Abstract
Background
Our laboratories have previously reported on the experimental infection of cattle with Mycobacterium avium subsp paratuberculosis (M. paratuberculosis) using an intratonsillar infection model. In addition, we have recently developed a partial protein array representing 92 M. paratuberculosis coding sequences. These combined tools have enabled a unique look at the temporal analysis of M. paratuberculosis antigens within the native host. The primary objective of this study was to identify M. paratuberculosis antigens detected by cattle early during infection. A secondary objective was to evaluate the humoral immune response in cattle during the initial year of infection.
Results
Sera from two experimentally infected cattle, taken pre-inoculation and at day 70, 194 and 321 post infection, identified dynamic antibody reactivity among antigens with some showing an increased response over time and others showing declining levels of reactivity over the same time period. A M. paratuberculosis specific protein, encoded by MAP0862, was strongly detected initially, but the antibody response became weaker with time. The most reactive protein was a putative surface antigen encoded by MAP1087. A second protein, MAP1204, implicated in virulence, was also strongly detected by day 70 in both cattle. Subsequent experiments showed that these two proteins were detected with sera from 5 of 9 naturally infected cattle in the subclinical stage of Johne's disease.
Conclusion
Collectively these results demonstrate that M. paratuberculosis proteins are detected by sera from experimentally infected cattle as early as 70 days after exposure. These data further suggest at least two antigens may be useful in the early diagnosis of M. paratuberculosis infections. Finally, the construction and use of a protein array in this pilot study has led to a novel approach for discovery of M. paratuberculosis antigens.
Background
Johne's disease is an economically significant intestinal disease caused by Mycobacterium avium subsp paratuberculosis (M. paratuberculosis). A recent survey estimated that 20%–40% of dairy herds in the United States are infected with M. paratuberculosis and producers lose $227 USD annually for each infected animal [1]. These costs are mostly attributed to the decreased milk production and weight loss resulting from the disease.
After M. paratuberculosis infection by ingestion of contaminated milk or grass containing fecal material from a shedding cow, there is a lengthy subclinical phase that can last for several years. During this stage the cows may appear healthy, but can intermittently shed low numbers of mycobacteria in the feces, enabling transmission to other animals including wildlife species. A major challenge in controlling Johne's disease is the ability to detect infected cattle prior to appearance of disease signs, such as diarrhea and heavy fecal shedding of M. paratuberculosis. An unknown trigger, possibly stress during lactation or parturition, advances the disease from subclinical to clinical where disease signs such as weight loss and diarrhea become evident [2,3]. This trigger appears to coincide with a shift in immune function from a Th1 response to a Th2 response [4]. Current detection of subclinical animals depends on the timing and sensitivity of the test. Even the most sensitive culture-based tests will not detect M. paratuberculosis if a subclinically infected animal is not shedding bacilli at the time the fecal or milk sample is collected. M. paratuberculosis antigen induced interferon (IFN)-γ has been shown to be elevated in subclinical animals, but this cytokine declines in the clinical stage concomitant with an increase in M. paratuberculosis specific IL-10 production [5,6]. A comprehensive cytokine profile has been reported for both circulating monocytes and local tissues obtained from M. paratuberculosis-infected cattle [7].
With a few notable exceptions [8-10], there is very little data on antibody detection of M. paratuberculosis at early stages of infection in cattle. There are several reasons for this, but one in particular is that cattle that appear healthy are not routinely evaluated using serial test bleeds and analysis. Furthermore, there are numerous studies that show the cell-mediated immune response in cattle predominates during the early stages of infection and is responsible for the initial control of this infection [4,6,11]. However, despite the lack of data describing the temporal detection of specific antigens by host antibodies early post infection, these experiments are critical to gain a better understanding of the pathogenesis, diagnostics and vaccine strategies for Johne's disease. For example, the ideal diagnostic antigen would be detected early and remain easily detected throughout the course of the disease. Alternatively, a good vaccine candidate antigen might only be detectable by antibody at a specific stage of the disease. Thus far, no such antigen has been discovered for Johne's disease.
The recent literature has revealed an emphasis on developing consensus animal models for Johne's disease study. One publication [12] is a result of an international meeting, sponsored by the Johne's Disease Integrated Program in the United States, which had the goal of proposing consensus animal models for each of the commodity groups including sheep and cattle as well as farmed deer in New Zealand. Detailed methods including dose, route, length of time for disease signs to appear, etc. were determined [12]. A second communication, published independently around the same time, provided similar information [13]. Prior to these comprehensive animal model reviews, we published a study describing the intratonsillar route of infection with M. paratuberculosis in neonatal calves [9]. Serial test bleeds were collected over the course of the 321-day study and analyzed for T cell and B cell responses using a variety of immunological assays. Selected test bleeds from that study were used on the protein array in the current study. The health status of these calves was further monitored using additional tests, including fecal culture and IS900 PCR. Although M. paratuberculosis was detected in all calves by 271 days post challenge, no clinical signs were observed in any animal throughout the study [9]. However, an antibody response was observed by 134 days post infection using a lipoarabinomannan-based ELISA test. A second study that evaluated an experimental infection of goats observed an antibody response as soon as 180 days post infection [14].
The study described herein combines the intratonsillar infection model [9] with newly developed protein array tools to obtain a temporal picture of antigen detection during the initial year of infection in cattle. While the concept of protein arrays has been around nearly as long as the DNA array, there are very few protein arrays available. This is because the process of constructing these arrays involves the production of dozens of specific proteins, a rate limiting and labor-intensive step when compared to oligonucleotide synthesis used in constructing DNA arrays. Nonetheless, studies using bacterial protein arrays are beginning to appear in the literature [15-17] due to the power provided by this tool for parallel protein analyses. Our laboratory initially developed a 48 spot protein array consisting of purified recombinant proteins representing selected M. paratuberculosis gene products [18]. That array was expanded to a 96-spot array and used in this study.
Results
Construction of a partial M. paratuberculosis protein array
A 96-spot protein array was constructed on nitrocellulose filters using a dot blot apparatus to imprint the array. Represented among these spots are 92 M. paratuberculosis recombinant fusion proteins including hypothetical proteins (20), known antigens (6), membrane proteins (10), and proteins with no similarity in public sequence databases (25). The affinity tag present within these recombinant fusion proteins is the maltose binding protein (MBP). MBP was also expressed as a fusion with the E. coli LacZ alpha peptide and used as a control to determine antibody reactivity to this tag in each experiment. In addition, there are two spots that contain a sonicated whole cell extract of M. paratuberculosis and one spot with only spotting buffer as a negative control. A complete listing of the proteins present on the array along with their sizes, concentration and predicted function are listed in Table 1.
Table 1.
Mycobacterium avium subsp paratuberculosis proteins used in this study
| Protein namea | Concentration (mg/ml)b | Gene sizec | Amino acidsd | Predicted function |
| MAP sonicate | 1.0 | NAe | NA | NA |
| MAP0075 | 0.18 | 423 | 140 | conserved small membrane protein |
| MAP0087-his | 1.0 | 717 | 238 | probable secreted protein |
| MAP0105c | 1.24 | 1058 | 352/889 | hypothetical protein |
| MAP0182c | 0.67 | 411 | 136 | conserved hypothetical protein |
| MAP0216 | 1.66 | 1028 | 342/347 | antigen 85A, mycolyltransferase |
| MAP0261c | 0.49 | 465 | 154/161 | 19 kDa protein |
| MAP0389 | 2.04 | 947 | 315/337 | diarylpropane peroxidase (EC 1.11.1.14) [Nostoc] |
| MAP0736 | 5.73 | 744 | 247 | hypothetical protein |
| MAP0852 | 10.4 | 546 | 181 | no BLAST hits |
| MAP0853 | 4.04 | 660 | 219 | no BLAST hits |
| MAP0855 | 0.34 | 926 | 308/314 | no BLAST hits |
| MAP0857c | 0.70 | 318 | 105 | hypothetical protein |
| MAP0858 | 0.44 | 537 | 178/182 | no BLAST hits |
| MAP0859c | 0.66 | 609 | 202 | Mycobacterium phage L5 |
| MAP0860c | 0.04 | 885 | 294/296 | no BLAST hits |
| MAP0861 | 0.43 | 342 | 113 | no BLAST hits |
| MAP0862 | 0.48 | 989 | 329/360 | no BLAST hits |
| MAP0863 | 0.66 | 675 | 224 | no BLAST hits |
| MAP0864 | 7.42 | 426 | 141 | no BLAST hits |
| MAP0865 | 0.08 | 1272 | 423 | Hypothetical 30.9 kDa protein |
| MAP0866 | 3.07 | 804 | 267 | integrase |
| MAP0904 | 0.81 | 371 | 124/246 | conserved hypothetical protein |
| MAP0961c | 0.18 | 719 | 239/352 | hypothetical protein |
| MAP1087 | 0.43 | 441 | 146 | prob. peptide transp. system permease, N-terminal |
| MAP1121c | 0.32 | 498 | 165 | putative substrate binding protein |
| MAP1174c | 1.64 | 756 | 251 | glucose-6-phosphate 1-dehydrogenase |
| MAP1204 | 2.61 | 735 | 244 | putative exported p60 protein homologue |
| MAP1233 | 0.04 | 723 | 240 | conserved hypothetical protein |
| MAP1345 | 0.69 | 600 | 199 | no BLAST hits |
| MAP1388 | 0.08 | 396 | 131 | no BLAST hits |
| MAP1416c | 0.45 | 666 | 221 | no BLAST hits |
| MAP1417c | 0.54 | 435 | 144 | no BLAST hits |
| MAP1609c | 0.05 | 987 | 328/330 | antigen 85B, mycolyltransferase |
| MAP1636c | 1.12 | 425 | 141/157 | no BLAST hits |
| MAP1643 | 0.45 | 2277 | 759/762 | isocitrate lyase, [beta] module |
| MAP1655c | 0.93 | 381 | 126 | hypothetical protein |
| MAP1730c | 0.26 | 1023 | 340 | putative ATP/GTP-binding protein |
| MAP1931c | 0.32 | 1104 | 367 | anthranilate phosphoribosyltransferase |
| MAP2077c | 1.66 | 330 | 109 | hypothetical protein |
| MAP2116c | 0.27 | 1269 | 422 | cell invasion protein |
| MAP2121c | 7.14 | 887 | 295/307 | major membrane protein I |
| MAP2121c-his | 0.99 | 924 | 307 | major membrane protein I |
| MAP2151 | 2.79 | 438 | 145 | no BLAST hits |
| MAP2155 | 9.57 | 312 | 103 | similar to IS6110 |
| MAP2156 | 0.59 | 963 | 320 | putative transposase GEN: X52471) |
| MAP2157 | 0.38 | 1221 | 406 | IS900 |
| MAP2158 | 2.40 | 582 | 193 | no BLAST hits |
| MAP2182c | 0.26 | 435 | 144 | conserved hypothetical protein |
| MAP2231 | 0.31 | 1422 | 473 | PKS-associated protein, unknown function |
| MAP2360c | 0.13 | 534 | 177 | hypothetical protein |
| MAP2380 | 2.00 | 600 | 200/550 | acyl-CoA synthase |
| MAP2657 | 0.26 | 651 | 216 | putative oxidoreductase |
| MAP2663c | 0.03 | 510 | 169 | putative integral membrane protein |
| MAP2676c | 1.30 | 354 | 117 | hypothetical protein |
| MAP2734 | 0.37 | 1341 | 446 | putative dioxygenasesdiooxygenases |
| MAP2740 | 0.24 | 666 | 221 | putative membrane protein |
| MAP2751 | 0.70 | 582 | 193 | no BLAST hits |
| MAP2753 | 0.17 | 309 | 103/252 | hypothetical protein |
| MAP2761c | 0.03 | 692 | 230/238 | no BLAST hits |
| MAP2762c | 1.45 | 425 | 141/146 | no BLAST hits |
| MAP2764c | 0.10 | 290 | 96/149 | no BLAST hits |
| MAP2765c | 0.06 | 539 | 179/396 | no BLAST hits |
| MAP2767c | 0.42 | 473 | 157/183 | no BLAST hits |
| MAP2963c | 0.88 | 1499 | 499/874 | no BLAST hits |
| MAP3084c-his | 1.0 | 684 | 227 | probable secreted protein |
| MAP3121 | 0.38 | 855 | 284 | enoyl-CoA hydratase/isomerase superfamily |
| MAP3129 | 0.09 | 404 | 134/141 | hypothetical protein |
| MAP3131 | 0.30 | 1096 | 365/942 | conserved large membrane protein |
| MAP3155c | 1.22 | 273 | 90 | hypothetical protein |
| MAP3434 | 0.18 | 690 | 230/330 | possible membrane protein |
| MAP3437c | 0.29 | 843 | 279/280 | no BLAST hits |
| MAP3531c | 0.28 | 1034 | 344/352 | antigen 85C, mycolytransferase |
| MAP3734c | 0.19 | 1559 | 519/593 | putative ABC transporter ATP-binding protein |
| MAP3735c | 0.42 | 1181 | 393/429 | part of heavy metal tolerance protein |
| MAP3743 | 0.85 | 998 | 332/348 | hypothetical protein |
| MAP3751 | 0.11 | 879 | 293/979 | conserved large membrane protein |
| MAP3753 | 0.64 | 1353 | 450 | hypothetical protein |
| MAP3761c | 0.07 | 729 | 242 | conserved membrane protein |
| MAP3771 | 0.02 | 294 | 97 | 50S ribosomal protein L31 |
| MAP3817c | 0.65 | 939 | 312 | no BLAST hits |
| MAP3833c | 0.13 | 626 | 208/260 | hypothetical protein |
| MAP3840 | 0.10 | 1872 | 623 | 70 kD heat shock protein, chromosome replication |
| MAP3902c | 0.15 | 522 | 173 | Serine/threonine kinase (EC 2.7.1.37) [Myc... |
| MAP3903c | 0.42 | 531 | 176 | Serine/threonine kinase (EC 2.7.1.37) [Myc... |
| MAP3954 | 0.05 | 279 | 92 | conserved hypothetical protein |
| MAP4014 | 0.21 | 951 | 316 | probable 4-amino butyrate transporter, N-terminal |
| MAP4025 | 0.32 | 854 | 284/325 | cytochrome c-type biogenesis protein |
| MAP4129 | 0.26 | 965 | 321/336 | ABC transporter |
| MAP4198 | 0.13 | 1283 | 427/441 | SecY subunit of preprotein translocase |
| MAP4207c | 5.28 | 686 | 228/231 | probable ATP-binding transport protein |
| MAP4199 | 0.03 | 540/546 | 179/181 | probable adenylate kinase |
| MAP4228 | 0.05 | 222 | 73 | initiation factor IF-1 |
aProtein name corresponds to the genome project designation (Li et al., 2005). A second designation is included if known.
bProtein concentration is reported in as measured by Nanodrop spectrometry at 280nm.
cGene size is reported in base pairs (bp). In cases where only a portion of the M. avium subsp paratuberculosis gene was cloned, the number of bp present in the clone is reported first followed by the number of bp in the full-length M. avium subsp paratuberculosis gene.
dNumber of M. avium subsp paratuberculosis amino acids present in the fusion protein. In cases where only a portion of the M. avium subsp paratuberculosis protein is represented, the number of amino acids present is listed first followed by the total number of M. avium subsp paratuberculosis amino acids in the full-length protein.
eNA is not applicable.
The performance of the array was tested using a monoclonal antibody developed to the MBP tag as shown previously [18]. These experiments showed that all proteins containing the tag were readily detectable by immunoblot analysis (data not shown).
Humoral immune response of experimentally infected cattle
An analysis of serial bleeds collected from two experimentally infected calves was performed to temporally examine the humoral immune response to the arrayed M. paratuberculosis proteins. In general, the antibody response against the set of 92 proteins was remarkably similar between the two animals. One notable difference was that fewer proteins were recognized at day 70 in animal 5904 compared to 5902, as only 11 proteins compared to 27 proteins were detected, respectively (Figure 1 and Figure 2). However by day 194, animal 5904 had antibodies against more M. paratuberculosis proteins than did animal 5902. Serum from both animals detected similar numbers of proteins at the end of the study period (Figure 1). The pre-infection sera showed no detection of any of the recombinant proteins (Figure 2) and there was no detection of the MBP-LacZ protein for all time points suggesting that these calves do not have antibodies to the maltose binding protein (MBP) affinity purification tag.
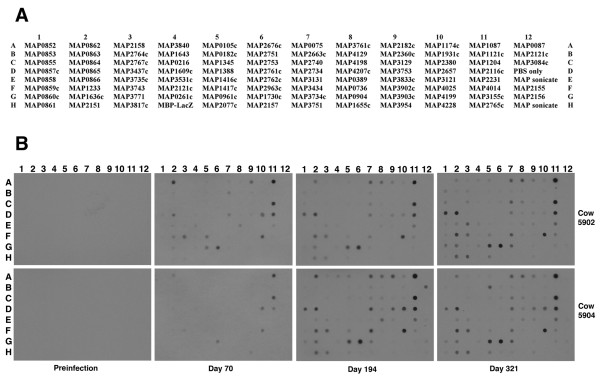
Figure 1.
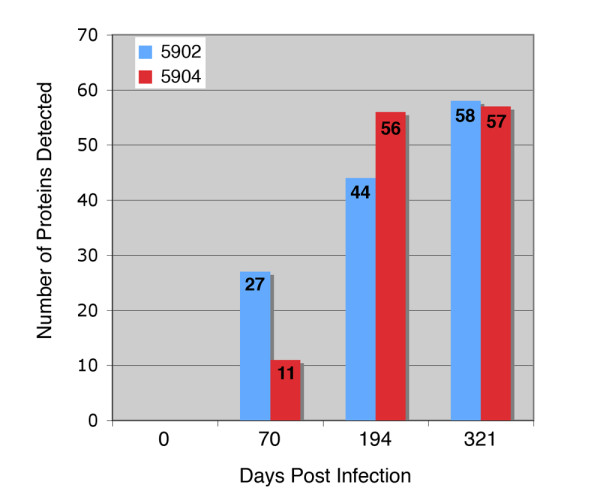
Total number of M. paratuberculosis recombinant proteins detected by sera from experimentally infected cattle. The graph shows the number of proteins on the 96-dot array (y-axis) that were detected at the day post infection (x-axis) for each calf. The total number of proteins detected is indicated on each bar. Note that similar numbers of proteins were detected by each animal at the end of the study period.
Figure 2.
Use of the dot blot protein array to obtain antibody reactivity profiles of experimentally infected calves. Shown are the spot assignments for the array (A) and dot blot arrays exposed to sera from experimentally infected calves (B). The time point for when each serum sample was collected is indicated in the margin beneath the images. The animal number is listed in the right margin. A whole cell lysate representing a majority of the proteins produced by M. paratuberculosis is spotted in E12 and H12 for all dot blots. Three proteins present on the upper right corner of the array (in column 12) are polyhistidine tagged proteins (MAP0087, MAP2121c and MAP3084c). The remaining 89 spots contain MBP fusion proteins of M. paratuberculosis coding sequences. Note that the MAP2121c coding sequence is represented twice on the array; once as an MBP fusion (spot F4) and also a polyhistidine tagged protein (12B).
Temporal analysis of M. paratuberculosis proteins
The arrays exposed to cattle sera were analyzed by densitometry to obtain a quantitative measure of antibody reactivity (Table 2). When evaluating selected proteins over the course of the study, certain trends in reactivity appear to emerge for some proteins. For example, in both calves, antibody reactivity declined between day 194 and day 321 for a group of 11 proteins. This was especially true for MAP0862 and MAP2657, which showed the most severe decline among these 11 proteins (Table 2). The seven proteins that best fit into strong, moderate and weak response categories are shown in Figure 3. While strong and moderately detected proteins showed a wide range between the calves at days 70 and 194, this range was considerably reduced in the high category proteins at day 321 (Figure 3).
Table 2.
Spot intensities of proteins present on the array.a
| Cow 5902 | Cow 5904 | ||||||||
| Spot Locb | Protein | Day 0 | Day 70 | Day 194 | Day 321 | Day 0 | Day 70 | Day 194 | Day 321 |
| A7 | MAP0075 | 0 | 4.472 | 21.431 | 21.122 | 0 | 0 | 27.612 | 21.36 |
| A12 | MAP0087-his | 0.978 | 0 | 0 | 0 | 1.112 | 1.882 | 0 | 1.228 |
| A5 | MAP0105c | 0 | 6.929 | 3.867 | 5.096 | 1.186 | 2.321 | 10.106 | 1.813 |
| B5 | MAP0182c | 0 | 5.407 | 0 | 1.924 | 1.177 | 2.409 | 1.617 | 22.366 |
| C4 | MAP0216 | 0.652 | 3.779 | 1.296 | 1.395 | 2.175 | 3.444 | 0 | 0 |
| G4 | MAP0261c | 2.395 | 2.287 | 3.487 | 5.668 | 1.097 | 3.146 | 2.397 | 2.496 |
| E8 | MAP0389 | 0 | 7.809 | 3.163 | 1.817 | 0 | 1.533 | 16.751 | 5.529 |
| F8 | MAP0736 | 0.734 | 0 | 0 | 0 | 0 | 0 | 1.446 | 0 |
| A1 | MAP0852 | 2.257 | 3.747 | 2.908 | 1.147 | 2.564 | 4.661 | 2.709 | 1.537 |
| B1 | MAP0853 | 2.144 | 3.734 | 5.095 | 8.497 | 1.961 | 4.158 | 3.239 | 2.649 |
| C1 | MAP0855 | 2.538 | 3.387 | 4.552 | 7.898 | 1.716 | 2.248 | 2.132 | 0 |
| D1 | MAP0857c | 2.709 | 6.445 | 21.867 | 37.718 | 1.045 | 1.602 | 23.425 | 15.825 |
| E1 | MAP0858 | 3.005 | 3.882 | 1.775 | 3.999 | 1.978 | 0 | 0 | 0 |
| F1 | MAP0859c | 2.821 | 4.137 | 2.87 | 3.745 | 1.909 | 0 | 2.803 | 0 |
| G1 | MAP0860c | 2.996 | 0 | 5.531 | 12.743 | 1.592 | 0 | 7.669 | 6.676 |
| H1 | MAP0861 | 5.074c | 0 | 3.407 | 3.053 | 1.546 | 4.05 | 3.111 | 3.52 |
| A2 | MAP0862 | 1.795 | 40.036 | 16.196 | 7.997 | 2.796 | 11.451 | 30.918 | 12.422 |
| B2 | MAP0863 | 1.518 | 5.524 | 3.174 | 4.592 | 2.057 | 4.891 | 3.274 | 1.473 |
| C2 | MAP0864 | 1.255 | 3.44 | 1.781 | 1.726 | 2.016 | 3.56 | 0 | 0 |
| D2 | MAP0865 | 1.909 | 18.708 | 32.138 | 44.846 | 1.317 | 4.386 | 31.361 | 35.251 |
| E2 | MAP0866 | 2.346 | 7.051 | 2.67 | 7.116 | 1.315 | 3.561 | 4.052 | 6.009 |
| G8 | MAP0904 | 1.678 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| G5 | MAP0961c | 1.797 | 14.366 | 26.5 | 37.546 | 0.78 | 2.103 | 33.046 | 42.877 |
| A11 | MAP1087 | 0.753 | 53.179 | 51.634 | 70.968 | 0.874 | 33.469 | 75.392 | 68.964 |
| B11 | MAP1121c | 0.758 | 2.438 | 0 | 2.094 | 0.699 | 3.364 | 0 | 1.848 |
| A10 | MAP1174c | 0 | 2.301 | 3.155 | 4.411 | 0.623 | 1.921 | 5.81 | 4.955 |
| C11 | MAP1204 | 0.78 | 40.8 | 43.856 | 55.046 | 0 | 28.367 | 61.122 | 58.985 |
| F2 | MAP1233 | 2.568 | 12.375 | 22.507 | 32.493 | 1.932 | 3.212 | 24.157 | 30.447 |
| C5 | MAP1345 | 0 | 3.339 | 1.861 | 1.018 | 1.147 | 0 | 3.413 | 2.389 |
| D5 | MAP1388 | 0 | 2.281 | 0 | 0 | 0.429 | 2.835 | 0 | 0 |
| E5 | MAP1416c | 0 | 1.647 | 0 | 0 | 0.791 | 3.791 | 0 | 0 |
| F5 | MAP1417c | 1.052 | 14.882 | 4.095 | 6.212 | 0.955 | 3.979 | 7.614 | 3.521 |
| D4 | MAP1609c | 0.733 | 3.916 | 0 | 0 | 1.003 | 5.292 | 0 | 0 |
| G2 | MAP1636c | 3.286 | 6.944 | 13.249 | 17.272 | 1.701 | 0 | 17.226 | 12.06 |
| B4 | MAP1643 | 0 | 4.155 | 0 | 1.534 | 1.827 | 0 | 0 | 0 |
| H8 | MAP1655c | 2.788 | 0 | 0 | 0 | 0 | 2.049 | 0 | 0 |
| G6 | MAP1730c | 1.147 | 29.831 | 43.872 | 52.35 | 0 | 12.272 | 57.799 | 57.47 |
| B10 | MAP1931c | 0 | 2.917 | 0 | 2.217 | 0.511 | 2.886 | 2.077 | 3.339 |
| H5 | MAP2077c | 3.833 | 0 | 2.784 | 9.443 | 1.385 | 0 | 3.63 | 9.221 |
| D11 | MAP2116c | 1.418 | 29.88 | 33.215 | 42.633 | 0.568 | 11.24 | 51.336 | 47.918 |
| F4 | MAP2121c | 1.76 | 4.158 | 1.373 | 1.513 | 0.969 | 3.005 | 0 | 0 |
| B12 | MAP2121c-his | 1.247 | 0 | 0 | 0 | 0.889 | 2.172 | 21.226 | 2.155 |
| H2 | MAP2151 | 4.136 | 1.919 | 12.688 | 21.897 | 1.571 | 3.402 | 24.619 | 20.889 |
| F12 | MAP2155 | 2.434 | 0 | 1.385 | 0 | 1.251 | 2.039 | 0 | 2.105 |
| G12 | MAP2156 | 3.731 | 2.327 | 0 | 2.12 | 1.357 | 2.647 | 5.42 | 5.113 |
| H6 | MAP2157 | 3.504 | 0 | 0 | 0 | 0.751 | 1.532 | 1.51 | 2.683 |
| A3 | MAP2158 | 1.403 | 3.487 | 4.2 | 3.396 | 3.187 | 1.704 | 6.682 | 2.204 |
| A9 | MAP2182c | 0 | 19.101 | 7.349 | 8.933 | 0 | 0 | 27.935 | 12.096 |
| E11 | MAP2231 | 1.846 | 3.732 | 11.54 | 15.671 | 1.166 | 0 | 17.203 | 18.933 |
| B9 | MAP2360c | 0 | 3.168 | 0 | 0 | 0 | 2.766 | 0 | 0 |
| C10 | MAP2380 | 0.772 | 2.139 | 1.198 | 0 | 0 | 0 | 1.968 | 3.225 |
| D10 | MAP2657 | 0 | 22.372 | 8.29 | 3.68 | 0 | 15.937 | 39.178 | 25.032 |
| B7 | MAP2663c | 0 | 9.594 | 3.661 | 2.248 | 0.868 | 0 | 4.9 | 2.717 |
| A6 | MAP2676c | 0 | 3.291 | 1.406 | 0 | 0 | 0 | 2.036 | 0 |
| D7 | MAP2734 | 2.726 | 9.109 | 11.031 | 9.269 | 0 | 0 | 27.935 | 18.79 |
| C7 | MAP2740 | 0 | 1.736 | 6.447 | 18.061 | 0.507 | 0 | 6.295 | 10.804 |
| B6 | MAP2751 | 0 | 3.157 | 1.101 | 2.373 | 0.8 | 0 | 1.729 | 2.206 |
| C6 | MAP2753 | 0 | 1.909 | 0 | 2.176 | 0.624 | 0 | 1.701 | 2.585 |
| D6 | MAP2761c | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E6 | MAP2762c | 0 | 0 | 0 | 0 | 0 | 2.397 | 0 | 0 |
| B3 | MAP2764c | 1.472 | 2.739 | 1.969 | 5.874 | 2.13 | 5.442 | 2.039 | 1.955 |
| H11 | MAP2765c | 4.816 | 0 | 0 | 0 | 0 | 4.799 | 0 | 0 |
| C3 | MAP2767c | 1.257 | 3.507 | 2.303 | 7.019 | 1.982 | 4.347 | 1.92 | 2.722 |
| F6 | MAP2963c | 0 | 0 | 0 | 0 | 0 | 2.204 | 0 | 0 |
| C12 | MAP3084c-his | 1.313 | 0 | 0 | 0 | 0.999 | 2.026 | 0 | 1.982 |
| E10 | MAP3121 | 1.079 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C9 | MAP3129 | 1.406 | 2.366 | 0 | 1.206 | 0 | 0 | 1.731 | 2.865 |
| E7 | MAP3131 | 0 | 0 | 0 | 0 | 0 | 0 | 1.404 | 1.713 |
| G11 | MAP3155c | 2.783 | 2.553 | 2.725 | 3.841 | 1.435 | 2.237 | 2.226 | 4.865 |
| F7 | MAP3434 | 0.832 | 1.401 | 8.312 | 19.522 | 0.508 | 0 | 33.289 | 14.418 |
| D3 | MAP3437c | 1.674 | 3.321 | 1.29 | 1.587 | 1.359 | 1.965 | 0 | 0 |
| E4 | MAP3531c | 0.926 | 7.373 | 2.472 | 4.313 | 0.558 | 2.834 | 1.374 | 0 |
| G7 | MAP3734c | 1.085 | 2.238 | 2.719 | 17.809 | 0 | 0 | 10.914 | 20.81 |
| E3 | MAP3735c | 1.944 | 4.897 | 5.078 | 18.618 | 1.326 | 3.368 | 3.887 | 4.502 |
| F3 | MAP3743 | 2.113 | 23.874 | 16.282 | 16.773 | 1.28 | 6.953 | 21.309 | 22.067 |
| H7 | MAP3751 | 2.991 | 0 | 0 | 0 | 0 | 0 | 2.499 | 3.033 |
| D9 | MAP3753 | 0 | 2.912 | 7.043 | 3.212 | 0 | 0 | 18.95 | 15.247 |
| A8 | MAP3761c | 0 | 2.473 | 14.931 | 23.178 | 0 | 0 | 18.02 | 21.264 |
| G3 | MAP3771 | 2.81 | 2.229 | 1.344 | 2.528 | 0.953 | 1.997 | 0 | 0 |
| H3 | MAP3817c | 3.836 | 2.526 | 10.298 | 22.826 | 1.101 | 1.552 | 15.736 | 19.901 |
| E9 | MAP3833c | 1.078 | 0 | 2.07 | 2.755 | 0 | 0 | 3.676 | 4.636 |
| A4 | MAP3840 | 0.823 | 4.968 | 3.842 | 4.521 | 1.403 | 0 | 10.083 | 0 |
| F9 | MAP3902c | 1.095 | 0 | 0 | 0 | 0 | 2.376 | 0 | 0 |
| G9 | MAP3903c | 1.755 | 1.355 | 0 | 1.027 | 0 | 1.772 | 0 | 0 |
| H9 | MAP3954 | 3.426 | 0 | 0 | 1.969 | 0 | 7.584 | 1.612 | 3.629 |
| F11 | MAP4014 | 2.266 | 2.035 | 5.415 | 8.98 | 0.545 | 0 | 5.293 | 8.043 |
| F10 | MAP4025 | 1.474 | 25.139 | 34.876 | 39.022 | 0 | 13.413 | 49.655 | 47.012 |
| B8 | MAP4129 | 0 | 1.489 | 0 | 1.363 | 0 | 0 | 1.435 | 2.585 |
| C8 | MAP4198 | 0 | 1.536 | 1.517 | 6.817 | 0 | 0 | 5.431 | 6.175 |
| G10 | MAP4199 | 1.919 | 1.688 | 0 | 0 | 0.821 | 2.339 | 0 | 0 |
| D8 | MAP4207c | 1.981 | 1.691 | 1.872 | 1.364 | 0 | 0 | 3.699 | 5.632 |
| H10 | MAP4228 | 4.373 | 0 | 0 | 0 | 0 | 3.122 | 0 | 0 |
| H4 | MBP-LacZ | 4.128 | 0 | 0 | 1.722 | 1.303 | 0 | 0 | 0 |
| D12 | PBS bufferd | 1.676 | 0 | 1.236 | 0 | 0.857 | 2.322 | 0 | 2.003 |
| E12 | Sonicatee | 2.673 | 0 | 0 | 0 | 1.287 | 1.936 | 2.963 | 3.523 |
| H12 | Sonicate | 5.01 | 1.359 | 0 | 0 | 1.366 | 4.337 | 3.002 | 4.472 |
aListed are the intensity values for each spot arranged by ascending MAP number. Measurements were made using a consistent 1804 pixel area circle to record spot density using the Adobe Photoshop CS3 Extended application.
bLoc is location on the dot blot array.
cThe strongest intensity for each time point is shown in bold.
dPBS buffer in spot D12 represents the spotting buffer only control.
eSonicate is a sonicated whole cell lysate of M. avium subsp paratuberculosis.
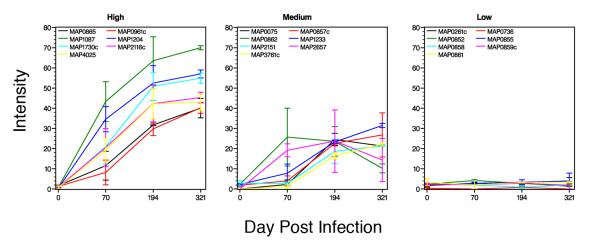
Figure 3.
Temporal trends of protein reactivity in experimentally infected cattle. The lines show the average intensity for each protein over the course of the study period. The range observed between the two calves is indicated by statistical bars. Kernel Density Estimation performed on the log-transformed maximum intensity scores indicated there were three distinct normally distributed peaks. These distributions are statistically significant and define the proteins belonging to the weak, moderate, or strong response groups. The graph labeled high contains the seven proteins most strongly detected by the cattle sera and the graph labeled low contains the seven proteins that showed the least reactivity with cattle sera. The seven proteins in the medium category are also representative of reactive antigens that either showed low reactivity initially or declining reactivity with time.
Identification of antigens detected early in M. paratuberculosis infection
Overall, the strongest antigen among this set of fusion proteins was encoded by MAP1087 (Figures 2 and 3). Antibody reactivity to this protein was observed in both animals by day 70 and remained the strongest detected protein throughout the course of the study (Figure 3 and Table 2). MAP1204, a putative p60 homolog, was also strongly detected by serum from both animals. Other antigens identified in these studies include MAP1730c, a putative ATP/GTP binding protein, and M. paratuberculosis hypothetical sequences MAP0865 and MAP0961c. MAP0865 contains the F57 DNA fragment which has previously been shown to be present uniquely in M. paratuberculosis [19] and resides on a 15.3 kb genomic island termed large sequence polymorphism 4 (LSP4) [20], making this protein an excellent candidate for use in a diagnostic test. Likewise, MAP0961c shows no sequence similarity in nucleotide databases and is present on a 13.5 kb segment of the genome termed LSP5 [20]. However, a similarity search of the SwissProt protein database reveals a glycosyltransferase domain that may be involved in cell envelope biogenesis. Furthermore, a corresponding ortholog, termed MUL_4677, was found in the recently published genome sequence of Mycobacterium ulcerans [21]. Therefore, caution must be used if this protein were to be considered for use in a diagnostic test. It is noteworthy that the whole cell sonicate antigen, which was spotted in duplicate on the protein array (12E and 12H), was not detected by either experimentally infected animal. This same antigen preparation reacted very strongly to anti-M. paratuberculosis sera from mice and rabbits previously [18]. In addition, two of three clinical cows showed an antibody response to the whole cell sonicate antigen [18] and hence it should be considered a potent preparation. This antigen preparation is likely similar to that used in the ELISA test.
Early antigens are detected by sera from naturally infected cattle in the subclinical stage of Johne's disease
A panel of sera collected from 9 cows in the subclinical stage of infection was analyzed by immunoblot against the two strongest antigens identified from the protein array experiments. These animals are housed at the National Animal Disease Center and are monitored four times a year for shedding of bacilli and immunological status as measured by ELISA assays for antibody and IFN-γ (Table 3). MAP1087 was detected in 4 of 9 serum samples (Figure 4, lanes 2–5) and very weakly reactive with another 2 serum samples (Figure 4, lanes 7 and 8). Three of the nine serum samples from the naturally infected cows detected MAP1204. Serum from cow 183 detected both MAP1087 and MAP1204 proteins. Likewise, serum from cow 113 detected both proteins although weakly for MAP1204. Neither antigen was detected by sera from three of the cows tested (Figure 4, lanes 1, 6, and 9). Finally, none of the sera from these animals detected the protein encoded by MAP0858, a protein that was also not detected by the experimentally infected cattle sera (Figure 2). These data suggest that MAP1087 and MAP1204 proteins may be detectable in M. paratuberculosis-infected animals prior to the clinical stage of disease. In conclusion, we hypothesize that while these antigens, if combined, would recognized by 66% of the subclinical cows, additional antigen(s) need to be included along with MAP1204 and MAP1087 in order to detect all nine subclinical cows.
Table 3.
Culture and immunological status of cattle used in this study.
| IFN-γ (mg/ml)c | ||||||
| Cow ID | Survey datea | Fecal culture (CFU)b | NS | ConA | MPS | ELISA (S/P ratio)d |
| 85 | 12/20/05 | C, C, C, C | 0.069 | 0.454 | 0.278 | 0.002 |
| 3/4/06 | 2, 2, 2 | 0.061 | 0.185 | 0.068 | ND | |
| 112 | 12/20/05 | C, C, 21, 1 | 0.07 | 0.218 | 0.064 | 0.027 |
| 4/10/06 | 2, 1, 1 | 0.063 | 1.83 | 0.09 | 0.014 | |
| 113 | 8/10/04 | C, C, C, C | 0.21 | 0.514 | 0.054 | 0.041 |
| 11/10/04 | 0, 3, 1 | 0.39 | 1.51 | 0.54 | 0.09 | |
| 117 | 12/17/04 | T, T, T, T | 0.053 | 0.143 | 0.097 | -0.052 |
| 3/1/05 | T, C, C | 0.039 | 0.174 | 0.046 | ND | |
| 183 | 12/20/05 | 0, 0, C, C | 0.054 | 0.241 | 0.145 | 1.42 |
| 4/7/06 | 3, 2, 0 | 0.089 | 1.134 | 0.194 | 0.981 | |
| 235 | 12/20/05 | 0, 0, 0, 0 | 0.068 | 0.362 | 0.114 | 0.039 |
| 3/4/06 | 0, 1, 1 | 0.05 | 0.35 | 0.064 | 0.021 | |
| 834 | 8/10/04 | 0, 1, 0, 1 | 0.033 | 0.336 | 0.142 | 0.87 |
| 11/11/04 | 0, 0, 0 | 0.044 | 0.806 | 0.076 | 0.648 | |
| 871 | 8/10/04 | C, C, C, C | 0.037 | 0.361 | 0.054 | 0.049 |
| 12/21/04 | C, 2, 1 | 0.043 | 0.189 | 0.064 | 0.121 | |
| 2715 | 12/20/05 | 2, 0, 1, 0 | 0.067 | 1.3 | 0.324 | 0.226 |
| 3/18/06 | 0, 0, 0 | 0.059 | 0.211 | 0.062 | 0.044 | |
aEach animal is surveyed quarterly to monitor progression of Johne's disease. The two most recent quarterly surveys are shown for each animal, but only sera collected on the most recent date was used for the immunoblot assay in Figure 4.
bFecal culture is reported as numbers of colony forming units (CFU) present on each HYEM slant (platings are in triplicate or quadruplicate). A "C" indicates the slant had bacterial growth other than M. paratuberculosis. A "T" designation means too many colonies to count on that slant.
cThe gamma interferon (IFN-γ) assay. Subclinical cows typically have a M. paratuberculosis sonicate (MPS) reading higher than the no stimulation (NS) reading, along with little or no colonies on fecal culture slants. The concanavalin A (ConA) reading is a positive control as cells are readily stimulated by this lectin protein.
dThe ELISA sample to positive (S/P) ratio is calculated from the optical density reading as described previously [11].
Figure 4.
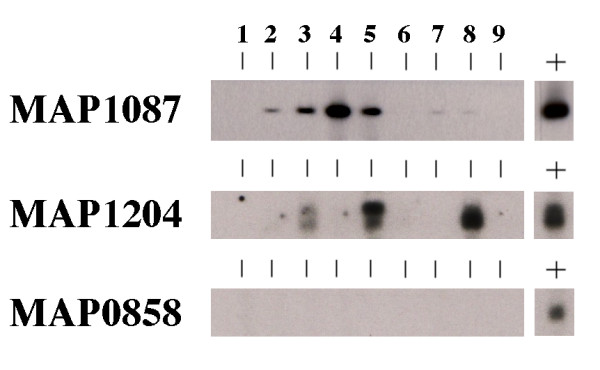
Slot immunoblot analysis of selected recombinant proteins with serum from cattle in the subclinical stage of infection. A preparative, single well gel was loaded with the protein indicated in the left margin and blotted onto nitrocellulose filters. The filters were placed into a slot blotting device and 1:500 dilutions of each cattle sera were loaded into independent slots. Each blot was further processed as described previously [29]. The positive control, indicated by the lane with a "+" sign above it, was probed with a monoclonal antibody developed against the MBP affinity tag as described previously [29]. These results demonstrate that MAP1087 and MAP1204 are detected by sera from naturally infected cattle in the subclinical phase of infection. Serum sample slot assignments: 1, cow 85; 2, cow 112; 3, cow 113; 4, cow 117; 5, cow 183; 6, cow 235; 7, cow 834; 8, cow 871 and 9, cow 2715.
Discussion
Diagnosis of Johne's disease is difficult because as many as 90% of infected animals may show no signs of disease during sample collection and there is a lack of sensitive and specific assays that detect M. paratuberculosis early during infection. Antigen-based diagnostic tests for Johne's disease currently use a complex mixture of antigens, such as PPD or mycobacterial membrane fractions, that can exhibit cross reactivity with other mycobacteria [22]. Results from this study further suggest that using complex protein mixtures such as a whole cell sonicate extract may not be as sensitive when compared to a few recombinant proteins. This may be due to the heterogeneity of the sonicated extract where each protein is present in very small quantities. New scientific breakthroughs are expected from the recently completed genome sequence of M. paratuberculosis as this information now enables researchers to select and characterize M. paratuberculosis sequences of interest. Our interest is in characterizing those sequences that are novel and specific to M. paratuberculosis in order to develop the best available diagnostic tools. This study represents the most comprehensive M. paratuberculosis antigen analysis on a temporal scale. Data from this study provide a first look at how specific antibody reactivity changes over time and in direct comparison to other proteins.
When a temporal analysis of serial bleeds from experimentally infected calves was undertaken, we discovered a 50-kDa protein that was detected by day 14 post-challenge and a 60-kDa protein was evident by day 42 [9]. However, these studies were performed with a whole cell extract as the antigen and thus the identity of these early antigens were never discovered. Nonetheless, these studies showed that a humoral immune response was initiated in calves within a short time period following exposure to M. paratuberculosis. The current study extends these initial findings by using a recently developed protein array to probe sera from these experimentally infected calves. The two proteins that elicited the strongest antibody reactivity were MAP1087, a probable peptide transport system permease and MAP1204, a p60 protein homolog. These proteins have a calculated size of 15.4 kDa and 25.4 kDa, respectively. Therefore, it is unlikely that the proteins encoded by MAP1087 and MAP1204 are the same 2 antigens detected in the initial study by Waters et al [9]. Although MAP1204 is much more conserved across the mycobacterial genus, both proteins are present as orthologs in at least the genome sequences of M. smegmatis and M. avium strain 104. This raises the potential for cross-reactivity if these antigens were incorporated into a diagnostic test for Johne's disease.
A M. paratuberculosis protein array was initially constructed from 43 recombinant proteins and tested with sera from a variety of host species [18]. The protein array has since been expanded to include a total of 92 recombinant proteins for the current study. Additional experiments are planned that will use the protein array to examine antibody profiles of subclinical cattle versus clinical cattle. This study focused specifically on antigens detected early following infection of cattle with M. paratuberculosis. Performing a test to detect M. paratuberculosis antigens when clinical signs of the disease are already evident in the host, make the control of this disease by test and cull strategies futile. An optimized serological test that incorporates antigens detected at early times post-infection is a critical tool that animal producers/farmers need to control Johne's disease. In this way infected animals can be identified prior to shedding and exposure of herd mates to the disease causing bacterium. Regarding uninfected cows, one key advantage of the experimental model used is that each cow had its own animal-specific negative control (the preinfection bleed). This type of control is not available in most Johne's disease studies. In summary, a highly specific test that could identify infected animals within herds may have a greater and more immediate impact on cattle industries than any other current management approach, including vaccination.
The ideal M. paratuberculosis protein antigen likely remains undiscovered simply because all the proteins produced by M. paratuberculosis are not represented on the current protein array. In this study, promising candidates have been found, but the genome sequence annotation suggests that M. paratuberculosis produces a total of 4,530 proteins [23]. Therefore, until a larger percentage of the genome has been evaluated in a parallel manner similar to this study, better antigens are still likely to be discovered. Another limitation to using a protein array for antigen discovery involves the identification of epitopes that may be formed by multiple protein complexes. This possibility cannot be revealed when proteins are produced and spotted in isolation such as on an array.
Our laboratory, in collaboration with others, has successfully completed the genome sequence of this significant veterinary pathogen [23] and have used these data to identify coding sequences specific to M. paratuberculosis. Those studies have revealed less than 40 coding sequences that are uniquely present in M. paratuberculosis [24,25]. This surprisingly small number of unique coding sequences is primarily due to the genetic similarity among members of the mycobacteria [26]. Among these coding sequences, one gene, designated ISMAP02, is present in six copies distributed randomly in the genome [25,27]. Furthermore, we have evaluated the expression products from several of these M. paratuberculosis-specific sequences for immunoreactivity with sera from animals exposed to M. paratuberculosis [25,28]. Those studies identified proteins encoded by MAP0862, MAP2963c and MAP3732c as potential diagnostic antigens. The newly constructed M. paratuberculosis protein array has many of these same coding sequences represented. Both MAP0862 and MAP2963c were detected in each animal in this study, but MAP3732c is not present on the array used in the current study. The antigens identified in this study should be further evaluated with naturally infected animals in field trials before incorporation into a diagnostic test for Johne's disease.
Conclusion
Of the 92 recombinant M. paratuberculosis proteins analyzed in this study, 2 emerged as potential antigens for the early detection of Johne's disease in cattle. These proteins are encoded by MAP1087 and MAP1204. The combination of MAP1087 and MAP1204 detected more subclinical cows than either protein alone, suggesting that the ideal detection antigen will comprise a mixture of more than one protein. Finally, this is a pilot scale study that demonstrates the method is effective for use in a large-scale antigen discovery project.
Methods
Animals
All cattle used in this study were housed at the National Animal Disease Center (NADC) and handled using institutional care guidelines set by the animal care and use committee. Experimentally infected calves were put in biosafety level 2 containment immediately following birth to prevent exposure to environmental mycobacteria. Naturally infected subclinical cattle are contained on a field pasture-barn set up on site at NADC. The experimentally infected animals consisted of male Holstein calves less than 1 year of age [9]. The health status and infection procedure have been reported previously [9]. Briefly, calves were infected with M. paratuberculosis by instillation of four weekly doses of approximately 4 × 106 CFU (in 0.2 ml PBS) into both tonsillar crypts weekly from 2 to 5 weeks of age. The subclinical cows used in this study comprised Holsteins, Brown Swiss and Guernsey with ages that ranged from 2–7 years.
Protein array content
The protein array consists of a 96-dot format of proteins spotted onto nitrocellulose. There are 92 total M. paratuberculosis proteins recombinantly produced in E. coli with 89 of these using the pMAL-c2 expression vector (New England Biolabs), and 3 using the pET vector. All expression clones were sequenced to confirm that the cloned insert matched the native M. paratuberculosis gene and was in-frame with expression signals built into the expression vector. One spot on the array contains the MBP/LacZ protein used as a control to assess antibody reactivity to the MBP affinity tag. This control protein comprises the 42-kDa maltose binding protein along with the 8-kDa LacZ alpha peptide. Two spots contain a sonicated whole cell extract of M. paratuberculosis prepared as described previously [9] and one spot contains the PBS spotting buffer control only.
Protein array production
PBS was used as the spotting buffer and diluent for all M. paratuberculosis proteins. All protein arrays were sequentially produced and no single array was used for more than one experiment. To print each array, the nitrocellulose membrane, pre-soaked in PBS, was placed over a rubber gasket such that it covered all wells of the Bio-dot 96-well manifold apparatus (BioRad, Hercules, CA). The top of the Bio-dot apparatus was then positioned over the membrane and fastened in each corner. Each well was pre-washed with 200 μl of PBS followed by a brief vacuum to pull the buffer through the membrane. Diluted working stocks of purified proteins were stored in deep 96-well plates with a 1-ml capacity per round bottom well (Nalgene Nunc International). Transfer of these proteins from the deep well plate to the assembled Bio-dot apparatus was performed using an 8-channel multi-pipette. Each well of the assembled 96-well dot blot apparatus was loaded with the appropriate protein at a final concentration of 3 μg/well as measured in a NanoDrop spectrophotometer (Thermo Fisher Scientific) at 280nm. The PBS-diluted samples were allowed to flow through the membrane by gravity flow. Each well was rinsed with 200 μl of PBS, 0.1% Tween-20 (TPBS). The vacuum was engaged to pull the rinse solution through and the nitrocellulose membrane was removed from the apparatus and placed in a Petri dish containing a blocking solution (TPBS-BSA; PBS with 2% bovine serum albumin, 0.1% Tween-20). After 1 hour in the blocking solution, the immunoblot assay was performed.
Immunoblot analysis of the protein array
Serum from M. paratuberculosis infected calves were diluted in the blocking buffer and exposed to spotted arrays for two hours at room temperature on a rocker platform. All cattle sera were diluted 1:500. After three washes in TPBS, the nitrocellulose array was incubated for 1.5 h with Horse Radish Peroxidase-conjugated anti-bovine IgG antibody (Pierce) diluted at 1:20,000 in TPBS-BSA. This was followed by a final three washes in TPBS. Assay development was with SuperSignal detection reagents (Pierce) and Kodak BioMax MR film.
Quantitative analysis of spot intensity
Spot intensity was measured using the Adobe Photoshop CS3 extended application. This version has the ability to record pixel gray values using the measurement scale of 1 pixel equals 1 pixel. Each spot was measured identically using a window area of 1804 pixels. Values were exported into a spreadsheet for further analysis. The background statistics were calculated by determining the mean and standard deviation of the 24 spots within each array that had the least signal intensity. Each intensity score was compared to the calculated background intensity. Values that were within two standard deviations of the background mean were set to zero and all other intensity values were subtracted from the background mean to give positive intensity values that were adjusted for the array background. To determine the reactivity distribution of all proteins in the complete set, the highest intensity score for each protein was examined as a way to determine if there was distinct stratification of maximal antibody response. Kernel Density Estimation performed on the log-transformed maximum intensity scores indicated there were three distinct normally distributed peaks. These distributions define the proteins belonging to the low, medium, or high reactivity groups presented in Figure 3.
Authors' contributions
JPB conceived of the study, printed the protein arrays, participated in study design and coordination and drafted the manuscript. DOB provided bioinformatics support, statistical analysis and assisted with densitometric analyses of protein arrays. WRW and MVP conceived the experimental calf challenge model and performed the immunological characterizations. JRS carried out the immunoassays and determined immunological status for the subclinical cattle. MLP participated in the design of the study and performed bioinformatics analyses. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are grateful to Janis Hansen for technical assistance. Portions of this study were funded by the USDA-CSREES-CAP grant (JDIP) and the USDA-Agricultural Research Service. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Contributor Information
John P Bannantine, Email: john.bannantine@ars.usda.gov.
Darrell O Bayles, Email: darrell.bayles@ars.usda.gov.
W Ray Waters, Email: ray.waters@ars.usda.gov.
Mitchell V Palmer, Email: mitchell.palmer@ars.usda.gov.
Judith R Stabel, Email: judy.stabel@ars.usda.gov.
Michael L Paustian, Email: michael.paustian@ars.usda.gov.
References
- Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med. 1999;40:179–192. doi: 10.1016/S0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- Stabel JR. Host responses to Mycobacterium avium subsp. paratuberculosis: a complex arsenal. Anim Health Res Rev. 2006;7:61–70. doi: 10.1017/S1466252307001168. [DOI] [PubMed] [Google Scholar]
- Stabel JR. Johne's disease: a hidden threat. J Dairy Sci. 1998;81:283–288. doi: 10.3168/jds.S0022-0302(98)75577-8. [DOI] [PubMed] [Google Scholar]
- Coussens PM. Model for immune responses to Mycobacterium avium subspecies paratuberculosis in cattle. Infect Immun. 2004;72:3089–3096. doi: 10.1128/IAI.72.6.3089-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabel JR. Production of gamma-interferon by peripheral blood mononuclear cells: an important diagnostic tool for detection of subclinical paratuberculosis. J Vet Diagn Invest. 1996;8:345–350. doi: 10.1177/104063879600800311. [DOI] [PubMed] [Google Scholar]
- Stabel JR. Transitions in immune responses to Mycobacterium paratuberculosis. Vet Microbiol. 2000;77:465–473. doi: 10.1016/S0378-1135(00)00331-X. [DOI] [PubMed] [Google Scholar]
- Coussens PM, Verman N, Coussens MA, Elftman MD, McNulty AM. Cytokine gene expression in peripheral blood mononuclear cells and tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis: evidence for an inherent proinflammatory gene expression pattern. Infect Immun. 2004;72:1409–1422. doi: 10.1128/IAI.72.3.1409-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda S, Bannantine JP, Waters WR, Mori Y, Whitlock RH, Scott MC, Speer CA. A Highly Sensitive and Subspecies-Specific Surface Antigen Enzyme – Linked Immunosorbent Assay for Diagnosis of Johne's Disease. Clin Vaccine Immunol. 2006;13:837–844. doi: 10.1128/CVI.00148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters WR, Miller JM, Palmer MV, Stabel JR, Jones DE, Koistinen KA, Steadham EM, Hamilton MJ, Davis WC, Bannantine JP. Early induction of humoral and cellular immune responses during experimental Mycobacterium avium subsp. paratuberculosis infection of calves. Infect Immun. 2003;71:5130–5138. doi: 10.1128/IAI.71.9.5130-5138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koets AP, Rutten VP, de Boer M, Bakker D, Valentin-Weigand P, van Eden W. Differential changes in heat shock protein-, lipoarabinomannan-, and purified protein derivative-specific immunoglobulin G1 and G2 isotype responses during bovine Mycobacterium avium subsp. paratuberculosis infection. Infect Immun. 2001;69:1492–1498. doi: 10.1128/IAI.69.3.1492-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo HC, Park YH, Hamilton MJ, Barrington GM, Davies CJ, Kim JB, Dahl JL, Waters WR, Davis WC. Analysis of the immune response to Mycobacterium avium subsp. paratuberculosis in experimentally infected calves. Infect Immun. 2004;72:6870–6883. doi: 10.1128/IAI.72.12.6870-6883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ME, 2nd, Stabel JR, Sweeney RW, Griffin F, Talaat AM, Bakker D, Benedictus G, Davis WC, de Lisle GW, Gardner IA, et al. Experimental challenge models for Johne's disease: a review and proposed international guidelines. Vet Microbiol. 2007;122:197–222. doi: 10.1016/j.vetmic.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Begg DJ, Whittington RJ. Experimental animal infection models for Johne's disease, an infectious enteropathy caused by Mycobacterium avium subsp. paratuberculosis. Vet J. 2007 doi: 10.1016/j.tvjl.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Munjal SK, Tripathi BN, Paliwal OP. Progressive immunopathological changes during early stages of experimental infection of goats with Mycobacterium avium subspecies paratuberculosis. Vet Pathol. 2005;42:427–436. doi: 10.1354/vp.42-4-427. [DOI] [PubMed] [Google Scholar]
- Groathouse NA, Amin A, Marques MA, Spencer JS, Gelber R, Knudson DL, Belisle JT, Brennan PJ, Slayden RA. Use of protein microarrays to define the humoral immune response in leprosy patients and identification of disease-state-specific antigenic profiles. Infect Immun. 2006;74:6458–6466. doi: 10.1128/IAI.00041-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman MB, McKevitt M, McLoughlin M, Perez C, Howell J, Weinstock GM, Norris SJ, Palzkill T. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J Clin Microbiol. 2006;44:888–891. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Jiang L, Song Q, Yang J, Chen Z, Guo Z, Zhou D, Du Z, Song Y, Wang J, et al. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect Immun. 2005;73:3734–3739. doi: 10.1128/IAI.73.6.3734-3739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Waters WR, Stabel JR, Palmer MV, Li L, Kapur V, Paustian ML. Development and use of a partial Mycobacterium avium subspecies paratuberculosis protein array. Proteomics. 2008;8:463–474. doi: 10.1002/pmic.200700644. [DOI] [PubMed] [Google Scholar]
- Poupart P, Coene M, Van Heuverswyn H, Cocito C. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J Clin Microbiol. 1993;31:1601–1605. doi: 10.1128/jcm.31.6.1601-1605.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semret M, Alexander DC, Turenne CY, de Haas P, Overduin P, van Soolingen D, Cousins D, Behr MA. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J Clin Microbiol. 2005;43:3704–3712. doi: 10.1128/JCM.43.8.3704-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris NB, Barletta RG. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin Microbiol Rev. 2001;14:489–512. doi: 10.1128/CMR.14.3.489-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA. 2005;102:12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Baechler E, Zhang Q, Li L, Kapur V. Genome Scale Comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium Reveals Potential Diagnostic Sequences. J Clin Microbiol. 2002;40:1303–1310. doi: 10.1128/JCM.40.4.1303-1310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paustian ML, Amonsin A, Kapur V, Bannantine JP. Characterization of Novel Coding Sequences Specific to Mycobacterium avium subsp. paratuberculosis: Implications for Diagnosis of Johne's Disease. J Clin Microbiol. 2004;42:2675–2681. doi: 10.1128/JCM.42.6.2675-2681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Zhang Q, Li LL, Kapur V. Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 2003;3:10. doi: 10.1186/1471-2180-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabel JR, Bannantine JP. Development of a nested PCR method targeting a unique multicopy element, ISMap02, for detection of Mycobacterium avium subsp. paratuberculosis in fecal samples. J Clin Microbiol. 2005;43:4744–4750. doi: 10.1128/JCM.43.9.4744-4750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Hansen JK, Paustian ML, Amonsin A, Li LL, Stabel JR, Kapur V. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol. 2004;42:106–114. doi: 10.1128/JCM.42.1.106-114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Radosevich TJ, Stabel JR, Sreevatsan S, Kapur V, Paustian ML. Development and Characterization of Monoclonal Antibodies and Aptamers against Major Antigens of Mycobacterium avium subsp. paratuberculosis. Clin Vaccine Immunol. 2007;14:518–526. doi: 10.1128/CVI.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]