Abstract
Congenital heart defects (CHDs) are among the most common birth defects in humans (incidence 8–10 per 1,000 live births). Although their etiology is often poorly understood, most are considered to arise from multifactorial influences, including environmental and genetic components, as well as from less common syndromic forms. We hypothesized that disturbances in left-right patterning could contribute to the pathogenesis of selected cardiac defects by interfering with the extrinsic cues leading to the proper looping and vessel remodeling of the normally asymmetrically developed heart and vessels. Here, we show that heterozygous loss-of-function mutations in the human GDF1 gene contribute to cardiac defects ranging from tetralogy of Fallot to transposition of the great arteries and that decreased TGF-β signaling provides a framework for understanding their pathogenesis. These findings implicate perturbations of the TGF-β signaling pathway in the causation of a major subclass of human CHDs.
Congenital heart defects (CHDs) are a major cause of morbidity and mortality in human newborns. Recent advances in our understanding of the molecular basis of embryonic patterning of the developing heart hold great promise for elucidating the underlying mechanisms of some of these cardiac malformations.1,2 Conceptually, these CHDs can be caused by intrinsically defective patterning within the developing heart tissues or, alternatively, by mispatterning due to the failure of instructive extrinsic cues during embryonic patterning.3 The linear heart tube typically loops to the right in vertebrate embryos, and this represents the first overt sign of the previously established molecular cascade governing asymmetric expression of genes—such as Nodal (MIM 601265) and NODAL, Leftyb (LEFTY1 [MIM 603037] and LEFTY2 [MIM +601877]), and Pitx2 (PITX2 [MIM 601542])—in the left lateral plate mesoderm (reviewed in the work of Hamada et al.4). These asymmetric factors provide left-right (L-R) positional information to developing organs, such as the lungs, heart, gut, and brain. In addition, CHDs are commonly seen in the multiorgan pathological spectrum of human laterality defects,5 some of which can be explained by attenuated TGF-β signaling.6 Furthermore, TGF-β signals, such as Nodal and Gdf1, have been shown to be essential for the establishment and maintenance of the asymmetric cues affecting organ morphogenesis.4,7–9
TGF-β ligands function in diverse roles during development, including the induction of mesendoderm and determination of the body axes. GDF1 (MIM *602880)10,11 is structurally related to and functionally homologous to Xenopus Vg112,13 and is within the activin-like subclass of TGF-β signaling molecules (activin/Nodal/Vg1). Signal transduction by this subfamily culminates in receptor-mediated phosphorylation of receptor-regulated R-Smad2 (SMAD2 [MIM 601366]) or R-Smad3 (SMAD3 [MIM 603109]) and in the assembly of R-Smad4 (SMAD4 [MIM 600993]) complexes together with cotranscription factors, such as FoxH1 (FOXH1 [MIM 603621]), onto target genes.14,15
Murine Gdf1 is upstream of a cascade of left-sided determinants that participate in the establishment and maintenance of L-R signals governing asymmetric organogenesis, including the heart and great vessels.8,9 Pitx2 is one such asymmetrically expressed downstream target of this L-R cascade, and, in mice, loss of function of this bicoid-class transcription factor by gene targeting causes transposition of the great arteries (TGA, the most common form of which is “dextro-looped TGA” [MIM #608808]), double-outlet right ventricle (DORV), persistent truncus arteriosus, and atrial isomerism.16 These observations strengthen the hypothesis that L-R patterning signals can directly or indirectly affect cardiac development and might help to explain CHD manifestations in humans.
Gdf1 is synthesized as a preproprotein that is posttranslationally processed in a regulated fashion into its mature form, containing the classic TGF-β structure (fig. 1), by tissue-specific convertases.17 Absence of Gdf1 in the mouse is associated with disturbances in L-R patterning, including situs inversus, right pulmonary isomerism, TGA, ventricular and atrial defects, and isomerisms.8,9 Since Gdf1 requires the coreceptor Cfc1 to activate the membrane receptors ActRIIB and ActRI,18 and since mutations in human CFC1 (MIM *605194) are associated with laterality defects6 and isolated TGA,19 we considered GDF1 a likely candidate gene for laterality/cardiac malformations. Therefore, we set out to determine whether a similar spectrum of human birth defects could be attributed to human GDF1.
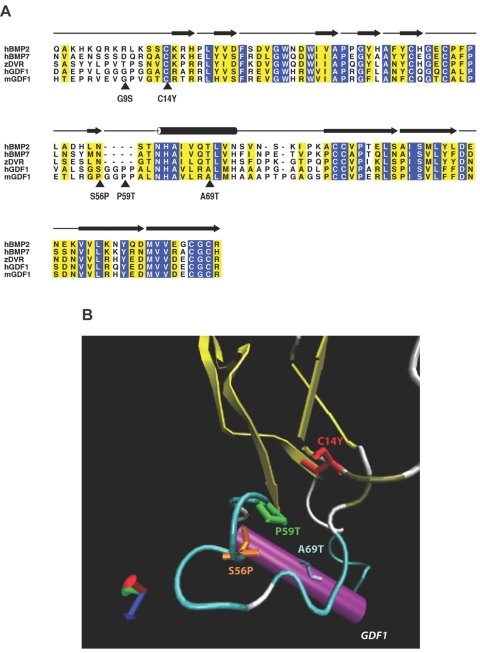
Figure 1. .
Structural analysis of GDF1 mutations. A, Alignment of human GDF1 (hGDF1) and structurally related members of the TGF-β family, including human BMP2 (hBMP2), human BMP7 (hBMP7), zebrafish DVR (zDVR), and murine Gdf1 (mGDF1). Amino acid residues showing absolute identity are shown with white letters against a blue background; those positions with conservative substitutions are shown against a yellow background. The positions of the five mutations considered in this study are indicated by arrowheads. The amino acid numbering is based on that of the mature processed ligand. The β-sheet elements of the winglike finger projections F1 and F2 are indicated by blackened arrows above the alignment, whereas the α-helical core is denoted by the cylinder above the alignment. B, Ribbon diagram of human GDF1, showing the position of the mutations clustered in the knuckle region of the monomer. (Note that the G9S variant lies within the poorly conserved N-terminal region of the protein where the crystal structure is unknown, precluding accurate modeling of this mutation.)
Material and Methods
Study Population
Our study population consisted of 375 unrelated individuals with a wide spectrum of congenital cardiovascular malformations, including TGA, tetralogy of Fallot (TOF), DORV, atrial septal defect (ASD), and interrupted aortic arch (IAA). Coded affected proband DNA samples were obtained from three centers (The Children’s Hospital of Philadelphia, Baylor College of Medicine, and Yale University School of Medicine) in accordance with the guidelines of the institutional review boards of each institution and with the approval of the National Human Genome Research Institute (NHGRI)/National Institutes of Health (NIH) Institutional Review Board (for subjects with cardiac malformations or holoprosencephaly [HPE]). Parents and siblings were not studied because of the preliminary nature of the research, because the links to patient identifiers were retained by the host institutions, and to maintain confidentiality. Additionally, we screened 198 unrelated individuals with HPE and 225 unrelated, unaffected controls (obtained as unrelated anonymous samples from the Coriell Institute for Medical Research: 125 individuals studied by denaturing high-pressure liquid chromatography [dHPLC] and 100 individuals by bidirectional DNA sequencing of the entire coding region of the gene). None of the disease-associated mutations detected in this study were observed in the remaining cardiac cohort (742 chromosomes), the HPE cohort (396 chromosomes), or the unaffected controls (450 chromosomes).
PCR Amplifications, Mutation Screening, and DNA Sequencing
The genomic organization of human GDF1 (GenBank accession number NM_001492) was characterized using nucleotide homology searches in the public database, with use of the BLASTN program. Oligo 4.1 was used to design primers: GDF1 exon 1, forward 5′-CCCTCAGCCCACTGGTCCC-3′ and reverse 5′-GGCCGAAGTTGCTAGTAGCCTGG-3′; exon 2a, forward 5′-AGCCCCAGCGTTCACCTTCCTCC-3′ and reverse 5′-CACCAGCAGCAGCGAGGCCTC-3′; and exon 2b, forward 5′-CCGCTTGGGCTCGCAACGC-3′ and reverse 5′-CCAAGGAGACCAGCGGAGCAGACC-3′. Amplification of genomic DNA was performed in a 30-μl reaction volume, with the use of 60–100 ng DNA template, 50 μM deoxynucleotide triphosphate, 0.25 μM of each primer, 3 μl of 10X PCR Amplification buffer (Invitrogen), 1.5 μl 10X Enhancer buffer (Invitrogen), and 0.3 μl Taq polymerase. All reactions were performed using a PTC-255 thermocycler (MJ Research). Typical PCR cycling parameters were 95°C for 4 min, followed by 30 cycles at 95°C, annealing at 62°C, extension at 72°C for 1 min, and a final extension step of 72°C for 5 min. One-half of the PCR product was used for dHPLC analysis (WAVE [Trangenomic]), and the remainder was retained for direct DNA sequencing.20 Amplicons displaying heterozygous profiles were purified using a High Pure PCR purification kit (Roche) and were bidirectionally sequenced using the BigDye version 3.1 terminator cycle sequencing kit in accordance with the manufacturer’s protocol (Applied Biosystems) on an ABI 3100 automated sequencer. Sequence variants likely to represent SNPs were also identified: L26L, A118V (rs4800063), V130I, V304V, and L259L. Commonly occurring duplications and deletions were 466-468dupGCG and 466-468delGCG, and common intervening sequence variations in intron 1 were IVS1+21G→A and IVS1+84G→A.
Site-Directed Mutagenesis and Construct Design
A chimeric construct of pCS2, BMP2 (Xenopus), and Gdf1 (P56S murine) was used in the zebrafish Gdf1 overexpression assay. Site-directed mutagenesis was performed, with minor modifications of the Transformer kit (Promega) by Transponics, to introduce the human missense changes onto the P56S murine Gdf1 protein backbone.
Structural Molecular Modeling
Molecular modeling for the TGF-β domain of GDF1 was generated using the MODELLER package,21 as implemented within Insight II (Accelrys). Modeling was based on the solved nuclear magnetic resonance structure of BMP7 (Protein Data Bank [PDB] 1LX5).22 The MODELLER program was run in fully automated mode with a high optimization level to construct a minimized three-dimensional model of the target sequence by satisfaction of spatial restraints extracted from the template PDB files. Three models were generated for each of the target sequences. The stereochemistry and geometry of the models were checked using PROCHECK.23 The model with the best parameters, as determined by PROCHECK, was used for building mutant models.
Zebrafish Gdf1 Overexpression Assays
Plasmids were linearized with SacII, and the sense strand was synthesized using the SP6 Message Machine kit (Ambion). Zebrafish embryos were dechorinated by pronase treatment and were injected between the one- and four-cell stages. For phenotypic analysis, the zebrafish embryos at 27 h were mounted in 2% methylcellulose and were photographed using a Zeiss M2Bio dissecting microscope. In situ hybridization was performed as described elsewhere with goosecoid (gsc) or no-tail (ntl) probes.24
Results
Analysis of the Mutational Spectrum of GDF1
We screened a cohort of 375 unrelated individuals affected with a wide spectrum of cardiovascular malformations, including TGA, DORV, TOF, and IAA. Eight different putative disease-causing mutations (table 1) were detected in only one proband per mutation (1 of 750 chromosomes) and neither in 450 unaffected control chromosomes nor in those of subjects with HPE (396 additional chromosomes). Hence, these sequence variants were highly correlated with the presence of cardiac malformations. Interestingly, we did not detect any deleterious changes among the HPE cases, consistent with our previous findings of the lack of CFC1 changes and the rarity of changes in CRIPTO in this population group.6,20
Table 1. .
Clinical and Molecular Findings in Patients with Cardiac Defects
| Patient | GDF1 Mutation | Laterality Defect(s) | Other Finding(s) |
| 1 | R68H | Atrioventricular canal with cleft mitral valve | Left superior vena cava to coronary sinus and coarctation of the aorta |
| 2 | G162D | TOF | … |
| 3 | C227X | TGA | … |
| 4 | G262S (G9S) | Ratelli type atrioventricular canal | Left–pulmonary artery stenosis, hypoplastic left lung and pulmonary veins, small secundum ASD, and trisomy 21 |
| 5 | C267Y (C14Y) | DORV | Left–pulmonary artery stenosis |
| 6 | S309P (S56P) | TOF | Ventricular septal defect, aortic root dilatation, and bicuspid stenotic pulmonary valve stenosis |
| 7 | P312T (P59T) | TOF | … |
| 8 | A318T (A69T) | TGA | … |
Two mutations, C227X and C267Y (also referred to as “C14Y”; see below), were considered likely loss-of-function mutations because of premature termination in the prodomain (C227X) or elimination of one of the six cysteines crucial for the formation of the cystine knot (fig. 1). Two additional mutations in the prodomain (R68H and G162D) could not be evaluated either structurally or functionally (table 1) (see below).
Since the crystal structure of BMP7 has been solved,22 we were able to perform a structural analysis of four of the missense changes in the TGF-β ligand domain (fig. 1A and 1B). This domain contains six cysteine residues that form three intramolecular disulfide bonds, resulting in what has been termed a “cystine-knot” conformation.25,26 A cysteine→tyrosine mutation at position 14 (C14Y) interferes with the ability of this network of disulfide bonds to form properly. The G9S variant lies within the poorly conserved N-terminal region, whose crystal structure is unknown. Interestingly, the S56P, P59T, and A69T mutations are clustered in a portion of the molecule called the “knuckle” region, which is implicated in dimerization and overall stability of the core of the monomer. No mutations have thus far been detected in the winglike finger projections that make hydrophobic contacts with the receptor heterodimer.
We set out to assess the functional effects of GDF1 mutations, using zebrafish as a model system. Previous studies have established the zebrafish as a powerful assay system in which to study the activity of TGF-β signaling components.6,18–20,27–31 A chimeric protein consisting of the mature region of murine Gdf1 and the Xenopus BMP2 prodomain has been shown elsewhere to induce mesodermal marker expression and patterning defects in zebrafish.18 Since P56 is the normal residue in the murine protein (and corresponds to the amino acid change in patient 6), we changed P56 to serine to humanize the murine GDF1. Mutant residues G9S, C14Y, S56P, P59T, and A69T were incorporated into this GDF1 derivative. We did not test the sixth mutation (C227X), since premature termination is not compatible with the production of an active ligand. Overexpression of Gdf1 led to a strong induction of the downstream targets ntl and gsc (figs. 2 and 3) and to developmental defects detected at later stages of development (fig. 4). Previous studies have established that the ectopic activation of Nodal/GDF1/activin signaling leads to the induction of downstream genes such as gsc and ntl and to defects in axis formation. Moreover, the extent of downstream gene expression reflects the activity of the pathways.6,18–20,27–34 In contrast, the mutants G9S, C14Y, S56P, P59T, and A69T were significantly attenuated in these activities (figs. 2–5). These results indicate that the human GDF1 mutations constitute hypomorphic or loss-of-function alleles.
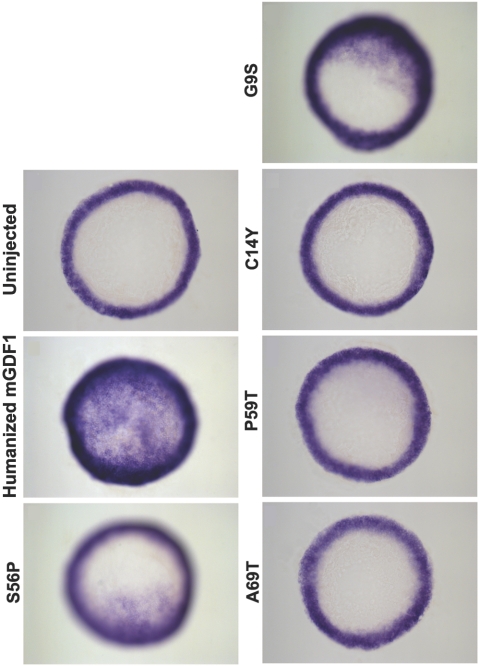
Figure 2. .
GDF1 activity assay by morphological phenotype. Twenty picograms of mRNA synthesized from humanized wild-type (WT) and mutant GDF1 constructs were injected at the one-cell stage. Embryos were allowed to develop for 27 h and were assayed for severity of developmental defects. Representative samples from injected clutches are shown. WT humanized GDF1 exhibited the severest developmental defects indicative of strongest activity. Mutant GDF1 constructs exhibited varying degrees of decreased activity compared with WT GDF1. Uninjected WT embryos are included for comparison. Twenty picograms of LacZ mRNA was injected as control and did not exhibit any phenotype (data not shown). mGDF1 = murine GDF1.
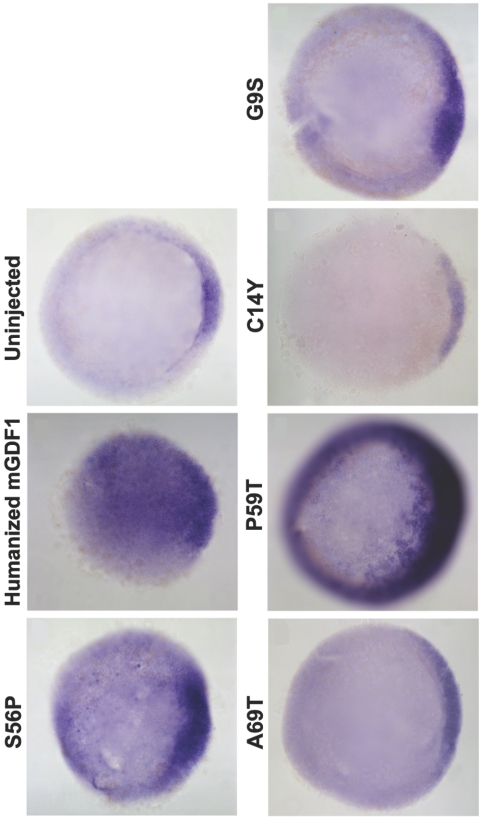
Figure 3. .
GDF1 activity assay by gsc induction. As in figure 2, injected embryos were fixed and stained for gsc. gsc is a higher-threshold target of GDF1; thus, higher amounts of mRNA (200 pg) were injected per embryo. mGDF1 = murine GDF1.
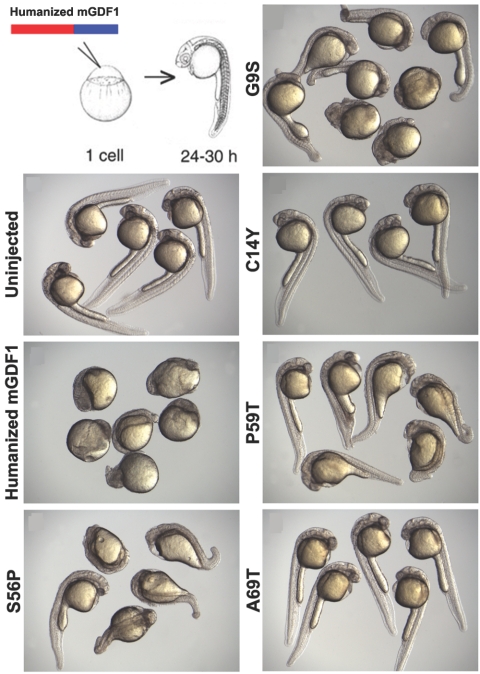
Figure 4. .
GDF1 activity assay by induction of ntl expression. Twenty picograms of mRNA synthesized from humanized wild-type (WT) and mutant GDF1 constructs were injected at the one-cell stage. Embryos were allowed to develop for 5 h and were fixed and stained for ntl, a downstream target of GDF1. Representative animal samples from injected clutches are shown. Mutant GDF1 constructs exhibited varying degrees of decreased activity compared with WT GDF1, as assessed by the extent of ectopic ntl induction. Stained samples from uninjected WT embryos are included for comparison. mGDF1 = murine GDF1.
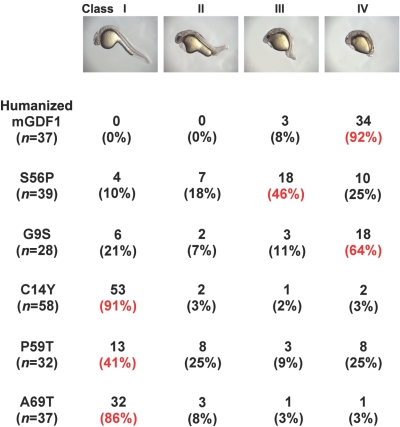
Figure 5. .
Classification and quantification of morphological phenotypes. Embryos aged 27 h injected with GDF1 constructs were sorted into classes I–IV (from wild type [WT] to those with the severest developmental defects) and were quantified. All mutant constructs exhibited varying degrees of decreased activity compared with WT humanized GDF1. mGDF1 = murine GDF1. Red font indicates the predominant class.
Discussion
The phenotypes observed with the GDF1 loss-of-function mutations are consistent with a model of disturbed L-R patterning, with incomplete establishment of left-sided identity. TGA (as for patients 3 and 8) and DORV (as for patient 5) can be seen in mice lacking Gdf1 or Cfc1.8,9,35,36 Similar defects are seen in humans with loss-of-function mutations in CFC119 (authors' unpublished data), the essential coreceptor for NODAL/GDF1. Interestingly, patients 4 and 5 manifest stenosis of the left pulmonary artery, suggestive of deficiency of left-sided anterior mesoderm. Patient 4 displays an atrioventricular-canal defect. Although this malformation is typically attributed to a primary disturbance in endocardial cushion development, as commonly seen in Down syndrome, the left–pulmonary artery stenosis and left-lung hypoplasia are not associated with Down syndrome atrioventricular septal defects. In addition, atrioventricular canal defects are commonly associated with the complex cardiovascular malformations seen in heterotaxy. These observations support the argument that the GDF1 mutation contributes in this case and acts in concert with the effects of the trisomy 21 as well. Finally, patients 6 and 7 have TOF, a complex malformation attributed to the primary malformations (the pulmonic stenosis with overriding aorta) and to the secondary associated features (the obligate ventricular septal defect and development of right ventricular hypertrophy). Our findings suggest that septation of the conotruncus and development of the aortic and pulmonic valves are affected by L-R patterning influences in humans. A specific role for Pitx2, whose expression may be altered by Gdf1 deficiency, has recently been established in the development of the second cardiac lineage myocardium in mice.37 The phenotypes of patients 1 and 2 are also compatible with L-R defects (e.g., TOF, AVC, and persistent left superior cava). However, additional functional studies will be needed to prove whether these mutations are also pathogenic.
An important observation that distinguishes these studies in humans from those described in mice is the presence of cardiac malformations in the context of heterozygous mutations in GDF1. Inbred strains of mice heterozygous for mutations in Gdf1 are described as phenotypically normal. In contrast, mice null for Gdf1 display extremes of heterotaxy not observed in our human subjects (e.g., right pulmonary isomerism). We propose that mutations in GDF1 constitute only one genetic susceptibility trait in an otherwise multifactorial disorder (i.e., CHD) with heterogeneous causes, including environmental and potentially additional genetic modifiers. None of our subjects had known signs of heterotaxy of abdominal organs, suggesting that L-R determination defects attributed to heterozygous mutations in GDF1 might affect some organs and not others.
Recent studies in mice suggest that it is best to view the total TGF-β signaling strength—and not just the contribution of a single factor—for a particular patterning event. With respect to axis formation in the mouse, a synergistic interaction was observed between Gdf1−/− and Nodal+/− genotypes, which was not seen in either genotype alone.38 Whereas Gdf1−/− mice exhibit cardiac and laterality defects but normal axis development, the reduction of a single copy of the Nodal gene reveals novel midline abnormalities, including HPE, in some cases. These findings reveal that there are different thresholds for TGF-β signaling required for different patterning functions. The observation that only the genotypes with the most-severe reductions in TGF-β signaling affect the midline with less pronounced reductions, leading to laterality and cardiac malformations, is entirely consistent with the paucity of detectable mutations we observed in our HPE cohort. By extension of this general concept, it is reasonable to postulate that reduction of effective GDF1 signaling in humans could result in cardiac malformations without the simultaneous manifestations of right-sided isomerism seen in Gdf1-null mice. Similar evidence for a spectrum of phenotypic effects attributed to gradations of attenuated Nodal signaling have been well described in the mouse and zebrafish.39–41
Dissecting the full range of genetic and environmental contributors to human CHD is a challenging problem. We hypothesize that complex disorders arise from the interplay between rare mutations and more-common genetic or environmental modifiers and that detected mutations must always be considered in the broader context of these interacting factors.42 Our studies suggest that mutations in GDF1 are likely to contribute to a distinct subclass of CHD, especially affecting the conotruncus, and a more targeted mutation search based on a more narrow spectrum of phenotypes may be indicated. It will be important to assess the penetrance of GDF1 mutations in family studies and to search for evidence of additional genetic and environmental modifiers. In this regard, mutations in the entire pathway of L-R determination may be considered candidates for CHD on the basis of a similar mechanism identified for human GDF1.
Acknowledgments
We thank the patients who participated in this research. This work was supported in part by the Division of Intramural Research of the NHGRI, NIH, and by grants from NIH and the Human Frontier Science Program (to A.F.S.) and by grant PSOHL74731 (to E.G.).
Web Resources
The accession number and URLs for data presented herein are as follows:
- BLASTN, http://www.ncbi.nlm.nih.gov/blast/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human GDF1 [accession number NM_001492])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NODAL, LEFTY1, LEFTY2, PITX2, GDF1, SMAD2, SMAD3, SMAD4, FOXH1, TGA, and CFC1)
- PDB, http://www.pdb.org/ (for 1LX5)
References
- 1.Brand T (2003) Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol 258:1–19 10.1016/S0012-1606(03)00112-X [DOI] [PubMed] [Google Scholar]
- 2.Burn J, Goodship J (1996) Developmental genetics of the heart. Curr Opin Genet Dev 6:322–325 10.1016/S0959-437X(96)80009-8 [DOI] [PubMed] [Google Scholar]
- 3.Harvey RP (2002) Patterning the vertebrate heart. Nat Rev Genet 3:544–556 10.1038/nrg843 [DOI] [PubMed] [Google Scholar]
- 4.Hamada H, Meno C, Watanabe D, Saijoh Y (2002) Establishment of vertebrate left-right asymmetry. Nat Rev Genet 3:103–113 10.1038/nrg732 [DOI] [PubMed] [Google Scholar]
- 5.Zhu L, Belmont JW, Ware SM (2006) Genetics of human heterotaxias. Eur J Hum Genet 14:17–25 [DOI] [PubMed] [Google Scholar]
- 6.Bamford RN, Roessler E, Burdine, RD, Saplakoglu U, dela Cruz J, Splitt M, Towbin J, Bowers P, Marino B, Schier AF, et al (2000) Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet 26:365–369 10.1038/81695 [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Milne N, Mochida K, Sakai Y, Saijo Y, Meno C, Hamada H (2003) Nodal signaling induces the midline barrier by activating nodal expression in the lateral plate. Development 130:1795–1804 10.1242/dev.00408 [DOI] [PubMed] [Google Scholar]
- 8.Rankin CT, Bunton T, Lawler AM, Lee S-J (2000) Regulation of left-right patterning in mice by growth/differentiation factor-1. Nat Genet 24:262–265 10.1038/73472 [DOI] [PubMed] [Google Scholar]
- 9.Wall NA, Craig EJ, Labosky PA, Kessler DS (2000) Mesoderm induction and reversal of left-right pattern by mouse GDF1, a Vg1-related gene. Dev Biol 227:495–509 10.1006/dbio.2000.9926 [DOI] [PubMed] [Google Scholar]
- 10.Lee S-J (1990) Identification of a novel member of the transforming growth factor-β superfamily. Mol Endocrinol 4:1034–1040 [DOI] [PubMed] [Google Scholar]
- 11.Lee S-J (1991) Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc Natl Acad Sci USA 88:4250–4254 10.1073/pnas.88.10.4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomsen GH, Melton DA (1993) Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell 74:433–441 10.1016/0092-8674(93)80045-G [DOI] [PubMed] [Google Scholar]
- 13.Birsoy B, Kofron M, Schiable K, Wylie C, Heasman J (2005) Vg1 is an essential signaling molecule in Xenopus development. Development 133:15–20 10.1242/dev.02144 [DOI] [PubMed] [Google Scholar]
- 14.Massague J (1998) TFGβ signal transduction. Ann Rev Biochem 67:753–791 10.1146/annurev.biochem.67.1.753 [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Massague J (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685–700 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- 16.Franco D, Campione M (2003) The role of pitx2 during cardiac development. Trends Cardiovasc Med 13:157–163 10.1016/S1050-1738(03)00039-2 [DOI] [PubMed] [Google Scholar]
- 17.Constam DB, Robertson EJ (2000) SPC4/PACE4 regulates a TGFβ signaling network during axis formation. Genes Dev 14:1146–1155 [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng SK, Olale F, Bennett JT, Brivanlou AH, Schier AF (2003) EGF-CFC proteins are essential coreceptors for the TGF-β signals Vg1 and GDF1. Genes Dev 17:31–36 10.1101/gad.1041203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldmuntz B, Bamford RN, Karkera JD, dela Cruz J, Roessler E, Muenke M (2002) CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet 70:776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Cruz J, Bamford RN, Burdine RD, Roessler E, Donnai D, Schier AF, Muenke M (2002) A loss of function mutation in the CFC domain of TDGF-1 is associated with human forebrain defects. Hum Genet 110:422–428 10.1007/s00439-002-0709-3 [DOI] [PubMed] [Google Scholar]
- 21.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23:318–326 10.1002/prot.340230306 [DOI] [PubMed] [Google Scholar]
- 22.Greenwald J, Groppe J, Gray P, Wlater E, Kwiatkowski W, Vale W, Choe S (2003) The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 11:605–617 10.1016/S1097-2765(03)00094-7 [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486 10.1007/BF00228148 [DOI] [PubMed] [Google Scholar]
- 24.Stachel SE, Grunwald DJ, Myers PZ (1993) Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development 117:1261–1274 [DOI] [PubMed] [Google Scholar]
- 25.MacDonald NO, Hendrickson WA (1993) A structural superfamily of growth factors containing a cystine knot motif. Cell 73:421–424 10.1016/0092-8674(93)90127-C [DOI] [PubMed] [Google Scholar]
- 26.Isaacs NW (1995) Cystine knots. Curr Opin Struct Biol 5:391–395 10.1016/0959-440X(95)80102-2 [DOI] [PubMed] [Google Scholar]
- 27.LeGood JA, Joubin K, Giraldez AJ, Ben-Haim N, Beck S, Chen Y, Schier AF, Constam DB (2005) Nodal stability determines signaling range. Curr Biol 15:31–36 10.1016/j.cub.2004.12.062 [DOI] [PubMed] [Google Scholar]
- 28.Cheng SK, Olale F, Birvanlou AH, Schier AF (2004) Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol 2:E30 10.1371/journal.pbio.0020030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michiotti G, Manco G, Parisi S, Lago CT, Rosa F, Persico MG (2001) Structure-function analysis of the EGF-CFC family member Cripto identifies residues essential for nodal signalling. Development 128:4501–4510 [DOI] [PubMed] [Google Scholar]
- 30.Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF (1999) The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97:121–132 10.1016/S0092-8674(00)80720-5 [DOI] [PubMed] [Google Scholar]
- 31.Toyama R, O’Connell ML, Wright CV, Kuehn MR, Dawid IB (1995) Nodal induces ectopic goosecoid and lim1 expression and axis duplication in zebrafish. Development 121:383–391 [DOI] [PubMed] [Google Scholar]
- 32.Heasman J (2006) Patterning the early Xenopus embryo. Development 133:1205–1217 10.1242/dev.02304 [DOI] [PubMed] [Google Scholar]
- 33.Schier AF (2003) Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol 19:589–621 10.1146/annurev.cellbio.19.041603.094522 [DOI] [PubMed] [Google Scholar]
- 34.Gurdon JB, Bourillot PY (2001) Morphogen gradient interpretation. Nature 413:797–803 10.1038/35101500 [DOI] [PubMed] [Google Scholar]
- 35.Gaio U, Schweickert A, Fischer A, Garratt AN, Muller T, Ozcelik C, Lankes W, Strehle M, Britsch S, Blum M, et al (1999) A role of the cryptic gene in the correct establishment of the left-right axis. Curr Biol 9:1339–1342 10.1016/S0960-9822(00)80059-7 [DOI] [PubMed] [Google Scholar]
- 36.Yan YT, Gritsman K, Ding J, Burdine RD, Corrales JD, Price SM, Talbot WS, Schier AF, Shen M (1999) Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev 13:2527–2537 10.1101/gad.13.19.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ai D, Liu W, Ma L, Dong F, Lu F-L, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, et al (2006) Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol 296:437–449 10.1016/j.ydbio.2006.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson O, Reissmann E, Jornvall H, Ibanez CF (2006) Synergistic interaction between Gdf1 and Nodal during anterior axis development. Dev Biol 293:370–381 10.1016/j.ydbio.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 39.Gritsman K, Talbot WS, Schier AF (2000) Nodal signaling patterns the organizer. Development 127:921–932 [DOI] [PubMed] [Google Scholar]
- 40.Norris DP, Brennan J, Bikoff EK, Robertson EJ (2002) The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129:3455–3468 [DOI] [PubMed] [Google Scholar]
- 41.Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ (2003) Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev 17:1646–1662 10.1101/gad.1100503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ming JE, Muenke M (2002) Multiple hits during early embryonic development: digenic disease and holoprosencephaly. Am J Hum Genet 71:1017–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]