be published elsewhere.
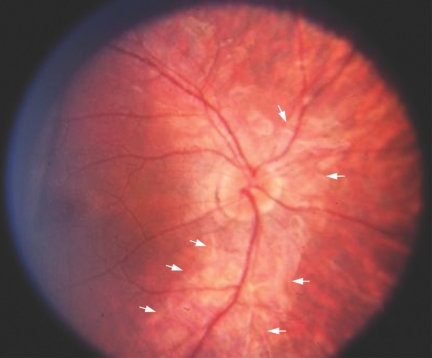
Figure 1. .
Color photograph of the right eye of a 10-year-old affected child (IV:3). At this age, a very obvious and unusual perivascular “cuff” of atrophy (arrows) around the superior and inferior vascular arcades is visible. The optic disc appears normal in color, although the retinal arterioles are narrow, and there was no pigmentary degeneration at this stage. The cuff of RPE atrophy was found in three other children in this family as well but was not found in the adults of this pedigree. At a later age, this feature apparently disappears.
Linkage analysis spanning the disease locus on chromosome 9p21 refined the genetic interval to 14 Mb between markers IVS4-46G (NP_060395.3) and AFMa153td9; this critical region contains 48 known and 5 novel (hypothetical) genes. The list of genes (in order from telomere to centromere of chromosome 9) that have been systematically screened includes NP_060395.3, RPS6, ASAH3L, SLC24A2, MllT3, KIAA1797, NP_001010915.1, IFNB1, IFNW1, IFNA21, IFNA4, IFNA14, IFNA10, IFNA17, IFNA5, KLHL9, IFNA6, NP_008831.2, IFNA2,IFNA8, IFNA1, Q8WTY6, NP_795372.1, MTAP, NSGX, NP_478102.1, CDKN2A, CDKN2B, DMRTA1, ELAVL2, TUSC1, NP_079104.2, PLAA, CCDC2, LRRC19, TEK,NP_065692.1, MOBKL2B, IFNK, NP_659442.2, NP_689783.1, Q5T776, ACO1, DDX58, TOPORS (MIM 609507), NDUFB6, TAF1L, and NP_997723.1 (Ensembl). All exons and their respective acceptor- and donor-splice sites were directly analyzed from the PCR products with the use of ABI BigDye terminator cycle sequencing kit v3.1 (Applied Biosystems) on an ABI 3100 Genetic Analyzer. Most of the genes encode proteins with well-documented functions. From this list of 53 genes, 2 were prioritized for immediate screening because of their specific expression and known retinal function: the gene encoding retinal cone Na-Ca+K-exchanger (SLC24A2 [MIM 609838]), which is abundantly expressed in cone photoreceptors and retinal ganglion cells,4 and the gene encoding embryonic-lethal abnormal visual RNA-binding protein involved in growth, differentiation, and posttranscriptional gene expression (ELAVL2 [MIM 601673]), which plays an important role in RNA processing.5 No disease-causing mutations were identified in these two genes. We then continued with our systematic mutation screening of all expressed genes from the candidate region. Several of the genes contained sequence changes that were also identified in healthy unrelated controls, confirming that the variant is benign.
Sequence analysis of one of the genes, TOPORS (MIM 609923, NM_005802), a 13-kb gene with three exons,6 revealed a heterozygous 1-bp insertion (c.2474_2475insA) in exon 3; this frameshift mutation segregates with the affected status and is predicted to result in a premature stop codon (p.Tyr825fs) (fig. 2A and 2B). We also identified a second TOPORS mutation, a 2-bp deletion (c.2552_2553delGA) in exon 3 leading to p.Arg851fs, in a small German family (fig. 2C and 2D). We did not detect these changes in a control panel of 200 individuals of white origin. Primer sequences for mutation analysis of all three exons of TOPORS are listed in table 1. The presence of two distinct heterozygous frameshift mutations in two independent families strongly supports the argument that mutations in TOPORS are responsible for RP31. Sequencing of the coding regions of all other genes in the RP31 critical region revealed only nonpathogenic sequence variants. The second TOPORS mutation was identified as a result of our screening of a panel of 65 unrelated German patients with adRP attending the Retina Clinic at University Eye Hospital, Tubingen. On the basis of these preliminary findings, it is likely that TOPORS mutations are a rare cause of retinal degeneration.
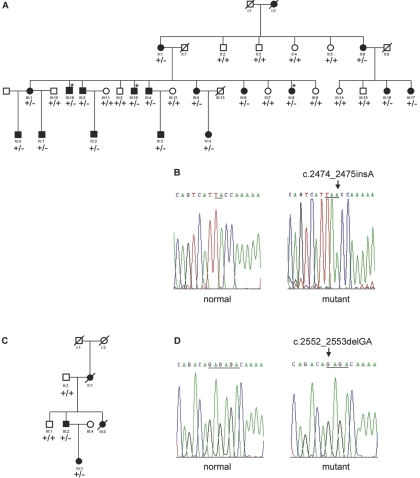
Figure 2. .
Pedigree structure and sequence analysis of TOPORS mutations. A and C, Canadian and German adRP-affected pedigrees, respectively, used in this study. Affected individuals are shown by blackened symbols, unaffected individuals are identified by unblackened symbols, and deceased individuals are indicated by a slash (/). Asymptomatic individuals are marked by an asterisk (*). Mutation segregation is shown on the pedigree as +/+ (normal) or +/− (affected). B, Electropherogram of the heterozygous mutation, c.2474_2475insA (p.Tyr825fs), in exon 3 of the TOPORS gene found in the Canadian family. D, Electropherogram of the heterozygous mutation found in the German family, c.2552_2553delGA (p.Arg851fs). Both mutations were identified after cloning of the amplified PCR product into pGEMT-Easy vector (Promega).
Table 1. .
Primers Used for PCR Amplification of TOPORS Exons
| Primer Sequence (5′→3′) |
||||
| Exon | Product Size (bp) |
Forward | Reverse | Temperature (C°) |
| 1 | 628 | CGTCAGGTTACCGTTGCC | ATATTGCATTGCAACTAG | 57 |
| 2 | 616 | GGCTGAGTACCAGTAAC | CACCAATCCACAGGTGCG | 61 |
| 3a | 847 | AGTAATGGGTCACTTAAG | AAGAACTGTAAGTTCACG | 50 |
| 3b | 901 | ATTCCTCAGTTTATGAGAC | TATCACCAGAACTGTAAG | 48 |
| 3c | 864 | CTTCTGACAGTTCAGATG | GAGCTCTGGACAGAGTCC | 56 |
| 3d | 914 | TAGCAGTTGGTCCAGAAG | TTGTAACTACATCTTTAG | 57 |
| 3e | 997 | CAGATCAAGGAGCCTGTCTAG | TAAGCTGCTAGCAGTATC | 60 |
The TOPORS gene encodes a major transcript of 3.1 kb, which produces a multidomain protein of 1,045 aa with an N-terminal RING-type zinc-finger domain (amino acids 103–141); five stretches enriched in proline, glutamic acid, serine, and threonine (PEST) residues, which are frequently characteristic of rapidly degraded proteins7; a bipartite nuclear localization signal; and a region rich in Arg-Ser (RS)8 (fig. 3A). RT-PCR analysis revealed a broad tissue expression of TOPORS in human tissues, including the retina (fig. 3B).
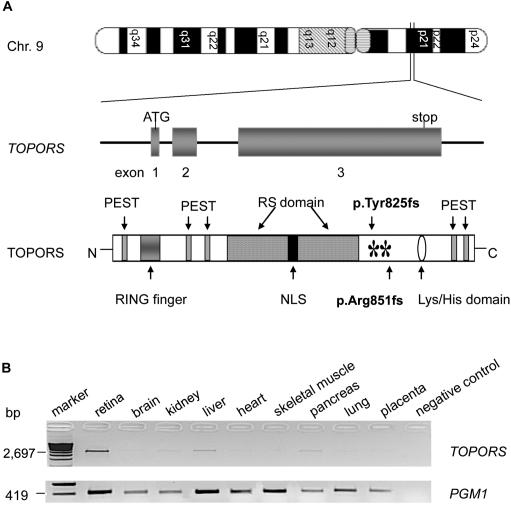
Figure 3. .
Chromosomal location, domain organization, and expression of TOPORS. A, Schematic representation of exon-intron structure of TOPORS and domain structure of its protein, including the RING-type zinc-finger domain, RS domain, nuclear localization signal (NLS), lysine-histidine domain, PEST region, N-amino-terminal (N), and carboxyl-terminal (C). Both mutations (p.Tyr825fs and p.Arg851fs) are shown with asterisks (*). B, RT-PCR analysis of TOPORS transcript in human tissues: PCR was performed with primers selected from exon 3 (with 28 cycles). A band of 2.69 kb was observed in all the tested QUICK-Clone cDNAs (Clontech) from retina, brain, kidney, liver, heart, skeletal muscle, pancreas, lung, and placenta. A ubiquitously expressed gene, PGM1, was used as a control.
To examine the effect of disease-associated mutations, we performed immunoblotting of whole lymphoblastoid cell lysates from both families in affected and unaffected individuals, using an N-terminus mouse monoclonal TOPORS antibody (Abnova). The full-length TOPORS-immunoreactive band of 150 kDa is present in the lymphocytes of control and affected individuals; however, the mutations are predicted to delete 221 aa (∼30 kDa) and 175 aa (∼25 kDa) from the C-terminus of the protein in the French Canadian and German families, respectively. As a result, the truncated protein is expected to have a theoretical molecular weight of 120–125 kDa. Our analysis did not detect the predicted truncated TOPORS-immunoreactive bands in patient lymphocytes, indicating that the mutant protein is relatively unstable (fig. 4). The absence of the truncated protein in patients from both families suggests haploinsufficiency, rather than a dominant negative effect, as the molecular basis of the disease.
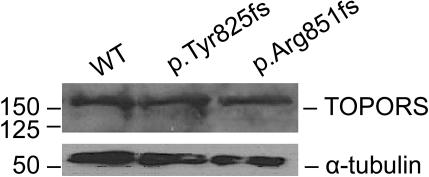
Figure 4. .
TOPORS expression in patient lymphoblastoid cell lines. Protein extracts from cell lines from one unaffected and two affected individuals from the Canadian and German families with p.Tyr825fs and p.Arg851fs mutations, respectively, were analyzed by SDS-PAGE and immunoblotting, with the use of anti-TOPORS antibody (Abnova). α-tubulin was used as a control. WT = wild type.
TOPORS was identified initially in a screen of proteins that bind to the N-terminus of topoisomerase I and then as a protein that interacts with p53.6,9 Like other RING domain–containing proteins, TOPORS is an E3 ubiquitin ligase with specific E2 enzymes, and it can ubiquitinate p53.10 It also undergoes modification by the small ubiquitin-like modifier SUMO-1.11 It is located in nuclear speckles that closely associate with promyelocytic leukemia nuclear bodies—nuclear compartments that have been implicated in transcription, DNA repair, viral defense, stress, cell-cycle regulation, proteolysis, and apoptosis.12,13 In Drosophila, Topors (dTopors) ubiquitinates and regulates the activity of the transcriptional repressor Hairy.14 It also interacts with proteins of the gypsy insulator protein complex, where its influence on insulator activity has been shown to depend on its ubiquitin ligase activity.15
TOPORS contains an RS domain, which is implicated in pre-mRNA splicing. Mutations in three other proteins involved in mRNA splicing (PRPF3, PRPC8, and PRPF31) are also associated with RP.16–18 By binding to specific sequences on pre-mRNA and interacting with other splicing factors via their RS domain, SR proteins mediate different intraspliceosomal contacts, thereby helping in splice-site selection and spliceosome assembly. Binding of SR proteins to exonic (intronic) splicing enhancers (silencers) helps in recruitment of U1 snRNP to the 5′ splice site and U2 snRNP to the branch point sequence.19 However, TOPORS nuclear speckles do not colocalize with nuclear splicing speckles that contain other SR proteins—for example, SC35.11 A recent genomewide survey revealed a large complexity of RS domain–containing proteins in metazoans with functions not only in pre-mRNA splicing but also in chromatin remodeling, transcription by RNA polymerase II, and cell-cycle progression. We therefore predict that TOPORS not only may be involved in mRNA processing but also may have a more complex array of functions in mammalian cells.20
It is possible that the process underlying RP31 pathogenesis might be distinct from those operating in other forms of adRP where mutations have previously been identified in structural proteins and in transcription and splicing factors. Elucidation of its function in retinal photoreceptors and, more important, studies of its expression in photoreceptors and protein interactions in retina may provide new insight into the molecular basis of retinal degenerations.
Finally, as stated above, the lack of mutant TOPORS protein in patients in the families reported here is highly suggestive of haploinsufficiency as the molecular basis of the disease and therefore makes rescue of the disease phenotype amenable to somatic gene therapy. Should haploinsufficiency explain the disease mechanism, targeted increase in the level of the wild-type protein in photoreceptor cells of patients should lead to rescue of the disease phenotype. With use of a viral vector (adeno-associated virus or lentivirus)–mediated gene-delivery system and subretinal injection, a functional copy of the gene can be delivered under the control of photoreceptor-specific promoters to achieve targeted expression in the appropriate cell type. Somatic-gene-therapy rescue has been achieved in several animal models of retinal degeneration, including the rds mouse, and may also prove to be a successful approach for this type of adRP.21,22
Acknowledgments
We thank the patients and their families for participating in this study. We thank C. Murga-Zamalloa and Beverly Scott for technical assistance. This work was supported by grants from The Foundation Fighting Blindness (to S.S.B.), European Union grants EVI-GENORET LSHG-CT-2005-512036 (to S.S.B., V.M., A.G., and C.P.H.) and RETNET MRTN-CT-2003-504003 (to S.S.B. and V.M.), a grant from Special Trustees of Moorfields Eye Hospital (to S.S.B.), National Institutes of Health grants EY007961 and EY007003 (to A.S.), Biotechnology and Biological Sciences Research Council grant BB/C514458/1 (to K.M.), Deutsche Forschungsgemeinschaft grant KFO134 (to B.W. and E.Z.), and grants from Research to Prevent Blindness (to R.K.K.), George M. O’Brien Kidney Research Foundation (to R.K.K.), Foundation Fighting Blindness-Canada (to R.K.K.), and Fonds de la Recherche en Santee du Quebec (to R.K.K.).
Web Resources
The URLs for data presented herein are as follows:
- Ensembl Human Genome Browser, http://www.ensembl.org/ (for marker and gene positions)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for 16 adRP genes, RP31, TOPORS, SLC24A2, and ELAVL2
- RetNet, http://www.sph.uth.tmc.edu/Retnet/
References
- 1.Rivolta C, Sharon D, DeAngelis MM, Dryja TP (2002) Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum Mol Genet 11:1219–1227 10.1093/hmg/11.10.1219 [DOI] [PubMed] [Google Scholar]
- 2.Hims MM, Diager SP, Inglehearn CF (2003) Retinitis pigmentosa: genes, proteins and prospects. Dev Ophthalmol 37:109–125 [DOI] [PubMed] [Google Scholar]
- 3.Papaioannou M, Chakarova CF, Prescott Q, Waseem N, Theis T, Lopez I, Gill B, Koenekoop RK, Bhattacharya SS (2005) A new locus (RP31) for autosomal dominant retinitis pigmentosa maps to chromosome 9p. Hum Genet 118:501–503 10.1007/s00439-005-0063-3 [DOI] [PubMed] [Google Scholar]
- 4.Sharon D, Yamamoto H, McGee TL, Rabe V, Szerencsei RT, Winkfein RJ, Prinsen CF, Barnes CS, Andreasson S, Fishman GA, et al (2002) Mutated alleles of the rod and cone Na-Ca+K-exchanger genes in patients with retinal diseases. Invest Ophthalmol Vis Sci 43:1971–1979 [PubMed] [Google Scholar]
- 5.Antic D, Keene JD (1997) Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet 61:273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haluska P, Saleem A, Rasheed Z, Ahmed F, Su EW, Liu LF, Rubin EH (1999) Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res 27:2538–2544 10.1093/nar/27.12.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weger S, Hammer E, Heilbronn R (2002) Topors, a p53 and topoisomerase I binding protein, interacts with the adeno-associated virus (AAV-2) Rep78/68 proteins and enhances AAV-2 gene expression. J Gen Virol 83:511–516 [DOI] [PubMed] [Google Scholar]
- 8.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96:11364–11369 10.1073/pnas.96.20.11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou R, Wen H, Ao SZ (1999) Identification of a novel gene encoding a p53-associated protein. Gene 235:93–101 10.1016/S0378-1119(99)00203-6 [DOI] [PubMed] [Google Scholar]
- 10.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH (2004) Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem 279:36440–36444 10.1074/jbc.C400300200 [DOI] [PubMed] [Google Scholar]
- 11.Weger S, Hammer E, Engstler M (2003) The DNA topoisomerase I binding protein topors as a novel cellular target for SUMO-1 modification: characterization of domains necessary for subcellular localization and sumolation. Exp Cell Res 290:13–27 10.1016/S0014-4827(03)00292-1 [DOI] [PubMed] [Google Scholar]
- 12.Rasheed ZA, Saleem A, Ravee Y, Pandolfi PP, Rubin EH (2002) The topoisomerase I-binding RING protein, topors, is associated with promyelocytic leukemia nuclear bodies. Exp Cell Res 277:152–160 10.1006/excr.2002.5550 [DOI] [PubMed] [Google Scholar]
- 13.Borden KL (2002) Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol 22:5259–5269 10.1128/MCB.22.15.5259-5269.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Secombe J, Parkhurst SM (2004) Drosophila Topors is a RING finger-containing protein that functions as a ubiquitin-protein isopeptide ligase for the hairy basic helix-loop-helix repressor protein. J Biol Chem 279:17126–17133 10.1074/jbc.M310097200 [DOI] [PubMed] [Google Scholar]
- 15.Capelson M, Corces VG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20:105–116 10.1016/j.molcel.2005.08.031 [DOI] [PubMed] [Google Scholar]
- 16.Chakarova CF, Hims MM, Bolz H, Abu-Safieh L, Patel RJ, Papaioannou MG, Inglehearn CF, Keen TJ, Willis C, Moore AT, et al (2002) Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet 11:87–92 10.1093/hmg/11.1.87 [DOI] [PubMed] [Google Scholar]
- 17.McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, et al (2001) Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 10:1555–1562 10.1093/hmg/10.15.1555 [DOI] [PubMed] [Google Scholar]
- 18.Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, et al (2001) A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 8:375–381 10.1016/S1097-2765(01)00305-7 [DOI] [PubMed] [Google Scholar]
- 19.Lorkovic ZJ, Lopato S, Pexa M, Lehner R, Barta A (2004) Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA splicing. J Biol Chem 279:33890–33898 10.1074/jbc.M400270200 [DOI] [PubMed] [Google Scholar]
- 20.Cazalla D, Newton K, Caceres JF (2005) A novel SR-related protein is required for the second step of pre-mRNA splicing. Mol Cell Biol 25:2969–2980 10.1128/MCB.25.8.2969-2980.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali RR, Sarra GM, Stephens C, Alwis MD, Bainbridge JW, Munro PM, Fauser S, Reichel MB, Kinnon C, Hunt DM, et al (2000) Restoration of photoreceptor ultrastructure and function in retinal degeneration slow mice by gene therapy. Nat Genet 25:306–310 10.1038/77068 [DOI] [PubMed] [Google Scholar]
- 22.Bainbridge JW, Tan MH, Ali RR (2006) Gene therapy progress and prospects: the eye. Gene Ther 13:1191–1197 10.1038/sj.gt.3302812 [DOI] [PubMed] [Google Scholar]