Abstract
Background
The yeast Paf1 protein complex is required for efficient transcription elongation by RNA polymerase II (RNA pol II), but the precise role of the complex has been unclear.
Results
Here we show that depletion of the Ctr9 or Paf1 component of the Paf1 complex delays the loss of histones from the GAL1 gene upon induction. This delay in histone removal is accompanied by a decrease in association of RNA pol II with GAL1 and altered distribution of the polymerase along the locus.
Conclusion
These observations may explain why initial induction of GAL transcripts is reduced in Ctr9- or Paf1-deficient cells, and is consistent with a model suggesting that the Paf1 complex and the histone modifications that it mediates increase efficiency of transcriptional elongation by promoting nucleosomal destabilization and histone removal.
Background
Transcription by RNA polymerase (pol) II is subject to complex regulation at the level of chromatin modification and assembly. Several classes of chromatin modifying proteins have been identified; these contribute to gene activation by promoting chromatin decondensation, thereby allowing access of RNA pol II to regulatory sequences and facilitating movement of RNA pol II and associated factors through transcription units. ATP-dependent remodeling factors, including SWI/SNF and NURF, disrupt nucleosome structure and increase DNA accessibility [1]. Histone acetyltransferases (GNAT and MYST families) promote chromatin opening, nucleosome fluidity and protein interactions by acetylating histones at specific lysine residues [2]. Histone methyltransferases (CARM1 and MLL-families) modify histones at lysine and arginine; methylation of particular residues, for example, on histone H3 at K4 and K36, is linked to active transcription and may mark recently transcribed regions [3].
Efficient transcriptional elongation through chromatin requires additional factors, including FACT, Spt6 and Asf1, which are required for histone eviction and histone redeposition [4-7]. Mutation of each of these factors is associated with inappropriate nucleosomal positioning and activation of cryptic promoters within coding regions [8-10]. FACT and Asf1 may facilitate elongation by stimulating the eviction of H2A/H2B or H3/H4 dimers, respectively, from chromatin.
The Paf1 complex, which was first identified in yeast as a functional and physical cluster of proteins including Paf1, Ctr9, Cdc73, Rtf1 and Leo1 [11-13], is also required for efficient transcriptional elongation through chromatin [14,15]. The Paf1 complex is associated with transcriptionally active RNA pol II and interacts with the transcription elongation factors Spt4, Spt5, FACT and TFIIS [11,13,16]. Paf1 and Rtf1 are required for monoubiquitylation of histone H2B by Rad6-Bre1 at K123 [17,18]. This modification, in turn, is essential for the di- and tri-methylation of histone H3 by the Set1 and Dot1 methyltransferases at K4 and K79, respectively [19-21]. Moreover, Paf1 and Rtf1 promote the recruitment of Set1 and Set2, a histone H3 K36 methyltransferase, to RNA pol II at sites of active transcription [22,23].
In this communication we show that depletion of the Ctr9 or Paf1 component of the Paf1 complex is associated with delayed loss of histones from the GAL1 gene upon induction, decreased association of RNA pol II with GAL1 and altered distribution of the polymerase along the GAL1 locus. These observations are consistent with the interpretation that the Paf1 complex enhances transcriptional elongation by promoting nucleosomal destabilization and histone removal.
Results
Correlation of transciption initiation frequency with occupancy by Ctr9 and Paf1
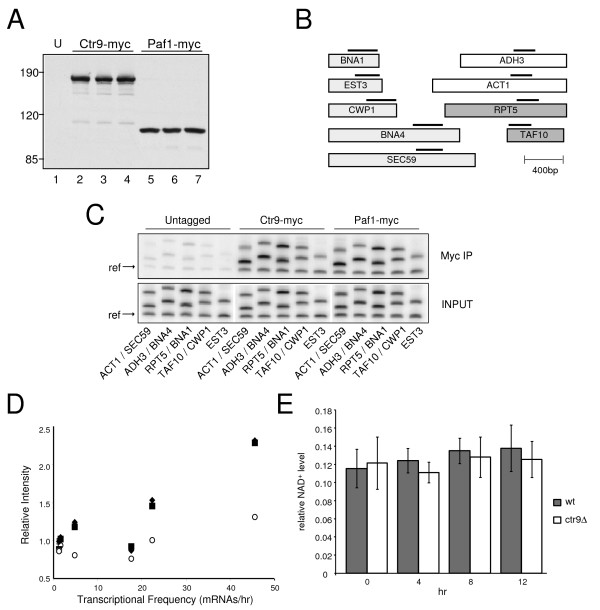
We began by examining association of Ctr9 and Paf1 with genes whose expression increased, decreased or remained unchanged upon depletion of Ctr9 or Paf1, as determined by microarray analysis ([24] and data not shown). Endogenous Ctr9 and Paf1 were each tagged at the carboxyl terminus with multiple c-myc epitopes; expression of tagged proteins was confirmed by immunoblotting (Fig. 1A). Association of tagged Paf1 and Ctr9 with specific open reading frames (ORFs) (Fig. 1B) was assayed by chromatin immunoprecipitation (ChIP).
Figure 1.
Gene occupancy by Ctr9 and Paf1. (A) Expression of endogenous, c-myc-tagged Ctr9 and Paf1 proteins. Protein from three Ctr9-myc-expressing isolates (HM198), three Paf1-myc-expressing isolates (HM199) or the untagged HM177 parental strain (U) were fractionated by SDS-PAGE. Protein was detected by immunoblotting with the 9E10 antibody. (B) Locations of gene-specific PCR probes. Each ORF is represented by a box. Light gray, genes whose expression decreased upon removal of Ctr9; dark gray, genes whose expression increased; white, genes whose expression remained unchanged. The positions and relative sizes of PCR products are indicated by dark bars above. (C) Assays of gene occupancy by ChIP. Association of Ctr9 and Paf1 with the regions indicated in (B) was monitored in cells maintained in YPD medium at 30°C. Upper panel, amplification of anti-myc immunoprecipitates; lower panel, amplification of input chromatin. The middle and upper bands in each lane represent amplified segments of the ORFs indicated (middle/upper) at bottom. The lowest band in each lane is an amplified fragment from the promoter region of ARN1, which was used as an internal reference [16]. (D) Correlation of transcriptional initiation frequency with gene occupancy by Ctr9 and Paf1. The relative signal intensities for each of six gene fragments associated with Ctr9-myc and Paf1-myc are plotted as a function of their transcriptional initiation frequencies as estimated in reference [25]. Diamonds, Paf1-myc; squares, Ctr9-myc; circles, untagged control. (E) Effect of acute Ctr9 depletion on intracellular NAD+ levels. Wild-type (HM167) and ctr9Δ (HM158) strains containing the Ctr9 expression plasmid pgCTR9M2 were initially grown in SC -leu + 2% galactose. Cells were then transferred to SC -leu + 2% glucose and maintained at 30°C. Relative intracellular levels of NAD+ were measured at 0, 4, 8 and 12 hours after switch to glucose. NAD+ levels are expressed on the Y axis as absorbance at 340 nm. Values represent mean and standard deviation (n = 3).
Contrary to expectation, occupancy of an ORF by either protein was not correlated with its change in expression upon depletion of Ctr9 (Fig. 1C). For example, association of Ctr9 and Paf1 was greatest with ACT1 and ADH3, two genes whose expression was unchanged in the ctr9Δ strain, while association of these proteins was lowest with EST3, whose expression decreased in response to Ctr9 depletion. Rather, the occupancy of a specific ORF by Ctr9 or Paf1 was positively correlated with the frequency of transcriptional initiation at that locus, as estimated by Young and coworkers ([25] and Fig. 1D). These findings suggest that the occupancy of a locus by the Paf1 complex is positively correlated with its occupancy by RNA pol II.
We next sought a common feature of transcriptional regulation among the genes in the downregulated set. A subset of BNA genes, which function in NAD+ biosynthesis, is down regulated in ctr9Δ cells (data not shown). These genes are coordinately repressed by the Sir2 homologue Hst1, a sensor of intracellular NAD+ [26]. This observation suggested that Ctr9 might function, directly or indirectly, in relief of repression by Hst1. Although the down regulation of BNA genes observed in ctr9Δ cells might have resulted from an increase in intracellular NAD+, no significant change in the amount of intracellular NAD+ was found (Fig 1E). Thus, the effect of Ctr9 depletion on expression of NAD+ biosynthetic genes was not likely to be mediated by NAD+ itself, but rather through a more direct effect on transcription. Together, these results raised the possibility that deleterious effects of Paf1 or Ctr9 depletion on transcription might be most readily observed at loci that are subject to fluctuation between active and silent states or that are capable of rapid induction.
Impaired transcription of telomeric genes upon loss of Ctr9
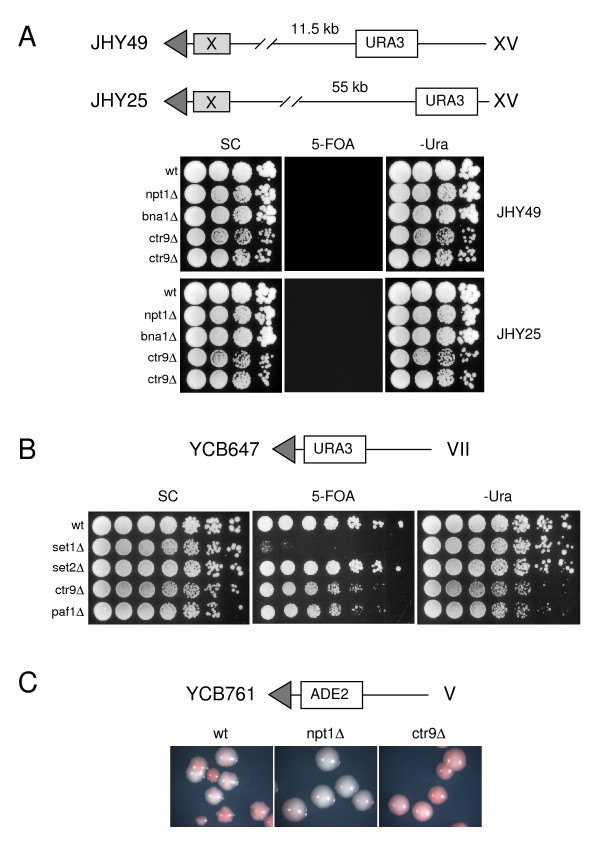
To test whether the Paf1 complex contributes to the establishment of transcriptionally permissive chromatin, we examined the effect of Ctr9 or Paf1 depletion on transcription of URA3 in its natural setting and at three other sites. Expression of URA3 from its natural locus was unaffected by loss of Ctr9 (data not shown). Additionally, expression of URA3 incorporated into sites not subject to telomeric silencing, either 11.5 or 55 kb downstream of the natural telomere on chromosome XV [27] was unperturbed (Fig. 2A). Wild-type and ctr9Δ cells, as well as the control strains npt1Δ and bna1Δ, which carry deletions in genes implicated in maintenance of silencing, grew similarly on plates lacking uracil, while growth was completely inhibited by 5-FOA. In contrast, when URA3 was integrated adjacent to an artificial telomere on chromosome VII [28], growth of Ctr9- or Paf1-deficient cells was inhibited on medium lacking uracil (Fig. 2B), indicating a context-dependent effect on URA3 expression. The growth of these cells was also impaired in the presence of 5-FOA, consistent with impairment of telomeric silencing, as previously noted [17,29], although the effects of CTR9 or PAF1 deletion were less severe than observed for a set1Δ strain (Fig. 2B).
Figure 2.
Position-dependent effects of CTR9 deletion on transcription and silencing. (A) The wild-type strains JHY49 and JHY25 each contain a single copy of URA3 integrated 11.5 Kb and 55 Kb, respectively, downstream of the natural left telomere of chromosome XV, as diagrammed above. Into these strains were introduced npt1Δ (HM193, HM190), bna1Δ (HM192, HM189) and ctr9Δ (HM191, HM188) deletions. Cells were diluted serially 10-fold and plated on SC, 5FOA or SC -ura as indicated. (B) The wild-type YCB647 strain carries a single copy of URA3 integrated into the left telomere of chromosome VII, as diagrammed above. Into this strain, set1Δ (HM206), set2Δ (HM207), ctr9Δ (HM180) or paf1Δ (HM205) deletions were introduced. Cells were diluted serially 5-fold and plated on SC, 5FOA, or SC -ura as indicated. (C) The wild-type YCB761 strain contains a single copy of ADE2 integrated into the right telomere of chromosome V as shown above. This and derivatives carrying npt1Δ (JS692) or ctr9Δ (HM182) deletions were plated on SC containing limiting adenine.
One interpretation of these results is that deletion of CTR9 or PAF1 impairs gene expression in the setting of an artificial telomere, while simultaneously reducing silencing. This interpretation was supported by examining expression of ADE2, integrated adjacent to an artificial telomere on chromosome V [28], in the presence or absence of Ctr9 (Fig. 2C). In this assay, cells expressing ADE2 appear white, while cells in which ADE2 is silenced appear red. On a wild-type background, as expected, sporadic loss of telomeric silencing at the ADE2 locus was associated with the appearance of red and white sectored colonies (Fig. 2C). Loss of NPT1, which encodes a component of the NAD+ salvage pathway, completely abolished silencing; as a result all colonies were white. On a ctr9Δ background, in contrast, colonies exhibited a uniform pink colour (Fig. 2C). This phenotype is consistent with expression of ADE2 at an intermediate level in most or all cells, as would be expected if loss of Ctr9 were to relieve telomeric silencing and at the same time to reduce transcriptional efficiency. These observations demonstrate position-dependent impairment of gene expression in the absence of Ctr9 and Paf1, and once again suggest that the Paf1 complex facilitates transcription through regions of chromatin that are undergoing a transition from inactive to active states. We proceeded to test this directly by examining the effects of Ctr9 and Paf1 depletion on induction of repressed loci.
Impaired induction of GAL genes in the absence of Ctr9 or Paf1
Ctr9 and Paf1 are required for monoubiquitylation of H2B at K123 [17,18], a process which in turn is necessary for efficient expression of GAL genes [30-32]. We examined the effects of Ctr9 or Paf1 depletion on induction of transcription at GAL loci. Strains were grown with raffinose as the sole carbon source before induction with galactose; this allowed us to bypass the process of derepression and focus solely on transcriptional activation.
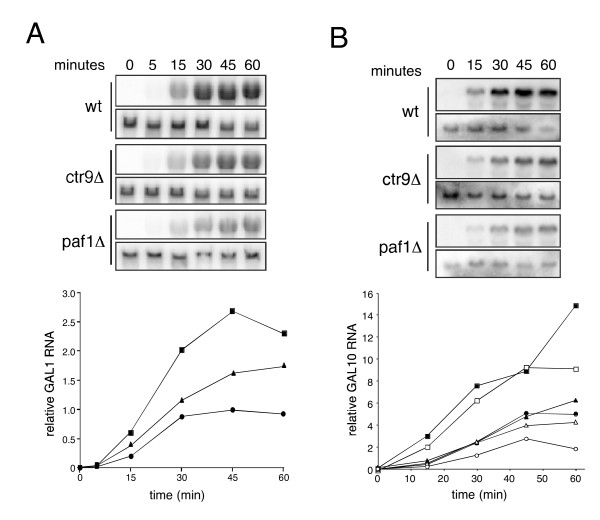
Induction of GAL1 RNA was impaired in both the ctr9Δ and paf1Δ strains, relative to wild-type (Fig. 3A). Deletion of CTR9 or PAF1 was associated with a 2- to 3-fold reduction in GAL1 RNA at 30 min after induction by galactose. We asked whether this decrease in RNA accumulation was accompanied by a change in transcriptional kinetics by assaying induction of GAL10 by galactose at 24°C. By inducing expression at a reduced temperature we hoped to unmask differences in the rate of transcript accumulation that might have been less pronounced at 30°C. We also used additional isolates of the wild-type, ctr9Δ and paf1Δ strains to militate against potential effects of secondary mutations cause by loss of Ctr9 or Paf1. Deletion of CTR9 or PAF1 had substantial effects on the initial accumulation of GAL10 transcripts, reducing RNA levels by about 5 fold at 15 min; by 30 min the difference in transcript levels between wild-type and the mutants was 3 fold and by 45 min this was reduced to only a 2-fold difference (Fig. 3B). We obtained similar results for GAL7 (data not shown). These results indicated that loss of the Paf1 complex is associated with delayed induction of GAL loci, consistent with preferential impairment of early transcription in the absence of the Paf1 complex, as has been described [14].
Figure 3.
Impaired induction of GAL loci in cells deficient in Paf1 or Ctr9. (A) Induction of GAL1 transcripts. Wild-type (HM200), ctr9Δ (HM201) and paf1Δ (HM202) strains were grown at 30°C in YEP supplemented with 2% raffinose. Galactose was added to a final concentration of 2%. RNA was isolated at 0, 5, 15, 30, 45 and 60 min after addition of galactose and GAL1 (upper panels) and ACT1 (lower panels) transcripts were detected by northern blotting (above). The GAL1 signal intensities were normalized to those of ACT1 and plotted as a function of time (below). Filled squares, wild-type; filled triangles, ctr9Δ ; filled circles, paf1Δ. (B) Induction of GAL10 transcripts. Isolates of wild-type (HM200), ctr9Δ (HM201) and paf1Δ (HM202) were grown at 24°C in media indicated above. Galactose was added as described and RNA was isolated at 0, 15, 30, 45 and 60 min after addition. GAL10 (upper panels) and ACT1 (lower panels) transcripts were detected by northern blotting (above). GAL10 signal intensities were normalized as in (A) and plotted together with data obtained from an independent experiment using a second set of wild-type, ctr9Δ and paf1Δ isolates. Filled and open squares, wild-type; filled and open triangles, ctr9Δ; filled and open circles, paf1Δ.
Loss of Ctr9 or Paf1 impairs displacement of histones upon GAL1 induction
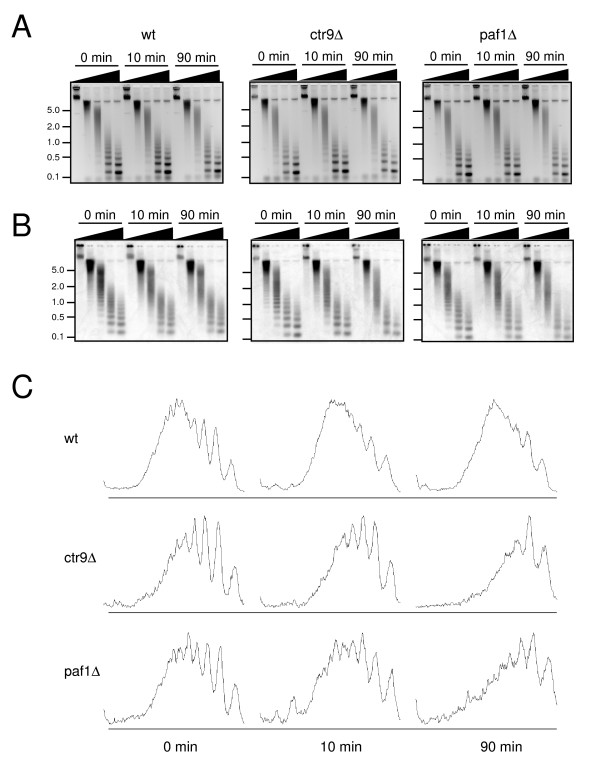
Induction of transcription at GAL loci is accompanied by loss of histones at promoter and coding regions [33-35]. We assessed the effects of CTR9 or PAF1 deletion on eviction of histones from the GAL1 locus upon induction. We first used micrococcal nuclease digestion of chromatin to examine nucleosomal occupancy and spacing before and after induction by galactose. Examination of bulk chromatin revealed no effect of CTR9 or PAF1 deletion on the general deposition or spacing of histones (Fig. 4A). Specific interrogation of the GAL1 locus, however, indicated that loss of either Ctr9 or Paf1 was associated with delayed displacement of histones from the gene. At 0 min, a discrete distribution of nucleosomal digestion products was observed in all samples. In the wild-type sample, this discrete distribution was largely lost by 10 min after galactose induction, and this pattern persisted at 90 min after induction (Fig. 4B and 4C). This suggested that in the wild-type strain, nucleosomes had been lost or their phasing disrupted by 10 min after induction. In contrast, discrete nucleosomal laddering was still observed at 10 min after galactose induction in the ctr9Δ and paf1Δ strains (Fig. 4B and 4C). By 90 min, nucleosome occupancy or phasing had been lost in all strains. The difference between wild-type and ctr9Δ or paf1Δ strains with respect to the kinetics of phased nucleosome loss was consistent with the interpretation that the Paf1 complex is required for the timely removal of histones from the GAL1 gene, or their displacement over the locus, upon induction of transcription. To distinguish between these possibilities, we used ChIP to determine whether depletion of Ctr9 or Paf1 affects the association of histones and RNA pol II with the GAL1 locus upon activation.
Figure 4.
Deletion of CTR9 or PAF1 impairs displacement of histones from the GAL1 locus upon induction. Wild-type (HM200), ctr9Δ (HM201) and paf1Δ (HM202) strains were grown at 24°C in YEP supplemented with 2% raffinose. Galactose was added to a final concentration of 2%. Nuclei were isolated at 0, 10 and 90 min after induction and digested with 0, 7, 15, 50 or 100 U of micrococcal nuclease. (A) Micrococcal nuclease digests of nuclei from wild-type (left), ctr9Δ (middle) and paf1Δ (right) strains were deproteinized and fractionated by agarose gel electrophoresis; DNA was detected by ethidium bromide staining. Inverse images of the stained gels are shown. Positions of markers and their sizes (in kb) are indicated at left. (B) Distribution of micrococcal nuclease digestion products at the GAL1 locus. DNA from gels in (A) was transferred to nylon and hybridized to a probe representing basepairs +422 to +925 of the GAL1 gene. Signal was detected by phosphorimaging. (C) Distributions of micrococcal nuclease products from the 50 U micrococcal nuclease reactions in (B) were quantified by densitometry for each strain at each time point. The signal intensities are plotted in arbitrary units.
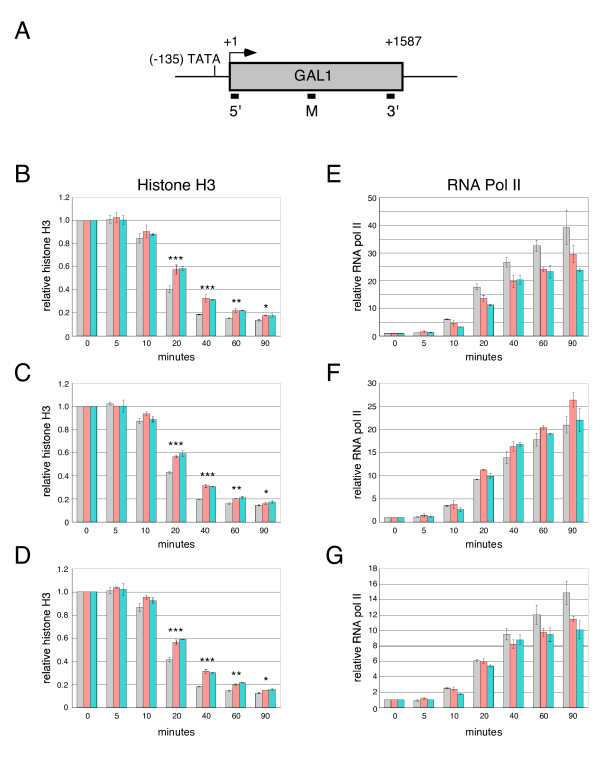
Association of histone H3 and RNA pol II with 5', middle and 3' regions of GAL1 (Fig. 5A) were examined in wild-type, ctr9Δ and paf1Δ strains maintained at 24°C before and at various times after induction by galactose. In wild-type cells, occupancy of GAL1 by histone H3 decreased sharply from 10 to 40 min after induction and remained low for the remainder of the time course (Fig. 5B – D). Deletion of either CTR9 or PAF1 impaired removal of histone H3 from the GAL1 locus (Fig. 5B – D). Before the 10 min time point, no statisically significant differences in histone H3 levels were detected among wild-type, ctr9Δ and paf1Δ samples. By 20 min, histone H3 occupancy was about 30% lower in wild-type cells than in the ctr9Δ and paf1Δ mutants and this difference increased to 40% by 40 min. These differences were determined to be highly significant (P < 0.0005). By 90 min after induction, depletion of histone H3 over the GAL1 locus in the ctr9Δ and paf1Δ cells began to approach that of wild-type, but nonetheless remained statistically higher (P < 0.04). These results suggest that the Paf1 complex promotes eviction of histones from the GAL1 locus during early stages of transcriptional induction. Moreover, these findings indicate that in the absence of the Paf1 complex, nucleosomes can be removed from the GAL1 locus upon activation, but with delayed kinetics relative to wild-type.
Figure 5.
Depletion of Ctr9 or Paf1 delays removal of histone H3 and alters the distribution of RNA pol II upon induction of GAL1 transcription. (A) Schematic representation of the GAL1 locus. The TATA region, translation start site and ORF are indicated. Black bars below indicate positions of PCR probes used in ChIP assays. (B – G) Association of histone H3 (B – D) or RNA pol II (E – G) with 5' (B, E), middle (C, F) and 3' (D, G) regions of the GAL1 locus as a function of time after induction. Wild-type (HM200), ctr9Δ (HM201) and paf1Δ (HM202) strains were grown at 24°C in YEP supplemented with 2% raffinose. Galactose was added to final concentration of 2%. Samples were taken at 0, 5, 10, 20, 40, 60 and 90 min after induction. Association of histone H3 (B – D) and Rpb3 (E – G) with 5' (B, E), middle (C, F) and 3' (D, G) regions of GAL1 were assayed by ChIP and real-time PCR for wild-type (gray), ctr9Δ (peach) and paf1Δ (turquoise) strains. All samples were also assayed with primers specific for an intergenic region (B – D) or FBA1 (E – G) as standards for normalization of the GAL1-specific signals. The normalized association of H3 or Rpb3 with each region of GAL1 at 0 min was set to 1; all other values represent the fold difference relative to the 0 min sample. Each value is the average of two experiments; error bars correspond to the standard errors of the mean. Statistical significance of pairwise comparisons between wild-type and ctr9Δ or paf1Δ strains was determined using a two-tailed t-test. ***, P < 0.0005; **, P < 0.004; *, P < 0.04.
In the nuclease digestion experiments we had observed more unphased DNA fragments of intermediate size in the wild-type sample than in the ctr9Δ and paf1Δ strains at 90 min (Fig. 4C). While this in principal could have reflected greater occupancy of the GAL1 locus by unphased nucleosomes in wild-type cells, our observation that removal of histones from GAL1 is more rapid in the wild-type than in ctr9Δ or paf1Δ strains argues against this point.
We observed an inverse relationship between histone displacement and the association of RNA pol II with the GAL1 locus. In wild-type cells, association of RNA pol II with GAL1 increased rapidly after addition of galactose (Fig. 5E – G). This association was apparent at 10 min after induction and continued to increase throughout the time course. The amount of RNA pol II at the 5' region was consistently greater than at the middle or 3' region of the gene, although the kinetics of polymerase association with these regions paralleled the kinetics observed for the 5' region (Fig. 5E – G). These findings are consistent with a report describing kinetics of association of RNA pol II with a galactose-inducible, synthetic locus [34].
Loss of Ctr9 or Paf1 was associated with changes in the association of RNA pol II with GAL1 and its distribution over the locus following induction. While the confidence limits determined for these differences were not below the significance threshold, similar trends were observed for the ctr9Δ and paf1Δ mutants. As with wild-type cells, association of RNA pol II with the 5' region of GAL1 was detectable in ctr9Δ and paf1Δ cells at 10 min after addition of galactose, but the amount of association was reduced (Fig. 5E). Throughout the time course, the amount of RNA pol II over the 5' region was consistently lower in the ctr9Δ and paf1Δ strains than in wild-type. Interestingly, loss of either Ctr9 or Paf1 was associated with a relative increase in the amount of RNA pol II associated with the middle region of the GAL1 locus (Fig. 5F), despite the reduction in the amount of polymerase associated with the 5' region. Only small differences in polymerase occupancy of the 3' region of GAL1 were observed among the strains (Fig. 5G). Deletion of CTR9 or PAF1 had similar effects on RNA pol II occupancy as well as histone displacement at the GAL10 locus (data not shown). These findings suggest that upon induction of transcription at GAL1 and GAL10 loci, the Paf1 complex promotes histone displacement and more efficient transcriptional elongation. We considered the possiblity that inappropriate internal initiations in the ctr9Δ and paf1Δ strains might contribute to alterations in the distribution of RNA pol II, but this seems unlikely as we observed no truncated GAL1 or GAL10 transcripts in hybridization assays of mutant cells using double-stranded probes, which would have detected transcripts from either strand (Fig. 3).
Discussion
It has been reported that the Paf1 complex associates with RNA pol II and that loss of the Ctr9 or Paf1 components of this complex impairs transcription of a subset of genes. We find that the amount of Paf1 associated with a particular gene is positively correlated with its rate of transcription initiation. The Paf1 complex supports specific histone modifications that are essential for maintenance of telomeric silencing, including monoubiquitylation of histone H2B and methylation of histone H3 at lysine 4. In agreement with previous reports, we detect a loss of telomeric silencing in CTR9 and PAF1 deletion mutants, but demonstrate in addition that the loss of these genes also impairs transcription of telomeric markers.
The Paf1 complex is likely required for efficient transcriptional elongation [15], but the underlying mechanism has remained unclear. A recent report indicates that Paf1 complex-dependent ubiquitylation of H2B is required for efficient transcription elongation in vitro, and further suggests that ubiquitylation of H2B permits FACT to efficiently displace H2A/H2B histone dimers [14]. Here we have shown that the loss of Ctr9 or Paf1 in vivo is associated with delayed removal of histones from the GAL1 locus upon induction of transcription. This delay in histone displacement may contribute to the decrease in association and altered distribution of RNA pol II that we observed in Ctr9- or Paf1-deficient cells. Our findings may also explain the relative delay in induction of GAL transcripts in ctr9Δ and paf1Δ strains, relative to wild-type.
We have considered the possibility that a reduction in recruitment of RNA pol II, rather than an impairment of histone eviction, might be responsible for the delay in histone displacement that we observed. In support of our interpretation are two reports documenting recruitment of the Paf1 complex to promoter regions after, but not before, initiation of transcription. In mammalian cells the Paf1 complex is recruited to the RARβ2 promoter after recruitment of RNA pol II and FACT [14]. Moreover, RNA pol II binds to the yeast ARG1 promoter in the absence of the Paf1 complex; subsequent recruitment of the Paf1 complex is dependent on prior binding of the Spt4 transcription elongation factor and phosphorylation of RNA pol II at Ser5 [36]. Lastly, association of Rad6 and the SAGA complex with the GAL1 promoter occurs independently of H2B monoubiquitylation [31,32]. Taken together, these observations suggest that recruitment of RNA pol II to promoter regions occurs prior to and independent of the actions of Ctr9 or Paf1. Our results are consistent with a direct role for the Paf1 complex in promoting histone eviction but do not exclude the possibility that Paf1 and Ctr9 exert their effects through one or more additional components of the elongating RNA pol II complex.
It has been proposed that newly transcribed genes undergo a so-called "pioneer round" of transcription in which RNA pol II functions in concert with specialized machinery for chromatin modification [37]. Based on our work and earlier findings we suggest that upon initial induction of GAL1, Gal4, Rad6/Bre1, SAGA and RNA pol II, among other factors, are recruited and initiate transcription [31]. Subsequent association of the Paf1 complex with RNA pol II would promote monoubiquitylation of H2B at K123 by Rad6/Bre1 as the polymerase complex traverses the coding sequence [32]. We imagine that ubiquitylation of K123 could confer upon FACT and Asf1 the ability to evict histones from the DNA in addition to the established roles of FACT and Asf1 in histone displacement and redeposition. Our results suggest that in the absence of the Paf1 complex histones are inefficiently removed and transcriptional elongation is impaired.
While it has been suggested that the Paf1 complex does not affect transcriptional elongation from a GAL1 promoter [38], this conclusion was based on observations made at 2.5 hr after galactose induction. Because our data indicate that the effects of Ctr9 and Paf1 depletion on histone eviction are most evident early after induction, it is not surprising that differences in elongation rates were not observed at a relatively late time point. Our results suggest that the Paf1 complex exerts its effects on histone displacement during the initial stages of transcriptional activation, and that at later times after induction, histones can be removed even in the absence of the Paf1 complex.
The Paf1 complex and the H2B monoubiquitylation that it promotes are clearly not universally required for gene expression [39]. Unlike FACT, the genes encoding components of the Paf1 complex and Rad6/Bre1 are inessential [18,40,41]. The available data suggest that the Paf1 complex enhances transcriptional efficiency in those settings in which environmental changes demand rapid transcriptional responses, such as induction of metabolic enzymes or stress response genes. In this regard, it is particularly interesting that one salient phenotype of paf1Δ and ctr9Δ strains is sensitivity to a broad range of environmental stressors [42].
Conclusion
In this communication we have shown that loss of either Paf1 or Ctr9 is associated with impaired induction of transcription at the GAL1 and GAL10 loci and altered recruitment and distribution of RNA pol II across the GAL1 locus. These effects are accompanied by delayed removal of histones from the GAL1 locus upon induction of transcription in Paf1- or Ctr9-deficient strains. Our findings suggest that the Paf1 complex and the histone modifications that it mediates increase the efficiency of transcriptional elongation by promoting destabilization of nucleosomes and histone removal.
Methods
Media, yeast strains and plasmids
Growth media were as described [43]. Glucose was the carbon source except where indicated. Yeast were maintained at 30°C unless noted. The strains used in this study were generated using standard techniques [44] and are listed in Table 1. Genetic manipulations were verified by Southern hybridization or PCR.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
| HM158 | Mat a his3Δ1 leu2Δ0 ura3Δ0 ctr9::kanMX4 [pgCTR9M2] |
| HM167 | Mat a his3Δ1 leu2Δ0 ura3Δ0 [pgCTR9M2] |
| HM177 | Mat a his3Δ1 leu2Δ0 ura3Δ0 |
| HM198 | Mat a his3Δ1 leu2Δ0 ura3Δ0 CTR9-13 Myc::kanMX6 |
| HM199 | Mat a his3Δ1 leu2Δ0 ura3Δ0 PAF1-13 Myc::kanMX6 |
| HM200 | Mat a his3Δ1 leu2Δ0 |
| HM201 | Mat a his3Δ1 leu2Δ0 ctr9Δ::kanMX4 |
| HM202 | Mat a his3Δ1 leu2Δ0 paf1Δ::kanMX4 |
| YCB647a | Mat a his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 leu2Δ::TRP1 ADH4::URA3-TEL |
| HM180 | Mat a his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 leu2Δ::TRP1 ctr9Δ::kanMX4 ADH4::URA3-TEL |
| HM205 | Mat a his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 leu2Δ::TRP1 paf1Δ::kanMX4 ADH4::URA3-TEL |
| HM206 | Mat a his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 leu2Δ::TRP1 set1Δ::kanMX4 ADH4::URA3-TEL |
| HM207 | Mat a his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 leu2Δ::TRP1 set2Δ::kanMX4 ADH4::URA3-TEL |
| YCB761a | Mat a ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 TEL-VR::ADE2 |
| HM182 | Mat a ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctr9Δ::kanMX4 TEL-VR::ADE2 |
| JS692a | Mat a ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 npt1Δ::kanMX4 TEL-VR::ADE2 |
| JHY25b | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15;AE – CAN1/ADE2/URA3 (XV) |
| HM188 | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15 ctr9Δ::kanMX4; |
| AE – CAN1/ADE2/URA3 (XV) | |
| HM189 | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15 bna1Δ::kanMX4; |
| AE – CAN1/ADE2/URA3 (XV) | |
| HM190 | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15 npt1Δ::kanMX4; |
| AE – CAN1/ADE2/URA3 (XV) | |
| JHY49b | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15; TELO – CAN1/ADE2/URA3 (XV) |
| HM191 | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15 ctr9Δ::kanMX4; |
| TELO – CAN1/ADE2/URA3 (XV) | |
| HM192 | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15 bna1Δ::kanMX4; |
| TELO – CAN1/ADE2/URA3 (XV) | |
| HM193 | Mat a ade2::hisG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::MET15 npt1Δ::kanMX4; |
| TELO – CAN1/ADE2/URA3 (XV) |
aRef 28.
bRef 27.
To construct a plasmid for expression of Ctr9, PCR was used to generate a DNA fragment encoding full-length CTR9, tagged at the carboxyl terminus with two c-myc epitopes; this was transferred to the p415-GAL1 expression vector to generate pgCTR9M2 [45].
Measurement of intracellular NAD+
Strains were grown in SC -leu + 2% galactose to an O.D. of 1.0. An aliquot of each culture was removed for the 0 hr time point. The remaining cells were transferred to SC -leu + 2% glucose. At 4, 8 and 12 hr, intracellular NAD+ were assayed as described [46].
Chromatin immunoprecipitation
Strains expressing endogenous, c-myc-tagged Ctr9 or Paf1, and the untagged parental strain, were grown in YPD. ChIP was performed as described [47]; cultures were treated with 1% formaldehyde for 30 min. DNA fragments were immunoprecipitated with the 9E10 anti-c-myc antibody. Association of c-myc-tagged Ctr9 and Paf1 with specific DNA segments was monitored by PCR amplification. PCR reactions (25 μl) contained 1/50 of the precipitated DNA, 0.25 mM of each dNTP, 12.5 pmol of each primer, 0.75 μCi [32P]-dCTP and 1.6 U Taq polymerase (Roche). Reactions were incubated for 4 min at 95°C, followed by 22 amplification cycles (40 sec at 95°C, 40 sec at 52°C, 45 sec at 72°C) and a 5 min extension step at 72°C.
The following gene segments (defined relative to translation start sites) were assayed for the presence of Ctr9 and Paf1: ACT1 (+1516 to +1737 bp), ADH3 (+569 to +818 bp), BNA1 (+201 to +522 bp), BNA4 (+1889 to +2225 bp), CWP1 (+1401 to +1719 bp), EST3 (+268 to +534 bp), RPT5 (+758 to +994 bp), SEC59 (+918 to +1206 bp) and TAF10 (+8 to +243 bp). A fragment from the ARN1 promoter (-260 to -83 bp) was used as a reference.
For ChIP assays after galactose induction, cells were grown at 24°C to an O.D. of 0.8 – 0.9 in YEP with 2% raffinose. Cultures were induced with 2% galactose and samples were removed at 0, 5, 10, 20, 40, 60 and 90 min post-induction. Chromatin was prepared as described above. DNA was precipitated using the anti-Rpb3 antibody 1Y26 (Neoclone) and an anti-histone H3 antibody (Abcam). Specific DNA was detected by real time PCR (ABI). Sequences from 5' (+17 to +80), middle (+633 to +696) and 3' (+1374 to +1440) regions of GAL1 were detected with SYBR Green (ABI) using standard amplification conditions (40 cycles; 95°C for 15 s, 60°C for 1 min). A portion of the FBA1 gene and an untranscribed, intergenic region (between NRG1 and HEM13) were amplified as controls. Precipitation of each region of GAL1 was normalized to that of the FBA1 (RNA pol II) or intergenic (H3) controls and expressed relative to the amount of association prior to galactose induction.
Assays of telomeric silencing
URA3 strains were grown on SC plates. Serial dilutions were spotted onto SC, SC -ura, and 5 FOA (0.1%) plates. To assay ADE2 silencing, strains were grown on SC plates. About 200 cells from each strain were plated on SC containing limiting adenine.
GAL induction assays
Strains were grown at 30°C to O.D. 0.4 – 0.5 in YEP supplemented with 2% raffinose. An aliquot of each culture (10 O.D. units) was removed and frozen at -80°C. To the remainder of the cultures, galactose was added to 2%. At 5, 15, 30, 45 and 60 min after addition of galactose, cells (10 O.D. units) were sampled. A second induction assay was repeated as above with minor changes. Cells were grown at 24°C to an O.D. of 0.6. Samples were removed at 15, 30, 45 and 60 min after addition of galactose. GAL1, GAL10 and ACT1 transcripts were assayed by northern hybridization.
Micrococcal nuclease digestion
Digest of genomic chromatin with micrococcal nuclease was performed as described with the minor modifications indicated below [48]. Yeast strains were grown at 24°C to an O.D. of 0.8 – 0.9 in YEP supplemented with 2% raffinose. Cultures were induced with 2% galactose for 0, 10 and 90 min. The preincubation solution consisted of 0.7 M β-mercaptoethanol, 10 mM EDTA (pH 8.0). Spheroplasts were prepared by addition of 3.5 mg of zymolase-100T (Seikagaku) per g of cells and the suspension was shaken at 37°C for 10 min. Samples were spun at 48,000 × g at 4°C for 30 min. Samples were digested with 0, 7, 15, 50 or 100 U micrococcal nuclease (USB). After deproteinization, samples (20 μg) were resolved on 1.1% agarose and bulk DNA was detected with ethidium bromide. GAL1 DNA was detected by Southern hybridization.
Authors' contributions
HAM conceived and executed all experiments and drafted the manuscript. SD helped to conceive and design the study, analyzed data and drafted the manuscript. Both authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by the Howard Hughes Medical Institute and grant RO1HL07391.
Contributor Information
Heather A Marton, Email: hmarton@jhmi.edu.
Stephen Desiderio, Email: sdesider@jhmi.edu.
References
- Johnson CN, Adkins NL, Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol. 2005;83:405–417. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- Verdone L, Caserta M, Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin Modifications by Methylation and Ubiquitination: Implications in the Regulation of Gene Expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Speer JL, Lindstrom DL. Transcript elongation on a nucleoprotein template. Biochim Biophys Acta. 2002;1577:276–286. doi: 10.1016/s0167-4781(02)00458-x. [DOI] [PubMed] [Google Scholar]
- Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J Biol Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. Embo J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/S1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/S0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/S0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke J. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/S1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. Embo J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. Embo J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Shadle T, Bhaumik SR. Functional analysis of H2B-Lys-123 ubiquitination in regulation of H3-Lys-4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J Biol Chem. 2006;281:19045–19054. doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- Malone EA, Clark CD, Chiang A, Winston F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5710–5717. doi: 10.1128/mcb.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD(+) salvage pathway. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics 2000: A Cold Spring Harbor Laboratory Course. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Bernardi F, Koller T, Thoma F. The ade6 gene of the fission yeast Schizosaccharomyces pombe has the same chromatin structure in the chromosome and in plasmids. Yeast. 1991;7:547–558. doi: 10.1002/yea.320070603. [DOI] [PubMed] [Google Scholar]