Abstract
H3 phosphorylation has been correlated with mitosis temporally in mammalian cells and spatially in ciliated protozoa. In logarithmically growing Tetrahymena thermophila cells, for example, H3 phosphorylation can be detected in germline micronuclei that divide mitotically but not in somatic macronuclei that divide amitotically. Here, we demonstrate that micronuclear H3 phosphorylation occurs at a single site (Ser-10) in the amino-terminal domain of histone H3, the same site phosphorylated during mitosis in mammalian cells. Using an antibody specific for Ser-10 phosphorylated H3, we show that, in Tetrahymena, this modification is correlated with mitotic and meiotic divisions of micronuclei in a fashion that closely coincides with chromosome condensation. Our data suggest that H3 phosphorylation at Ser-10 is a highly conserved event among eukaryotes and is likely involved in both mitotic and meiotic chromosome condensation.
In eukaryotic cells, DNA is closely associated with histone proteins in the form of chromatin, packaging DNA in a way that remains only partially understood. Although the structure of the nucleosome core is now known in considerable detail (1), how higher order chromatin structures are folded and unfolded to accommodate processes such as transcription, replication, and chromosome segregation remains unclear. During mitosis, DNA is compacted nearly 10,000-fold to ensure proper segregation of the genetic material to daughter cells. Faithful segregation of sister chromatids requires proper condensation of the chromatin during entry into mitosis and decondensation of the fiber during exit from mitosis and is essential for the viability of the cells.
Specific posttranslational modifications of histones, particularly acetylation and phosphorylation, correlate well with dynamic aspects of the folding and unfolding of the chromatin fiber (2). For example, hyperphosphorylation of linker histone H1 is temporally coupled with entry into mitosis and has often been presumed to function in mitotic chromatin condensation (3, 4). However, recent experiments show that chromatin condensation can occur in vivo (5) or in vitro (6, 7) in the absence of H1. Furthermore, H1 hyperphosphorylation does not occur in premature chromatin condensation induced by fostriecin (8) or okadaic acid (9). Therefore, the exact function of H1 hyperphosphorylation in mitosis remains unclear.
In contrast to H1 hyperphosphorylation, site-specific phosphorylation of core histone H3 at serine 10 seems to occur exclusively during mitosis in mammalian cells (10, 11). Moreover, fostriecin and okadaic acid, which initiate premature chromatin condensation in cell cultures, also induce H3 phosphorylation (8, 9). Similarly, vanadate-induced dephosphorylation of H3 correlates with chromatin decondensation and the rescue of a mitotic mutant that otherwise fails to initiate postmitotic chromatin decondensation (12). Recent studies, using an antibody selective for the Ser-10 phosphorylated H3 amino terminus, have documented a tight correlation between H3 phosphorylation and mitotic chromatin condensation in mammalian cells (13). Taken together, the above data suggest that H3 phosphorylation plays an important, yet poorly understood, role in mitotic chromatin condensation.
Like most ciliated protozoa, Tetrahymena thermophila cells contain two nuclei: a macronucleus and a micronucleus. In vegetative cells, macronuclei are transcriptionally active, highly endoreplicated, and divide amitotically. In contrast, micronuclei are inactive, germ-line nuclei that are diploid and divide mitotically (14). Consistent with the hypothesis that H3 phosphorylation is mechanistically linked to chromosome condensation, H3 phosphorylation has been found to occur only in micronuclei, but not in macronuclei of logarithmically growing vegetative cells (15). In this paper, we demonstrate that micronuclear H3 is phosphorylated at a single site within its amino-terminal domain, Ser-10, as shown previously for mammalian cells (10, 11). In addition, using an antibody highly specific for H3 phosphorylated at this residue, we find that H3 phosphorylation is temporally correlated with mitosis in Tetrahymena in a fashion that closely coincides with chromosome condensation. We also extend the association between H3 phosphorylation and chromosome condensation to meiotic chromosomes by analyzing micronuclear meiosis during the sexual process of conjugation. Our data argue that Ser-10 H3 phosphorylation is a highly conserved event among eukaryotes and support the hypothesis that this modification is involved in a pathway of higher order chromatin folding and/or unfolding.
MATERIALS AND METHODS
Cell Culture and [32P]Orthophosphate Labeling.
T. thermophila strain CU428 was grown in 1% proteose peptone as described previously (16). Where indicated, cells were labeled continuously during vegetative growth in proteose peptone in the presence of 10 μCi/ml [32P]orthophosphate. For conjugation, strains CU427 and CU428 (obtained from P. Bruns, Cornell University, Ithaca, NY) were used. Conjugation was induced according to Bruns and Brussard (17) with modifications described by Allis and Dennison (18).
Preparation of Nuclei and Nuclear Proteins.
Macro- and micronuclei were isolated from Tetrahymena as described by Gorovsky et al. (16), except that the nucleus isolation buffer contained 1 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium butyrate, and 200 μM chloromercuriphenylsulfonic acid, but not spermidine. Where indicated, macro- and micronuclei were further purified by sedimentation at unit gravity according to Allis and Dennison (18). H3 was purified from sulfuric acid extracts of micronuclei by reverse-phase-HPLC using a C8 column, as described previously (19).
Electrophoresis and Immunoblotting.
SDS/PAGE (20) and immunoblotting analyses (21) were performed as described previously. Phosphorylated H3 (Ser-10) antibody was generated and characterized as described by Hendzel et al. (13) and is available from Upstate Biotechnology (Lake Placid, NY). General (control) H3 antibody was generated against reverse-phase-HPLC purified Tetrahymena H3 (C.D.A., unpublished data). Crude phosphorylated H3 antiserum was routinely preincubated with an unphosphorylated H3 peptide (ARTKQTARKSTGGKAPRKQLC) to block contaminating antibodies that react with the proteolytically processed form of H3 (H3F) in micronuclei (22, 23).
Indirect Immunofluorescence Analyses.
Growing or conjugating cells were fixed and processed for indirect immunofluorescence as described previously (24). The phosphorylated H3 antiserum, pretreated as described above, was typically used at a dilution of 1:500 and detected with a rhodamine-conjugated secondary antibody. Cells were also stained with the DNA-specific dye, diamidinophenolindole (DAPI) at 0.3 μg/ml in Tris-buffered saline (TBS).
Enzymatic Treatment.
Where appropriate, HPLC-purified H3 was incubated with bacterial alkaline phosphatase (Worthington) as described previously (25) except that the enzyme preparation was not boiled before use.
Protein Microsequencing.
HPLC-purified micronuclear H3 (both H3S and H3F) was sequenced from the N terminus in an Applied Biosystems model 477A protein sequencer with an in-line 120A phenylthiohydantoin-analyzer (Applied Biosystems) using optimized cycles. Instead of butyl chloride, 90% methanol containing phosphoric acid (15 μl/100 ml) was used to extract the cleaved amino acids. After conversion, 50% of the sample was transferred to the HPLC for phenylthiohydantoin-amino acid identification, and the other 50% was collected for determination of radioactivity by scintillation counting.
RESULTS
Determination of Mitosis-Related H3 Phosphorylation Site(s).
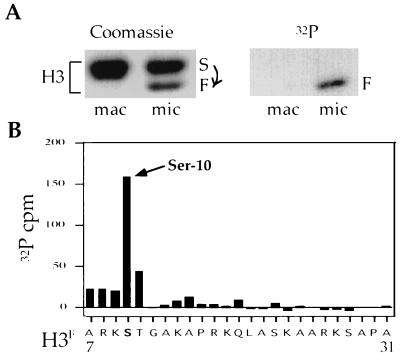
Previous work has shown that, in Tetrahymena, H3 phosphorylation occurs only in mitotic micronuclei and not in amitotic macronuclei (15). To identify H3 phosphorylation site(s) used during mitosis, macro- and micronuclei were prepared from mid-log-phase cultures grown continuously in [32P]orthophosphate, and H3 was extracted from both nuclei and purified by reverse-phase HPLC. Consistent with previous findings (22, 23), two electrophoretically distinct forms of H3 were observed in micronuclei. The slower migrating species, H3S, is identical to macronuclear H3. In contrast, the faster species, H3F, is derived from H3S by a proteolytic cleavage that removes the first six amino acids of H3S, generating an isoform that is unique to micronuclei (23). As shown in Fig. 1A, H3 phosphorylation occurs only in micronuclei and is specific to micronuclear H3F (15).
Figure 1.
There is a single phosphorylation site, Ser-10, in the N terminus of Tetrahymena micronuclear H3. (A) HPLC-purified H3 from either macro- or micronuclei of logarithmically growing Tetrahymena cells labeled in vivo by [32P]orthophosphate was resolved on a 12% SDS/PAGE gel and examined by Coomassie blue staining (Left) or autoradiography (Right). (B) In vivo phosphorylated micronuclear H3 was subjected to automated sequencing from the N terminus, and 32P radioactivity eluted from each sequencing cycle was collected and counted. Note that an identical conclusion was reported previously by us as an unpublished observation (26).
HPLC-purified micronuclear H3 (including H3S and H3F) was then subjected to N-terminal sequencing, and the 32P radioactivity eluted at each cycle of automated sequencing was determined. As shown in Fig. 1B, in the first 25 amino acids of H3F (note: cycle 1 = amino acid 7 because of the proteolytic processing leading to H3F, see ref. 23), only the serine corresponding to position 10 (Ser-10) has a significant amount of 32P radioactivity. These data indicate that there is a single phosphorylation site, Ser-10, in the amino terminus of micronuclear H3.
Specificity of the Ser-10 Phosphorylated H3 Antibody in Tetrahymena.
Recently, we generated a polyclonal antibody against a synthetic peptide that mimics the H3 Ser-10 phosphorylation epitope. In mammalian cells, this antibody is highly specific for mitotic H3 (13). To further characterize this antibody, we investigated its specificity using the unique biology offered by the Tetrahymena system.
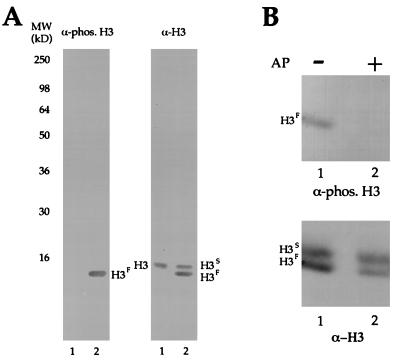
Total proteins from macro- and micronuclei were separated by SDS/PAGE, blotted, and probed with the phosphorylated H3 (Fig. 2A, Left) and general H3 (Fig. 2A, Right) antibodies. As expected, the phosphorylated H3 antibody recognizes micronuclear H3F (presumably phosphorylated) but fails to recognize similar amounts of nonphosphorylated micronuclear H3S or macronuclear H3. Specificity of the antibody was further demonstrated by in vitro dephosphorylation of Tetrahymena micronuclear H3F before immunodetection. As shown in Fig. 2B, when HPLC-purified micronuclear H3F was treated with alkaline phosphatase, reactivity with the phosphorylated H3 antibody was completely eliminated.
Figure 2.
The phosphorylated H3 antibody is specific for phosphorylated micronuclear H3F. (A) Total proteins from either macro- (lane 1) or micronuclei (lane 2) of logarithmically growing Tetrahymena cells were resolved on a 12% SDS/PAGE gel, transferred to a nitrocellulose membrane, and probed by either phosphorylated H3 antibody (Left) or general H3 antibody (Right). (B) HPLC-purified H3 from micronuclei of logarithmically growing Tetrahymena cells was incubated in the presence (lane 2) or absence (lane 1) of alkaline phosphatase (AP) and subjected to immunoblotting by either phosphorylated H3 antibody (Upper) or general H3 antibody (Lower).
Temporal Correlation between H3 Phosphorylation and Mitosis in Tetrahymena.
Previously, it has been shown that, in Tetrahymena, H3 phosphorylation occurs only in mitotic micronuclei and not in amitotic macronuclei (15). However, the temporal relationship between H3 phosphorylation and micronuclear mitosis is unknown. To determine this relationship, the phosphorylated H3 antibody was used to stain Tetrahymena in situ. A population of logarithmically growing cells stained with the DNA-specific dye DAPI is shown in Fig. 3A. The same cells stained with the phosphorylated H3 antibody are shown in Fig. 3B. Consistent with previous biochemical results obtained from isolated nuclei (15), the phosphorylated H3 antibody does not stain amitotic macronuclei (Fig. 3B, arrowheads). Curiously, only some micronuclei are stained. This antibody stains mitotic micronuclei (Fig. 3B, closed arrows), which have a characteristic “football” shape; round interphase micronuclei (Fig. 3B, open arrows) are not stained. This result demonstrates that H3 phosphorylation at Ser-10 is a highly conserved event closely associated with mitosis in organisms ranging from ciliates to mammals.
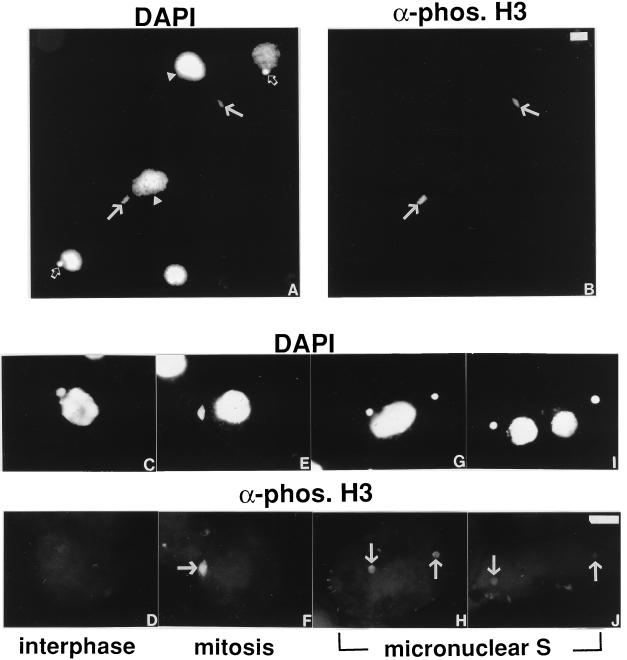
Figure 3.
H3 phosphorylation is temporally correlated with mitosis in Tetrahymena. Logarithmically growing Tetrahymena cells were stained with the phosphorylated H3 antibody (B, D, F, H, and J) and the DNA-specific dye DAPI (A, C, E, G, and I). In A and B, a random population of cells is shown in low magnification. The closed arrows denote mitotic micronuclei; the open arrows denote interphase micronuclei, and the arrowheads denote macronuclei. The bar represents 10 μm. In C, D, E, F, G, H, I, and J, cells with micronuclei in interphase, mitosis, and S phase are shown in high magnification. The arrows denote micronuclei. The bar represents 10 μm.
The above result suggests that, in Tetrahymena, H3 phosphorylation is correlated with mitosis not only spatially but also temporally. To investigate this relationship further, we examined H3 phosphorylation throughout the Tetrahymena cell cycle with particular emphasis on the micronuclear division cycle (i.e., entry and exit from mitosis).
The Tetrahymena cell cycle is somewhat unique in that the micronuclei and the macronuclei divide by different mechanisms (i.e., mitotically versus amitotically) and at different times in the cell cycle. Cell cycle stages can be determined by a combination of morphological criteria (micronuclear division, macronuclear division, cytokinesis, etc.) and cell size (for details, see refs. 27 and 28). In the majority of cells in a logarithmically growing population, the micronuclei rest in a pocket-like recess in the macronuclear surface, indicative of interphase macro- and micronuclei (Fig. 3C). As the micronuclei move away from the macronuclei, micronuclear mitosis initiates, and soon thereafter, the mitotic micronuclei display a characteristic football shape (Fig. 3E). Soon after the micronuclei finish dividing and macronuclear division initiates, micronuclear S phase begins (Fig. 3G). Micronuclear S phase continues for a short time after macronuclear division and cytokinesis finish; typically, these cells are small “half-size” cells and have small macronuclei (Fig. 3I). Correspondingly, interphase micronuclei are not stained by the phosphorylated H3 antibody (Fig. 3D). The staining becomes intensified as the micronuclei enter mitosis and acquire a characteristic football shape (Fig. 3F). The staining begins to diminish as the micronuclei exit mitosis and enter S phase (Fig. 3H). Staining of micronuclei with this antibody disappears close to when cytokinesis is finished and two daughter cells are born (Fig. 3J). Collectively, these data suggest that, as in mammalian cells (13), the singular phosphorylation of H3 at Ser-10 is also temporally correlated to mitosis.
Correlation between H3 Phosphorylation and Meiosis in Tetrahymena.
As far as we are aware, the possible association between H3 phosphorylation and meiotic chromosome condensation has not been reported. To investigate a possible relationship, we used the sexual pathway of the ciliate life cycle, conjugation. In brief, after pair formation in Tetrahymena, three prezygotic nuclear divisions occur (meiosis I, meiosis II, and a third prezygotic mitosis). Pronuclear exchange and fusion (karyogamy), two postzygotic nuclear mitoses, and differentiation of micronuclei and macronuclei then occur (for details, see refs. 29–31).
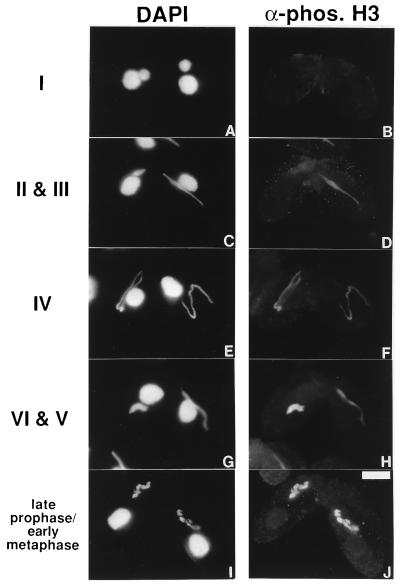
Remarkable changes in micronuclear morphology occur during Tetrahymena meiotic prophase (29–31). In stage I, the micronuclei become enlarged anterior to the macronuclei (Fig. 4A). The micronuclei then become elongated into a spindle- shaped crescent and enter stage II (left cell, Fig. 4C). In stage III, the micronuclei continue their elongation and become a thread with uneven thickness (right cell, Fig. 4C). In stage IV, the thread extends to the maximum length with uniform thickness (Fig. 4E). In stage V, the crescent shortens and forms a thicker rod-like structure (right cell, Fig. 4G). In stage VI, the thick-rod structure begins to disintegrate, and individual chromosomes begin to be visible (left cell, Fig. 4G). They soon enter diakinesis, and condensed chromosomal bivalents become apparent (Fig. 4I). Corresponding staining with the phosphorylated H3 antibody, as shown in Fig. 4 (Right), shows that H3 phosphorylation is absent in stages I and II, initiates in stage III, and increases through the rest of meiotic prophase until the condensed chromosomal bivalents are strongly stained.
Figure 4.
H3 phosphorylation during Tetrahymena meiotic prophase. Early conjugating Tetrahymena cells were stained with the phosphorylated H3 antibody (B, D, F, H, and J) and DNA-specific dye DAPI (A, C, E, G, and I). The bar represents 10 μm. Numbered stages correspond to those described in ref. 31.
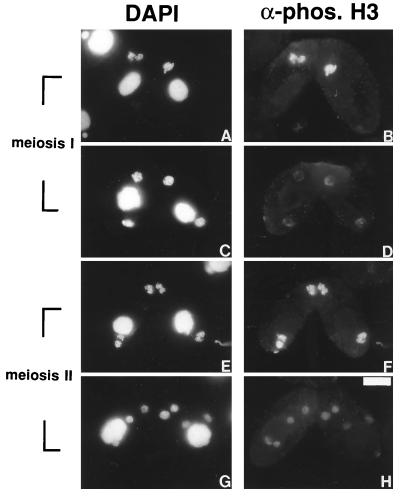
As micronuclei enter metaphase of meiosis I, the staining becomes maximal (right cell, Fig. 5 A and B). The staining begins to diminish in anaphase (left cell, Fig. 5 A and B) and fades away as the micronuclei exit meiosis I and the chromatin becomes decondensed (Fig. 5 C and D). The staining increases again as micronuclei enter the second division of meiosis (Fig. 5 E and F) and decreases as they exit meiosis, and the chromatin becomes decondensed (Fig. 5 G and H). As expected, H3 phosphorylation occurs during all mitotic divisions (pre- and postzygotic) during conjugation (data not shown and ref. 32). Thus, as for the mitotic divisions during vegetative growth, H3 phosphorylation, most likely at Ser-10, is closely correlated with chromosome condensation during both meiotic divisions. These results argue strongly that H3 phosphorylation plays a fundamental role in nuclear division-associated chromosome condensation.
Figure 5.
H3 phosphorylation is correlated with meiotic chromatin condensation in Tetrahymena. Late conjugating Tetrahymena cells were stained with the phosphorylated H3 antibody (B, D, F, and H) and DNA-specific dye DAPI (A, C, E, and G). The bar represents 10 μm.
DISCUSSION
H3 phosphorylation has been known to correlate closely with mitosis in mammalian cells (10, 11). However, the universality of this modification in facilitating chromosome condensation has not been well appreciated. In the ciliated protozoan Tetrahymena, H3 phosphorylation has been found to occur only in micronuclei, which divide mitotically, and not in macronuclei, which divide amitotically (15). Here, we demonstrate that Tetrahymena micronuclear H3 phosphorylation occurs at Ser-10, the same site targeted for phosphorylation in mammalian cells (10, 11). In addition, using an antibody specific for Ser-10 phosphorylated H3, we demonstrate that H3 phosphorylation is temporally correlated with micronuclear mitosis in Tetrahymena. These results, combined with our recent findings that mitotic H3 phosphorylation also occurs in fungi (Aspergillus nidulans), worms (Caenorhabditis elegans), and flies (Drosophila melanogaster) (38), strongly suggest that phosphorylation of H3 at Ser-10 is highly conserved among eukaryotes and plays an important, yet unclear, role in mitotic chromosome condensation.
Previously, there has been no report on the relationship between H3 phosphorylation and meiosis. Here, we demonstrate that, in Tetrahymena, H3 phosphorylation is temporally correlated with meiosis in a fashion that closely coincides with chromosome condensation. However, unlike mitosis, in which H3 phosphorylation occurs in late G2 before prophase (13), during Tetrahymena meiosis, the chromatin is not stained by the phosphorylated H3 antibody until stage III of meiotic prophase (Fig. 4), which corresponds to the zygotene stage of metazoan meiotic prophase (29, 30). This result suggests that the chromatin condensation observed in leptotene stage or stage I of Tetrahymena meiotic prophase (29, 30) is mechanistically different from chromatin condensation that occurs during meiotic metaphase or mitosis (similar results in maize have also been obtained by E. Kaszas and W. Z. Cande, personal communication).
Several differences exist between H3 phosphorylation in mammalian cells and in Tetrahymena. First, in micronuclei, a novel proteolytic processing mechanism occurs that cleaves H3S into H3F (22, 23), and only H3F is phosphorylated (15). However, H3F is not required for mitotic or meiotic chromosome condensation or for phosphorylation, because micronuclei undergo meiosis and mitosis without processing H3S to H3F, and H3S is phosphorylated during conjugation (32). Thus, although the significance of the proteolytic processing of H3S to H3F remains unknown, it is not invariably correlated with H3 phosphorylation or mitosis/meiosis. Second, in mammalian cells, H3 dephosphorylation, as detected by antibody staining, is complete immediately before detectable chromosome decondensation in telophase (13). In contrast, in Tetrahymena, H3 phosphorylation persists into the micronuclear S phase. This persistence may reflect some role(s) for H3 phosphorylation in S phase. More likely, slow dephosphorylation in Tetrahymena could be correlated with the fact that, unlike other mitotic nuclei, the transcriptionally inactive micronuclei do not decondense fully and do not need to decondense rapidly to resume transcription after mitosis. We suspect that subtle differences will exist between the timing of H3 phosphorylation and dephosphorylation in different organisms (also see ref. 13).
Clearly, mitotic and meiotic chromatin condensation are highly regulated and mechanistically complex events. Nonetheless, remarkable progress has recently been made by using Xenopus cell-free egg extracts to identify and purify proteins that seem to play a central role in mitotic chromosome condensation (33). The conserved nature of these molecules/pathways is underscored by the identification of genes encoding homologous proteins in yeast, worms, and flies (reviewed in refs. 34 and 35). Despite this progress, the molecular relationship between H3 phosphorylation and mitotic chromatin condensation remains unknown. Two models could serve to link H3 phosphorylation to other proteins involved in chromosome condensation. Phosphorylation of H3 may cause a local and transient decondensation of chromatin by reducing the interaction between H3 amino termini and DNA. In this case, the local and transient decondensation of chromatin would act as a first step to facilitate the binding of other trans-acting chromatin condensation factors, such as topoisomerase II and SMC/XCAP proteins (see ref. 4). Such a mechanism could explain why H3 phosphorylation also accompanies immediate-early gene transcription in response to mitogen stimulation (36, 37). Alternatively, cell cycle-regulated site-specific phosphorylation of H3 tails could act as a “receptor” to recruit chromatin condensation factors, which then in turn trigger mitotic chromatin condensation.
Acknowledgments
Drs. C. Glover and G. Abraham were involved in preliminary experiments that suggested that Ser-10 is the major, if not exclusive, phosphorylation site in Tetrahymena micronuclear H3. We thank Drs. E. Kaszas and W. Z. Cande for sharing the unpublished results in maize with us. Support and assistance from Upstate Biotechnology (Lake Placid, NY) is also gratefully acknowledged. This research was supported by National Institutes of Health Grants GM40922 (to C.D.A.) and GM21793 (to M.A.G.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: DAPI, diamidinophenolindole.
References
- 1.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe A P. Chromatin: Structure and Function. San Diego: Academic; 1995. pp. 72–77. [Google Scholar]
- 3.Bradbury E M. BioEssays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 4.Roth S Y, Allis C D. Trends Biochem Sci. 1992;17:93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- 5.Shen X, Yu L, Weir J W, Gorovsky M A. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 6.Ohsumi K, Katagiri C, Kishimoto T. Science. 1993;262:2033–2035. doi: 10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- 7.Dasso M, Dimitrov S, Wolffe A P. Proc Natl Acad Sci USA. 1994;91:12477–12481. doi: 10.1073/pnas.91.26.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X W, Th’ng J P, Swank R A, Anderson H J, Tudan C, Bradbury E M, Roberge M. EMBO J. 1995;14:976–985. doi: 10.1002/j.1460-2075.1995.tb07078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ajiro K, Yoda K, Utsumi K, Nishikawa Y. J Biol Chem. 1996;271:13197–13201. doi: 10.1074/jbc.271.22.13197. [DOI] [PubMed] [Google Scholar]
- 10.Gurley L R, D’Anna J A, Barham S S, Deaven L L, Tobey R A. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 11.Paulson J R, Taylor S S. J Biol Chem. 1982;257:6064–6072. [PubMed] [Google Scholar]
- 12.Ajiro K, Yasuda H, Tsuji H. Eur J Biochem. 1996;241:923–930. doi: 10.1111/j.1432-1033.1996.00923.x. [DOI] [PubMed] [Google Scholar]
- 13.Hendzel M J, Wei Y, Mancini M A, Van Hooser A, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 14.Gorovsky M A, Glover C, Johmann C A, Keevert J B, Mathis D J, Samuelson M. Cold Spring Harbor Symp Quant Biol. 1978;42:493–503. doi: 10.1101/sqb.1978.042.01.052. [DOI] [PubMed] [Google Scholar]
- 15.Allis C D, Gorovsky M A. Biochemistry. 1981;20:3828–3833. doi: 10.1021/bi00516a025. [DOI] [PubMed] [Google Scholar]
- 16.Gorovsky M A, Yao M C, Keevert J B, Pleger G L. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 17.Bruns P J, Brussard T B. J Exp Zool. 1974;188:337–344. doi: 10.1002/jez.1401880309. [DOI] [PubMed] [Google Scholar]
- 18.Allis C D, Dennison D K. Dev Biol. 1982;93:519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- 19.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lin R, Leone J W, Cook R G, Allis C D. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allis C D, Glover C V, Gorovsky M A. Proc Natl Acad Sci USA. 1979;76:4857–4861. doi: 10.1073/pnas.76.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allis C D, Bowen J K, Abraham G N, Glover C V, Gorovsky M A. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 24.Wenkert D, Allis C D. J Cell Biol. 1984;98:2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glover C V, Vavra K J, Guttman S D, Gorovsky M A. Cell. 1981;23:73–77. doi: 10.1016/0092-8674(81)90271-3. [DOI] [PubMed] [Google Scholar]
- 26.Gorovsky M A. In: The Molecular Biology of Ciliated Protozoa. Gall J G, editor. Orlando: Academic; 1986. pp. 227–261. [Google Scholar]
- 27.Yu S M, Horowitz S, Gorovsky M A. Genes Dev. 1987;1:683–692. [Google Scholar]
- 28.Wu M, Allis C D, Gorovsky M A. Proc Natl Acad Sci USA. 1988;85:2205–2209. doi: 10.1073/pnas.85.7.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray C., Jr J Protozool. 1956;3:88–96. [Google Scholar]
- 30.Sugai T, Hiwatashi K. J Protozool. 1974;21:542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 31.Martindale D W, Allis C D, Bruns P J. Exp Cell Res. 1982;140:227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- 32.Allis C D, Wiggins J C. Exp Cell Res. 1984;153:287–298. doi: 10.1016/0014-4827(84)90601-3. [DOI] [PubMed] [Google Scholar]
- 33.Hirano T, Mitchison T J. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 34.Koshland D, Strunnikov A. Annu Rev Cell Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 35.Heck M M. Cell. 1997;91:5–8. doi: 10.1016/s0092-8674(01)80002-7. [DOI] [PubMed] [Google Scholar]
- 36.Mahadevan L C, Willis A C, Barratt M J. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- 37.Barratt M J, Hazzalin C A, Cano E, Mahadevan L C. Proc Natl Acad Sci USA. 1994;91:4781–4785. doi: 10.1073/pnas.91.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei, Y. & Allis, C. D. (1998) Trends Cell Biol., in press.