Abstract
The plasma membrane nanoscale distribution of H-ras is regulated by guanine nucleotide binding. To explore the structural basis of H-ras membrane organization, we combined molecular dynamic simulations and medium-throughput FRET measurements on live cells. We extracted a set of FRET values, termed a FRET vector, to describe the lateral segregation and orientation of H-ras with respect to a large set of nanodomain markers. We show that mutation of basic residues in helix α4 or the hypervariable region (HVR) selectively alter the FRET vectors of GTP- or GDP-loaded H-ras, demonstrating a critical role for these residues in stabilizing GTP- or GDP-H-ras interactions with the plasma membrane. By a similar analysis, we find that the β2–β3 loop and helix α5 are involved in a novel conformational switch that operates through helix α4 and the HVR to reorient the H-ras G-domain with respect to the plasma membrane. Perturbation of these switch elements enhances MAPK activation by stabilizing GTP-H-ras in a more productive signalling conformation. The results illustrate how the plasma membrane spatially constrains signalling conformations by acting as a semi-neutral interaction partner.
Keywords: FRET, microdomain, nanocluster, plasma membrane, Ras
Introduction
Ras proteins are small GTPases that operate on the inner plasma membrane and other cellular membranes to regulate cell growth, proliferation and differentiation (Hancock, 2003). The mammalian Ras proteins H-, N- and K-ras comprise a near identical G-domain (residues 1–165), which binds guanine nucleotides, and interacts with effector proteins and exchange factors, but have markedly different C-terminal plasma membrane anchors. The anchors are attached as post-translational modifications of the hypervariable region (HVR, residues 166–188/9), which exhibits <15% homology between Ras isoforms (Hancock, 2003). The minimal Ras membrane anchors comprise a common C-terminal S-farnesyl cysteine carboxy methylester operating in concert with adjacent S-palmitoyl cysteine residues in N- and H-ras, or a polylysine domain in K-ras (Hancock et al, 1989, 1990; Hancock and Parton, 2005). These anchors are connected to the G-domain by the linker region comprising the remaining HVR sequence.
The classical Ras isoforms H-, N- and K-ras are nonrandomly arrayed on the plasma membrane in largely non-overlapping dynamic nanoclusters (Prior et al, 2003a; Rotblat et al, 2004; Hancock and Parton, 2005; Plowman and Hancock, 2005). This spatial organization is critical for Ras signal transduction because Ras nanoclusters allow cells to use digital signalling across the plasma membrane to generate high-fidelity signal transmission (Tian et al, 2007). The minimal C-terminal lipid anchors of Ras proteins are sufficient to drive nanoclustering on the plasma membrane (Prior et al, 2003a; Plowman and Hancock, 2005); however, full-length H- and N-ras proteins exhibit guanine nucleotide-dependent changes in lateral segregation (Hancock, 2003; Hancock and Parton, 2005). This is best characterized for H-ras, which operates in different types of nanocluster when GDP and GTP loaded. It has been shown previously that the di-palmitoylated and farnesylated C-terminal HVR and the G-domain are required for GTP-dependent changes in H-ras lateral segregation (Prior and Hancock, 2001; Prior et al, 2003a; Rotblat et al, 2004; Roy et al, 2005), but precisely how the interactions of the HVR with the lipid bilayer of the plasma membrane are controlled by conformational changes in the G-domain is unknown.
Results and discussion
H- and K-ras membrane anchors exhibit different lateral segregation FRET vectors
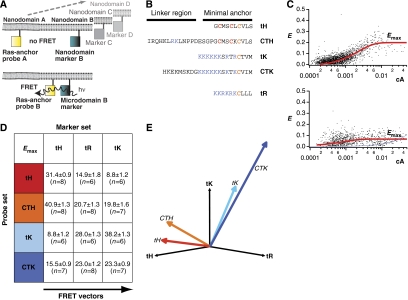
To explore the structural basis of guanine nucleotide-regulated membrane organization of H-ras in live cells, we used a novel FRET assay that can map any arbitrary number of probe molecules relative to a given set of markers (Abankwa and Vogel, 2007). We used mCFP- or mCit-tagged Ras constructs and complementary tagged nanodomain markers (Figure 1A and B; Supplementary Figure S1B and C). FRET on a 2D membrane depends on the acceptor surface density (Wolber and Hudson, 1979; Berney and Danuser, 2003; Kiskowski and Kenworthy, 2007), we therefore analysed this dependence for each FRET pair that was transiently coexpressed in BHK cells and determined Emax, which is the FRET efficiency reached at high acceptor concentrations (equation (1); Figure 1C) (Abankwa and Vogel, 2007). To describe the lateral segregation of a protein, we introduced the FRET vector, which is the set of Emax values of FRET pairs of a protein and various nanodomain markers (Figure 1A and E). We first determined FRET vectors for the minimal membrane anchors of H- and K-ras4B (tH and tK, respectively) and their complete HVR counterparts (CTH and CTK, respectively) in relation to three nanodomain markers, tH, tK and the minimal membrane anchor of Rac-1, tR (Abankwa and Vogel, 2007) (Figure 1A and B). All four membrane anchors had different FRET vectors, suggesting that they are laterally segregated (Figure 1D and E), consistent with previous EM analysis (Prior et al, 2003a; Rotblat et al, 2004; Plowman and Hancock, 2005). Therefore, FRET vectors can capture subtle differences in lateral segregation that in this case are associated with amino-acid sequences in the HVR.
Figure 1.
FRET vectors describe the lateral segregation of Ras proteins. (A) The extent of FRET between pairs of mCFP (blue squares) and mCit (yellow squares)-labelled Ras probes and complementary labelled nanodomain markers was measured. The nanodomain probing constructs were either Ras-derived membrane anchors or full-length H-Ras mutants (Supplementary Figures S1C and S4). High FRET values are expected for proteins that co-cluster, whereas a random distribution will result in no or very little FRET. In addition, markers of different lengths enable detection of conformational changes (Supplementary Figure S1B). The nanodomain markers C and D indicate that a set of nanodomain markers is used sequentially to characterize the lateral segregation of the Ras-derived probes by a set of FRET values. (B) Amino-acid sequences of Ras membrane anchors used for FRET experiments. (C) All mCFP/mCit-tagged proteins showed predominant localization to the plasma membrane and were homogeneously distributed when imaged by confocal microscopy (Supplementary Figure S4). We, therefore, increased the throughput of the analysis of >140 FRET pairs, by measuring fluorescence signals in a cytometer. The dependence of the FRET efficiency, E, on the normalized acceptor surface concentration, cA, at ∼1:1 donor–acceptor ratio was analysed using equation (1) (red curves), which yielded the characteristic FRET value, Emax. The Emax for most FRET pairs was higher than expected for randomly distributed donor and acceptor species (lower panel, calculated curve in blue), indicative of nanoclustering (Abankwa and Vogel, 2007). Data points are calculated from single cells, the example plots shown are mCFP-H-rasG12V/mCit-tH (upper panel) and mCFP-K-rasG12V/mCit-tH (lower panel). (D) The sample matrix shows Emax values (±s.e.m. and number of independent experiments n) for Ras membrane anchor probe and marker FRET pairs. The lateral segregation of each Ras membrane anchor probe is described by the FRET vector, given in this example by the set of three Emax values in each row. (E) FRET vectors can be plotted in a nanodomain marker 'space'. The direction of a vector describes the lateral segregation of a probe. Thus, the more similar the direction of FRET vectors, the more similar is the lateral segregation of the marker probes. The colouring matches that in (D).
When mapped in 3D the FRET vectors of H-ras-derived anchors tH and CTH showed the same directionality (Figure 1E), with a correlation coefficient, r=0.99 (Supplementary Figure S1A). Similarly, FRET vectors of K-ras4B-derived anchors tK and CTK were highly correlated (r=0.96). In contrast, FRET vectors of different isoforms were only moderately correlated (Figure 1E). Thus, biochemically similar membrane anchors, for example, tH and CTH, have similar FRET vectors, whereas biochemically different membrane anchors, such as tH and tK do not. The similarity of the lateral segregation of the H-ras-derived membrane anchors may be rationalized in the light of recent molecular dynamics (MD) simulations (Gorfe et al, 2007a, 2007b), which show that the membrane contacts and penetration depth of the isolated CTH and tH anchors are very similar. It is, therefore, conceivable that this similar mode of membrane anchorage is the basis for their similar lateral segregation, which we measure as highly correlated FRET vectors. To examine the relationship between EM nanoclustering parameters and Emax, we considered individual Emax values from the FRET vectors. Emax values decreased in the order Emax(CTH/CTH)(=52±4, n=10)>Emax(tK/tK)>Emax(tH/tH) (Figure 1E), which correlate with the decreasing density and increasing radius of the corresponding nanoclusters measured by EM (Prior et al, 2003a; Rotblat et al, 2004; Plowman et al, 2005). Moreover, the smallest value was Emax(tH/tK)=9±3 (Figure 1D), which represents the expected value for unclustered fluorophores (Figure 1C). This Emax value is again consistent with EM analysis, which shows that tK and tH are in spatially segregated nanodomains (Prior et al, 2003a). Therefore, any given Emax value captures nanoclustering data on a probe–marker FRET pair.
Basic residues in the HVR and helix α4 stabilize H-ras membrane interactions
To gain more insight into the guanine nucleotide dependence of H-ras membrane anchorage, we recently simulated GTP- and GDP-loaded H-ras bound to a membrane bilayer using MD (Gorfe et al, 2007b). The models we obtained showed specific contacts of basic residues on helix α4 and in the HVR that stabilized GTP-H-ras in a different orientation with respect to the plane of the membrane from GDP-H-ras (Gorfe et al, 2007b) (Figure 2A and B). To capture changes in H-ras orientation, we expanded the FRET assay by constructing H-ras-derived nanodomain markers with different lengths of linker sequence between the anchor and fluorophore (Supplementary Figures S1B, C and S4). We reasoned that these additional probes should detect orientation changes without the necessity to interpret FRET values in absolute terms, which would require precise knowledge of the distribution and average geometrical arrangement of donor and acceptor fluorophores in a nanocluster (Nazarov et al, 2006, 2007). To maximize the discriminatory power of the FRET assay, we used eight nanodomain markers yielding eight-tuple FRET vectors to integrate orientation and lateral segregation information that together define H-ras membrane organization (Supplementary Figure S1B and C).
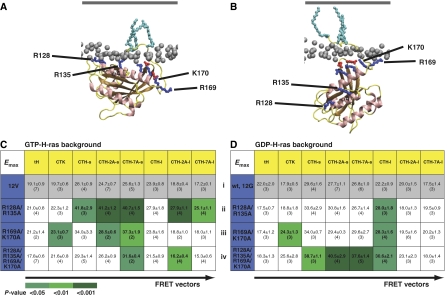
Figure 2.
Mutation of basic residues in helix α4 or the HVR has differential effects on the FRET vectors of GTP- and GDP-H-ras. MD simulations (Gorfe et al, 2007b) suggest GTP- and GDP-H-ras have different membrane interactions and orientations of the G-domain with respect to the plane of the membrane. (A) The GTP conformation is stabilized by membrane contacts of R128 and R135 on helix α4. (B) These contacts are lost in GDP-H-ras, which is stabilized by contacts of residues R169 and K170 in the HVR. Phosphorous atoms of lipid head groups of the inner membrane leaflet are shown in grey, the outer leaflet is shown as a grey line (not to scale) and H-ras lipid anchors are in light blue. Important basic residues in H-ras are shown in dark blue and acidic residues in red. (C, D) The matrices show eight-tuple FRET vectors for GTP- and GDP-H-ras with mutations at the indicated residues. Each FRET vector is the set of Emax values of a mCFP-tagged H-ras mutant (blue) and eight mCit-tagged nanodomain markers (yellow). The matrix shows Emax values±s.e.m. and number of independent experiments in brackets. Emax values of each mutant–marker pair were compared with the corresponding value in the cognate wild-type background (grey FRET vectors) using t-tests; significant differences are shown by green shading. The matrices show different patterns of altered Emax values in the GTP and GDP background.
For the remainder of the study, we compared the effect of specific mutations on the membrane organization of fluorescently tagged H-rasG12V, which is constitutively GTP loaded (hereafter, GTP-H-ras) and fluorescently tagged wild-type H-ras under serum-starved conditions, which is >95% GDP loaded (hereafter, GDP-H-ras) (Prior et al, 2001). We first introduced alanine substitutions at residues R128 and R135 in helix α4, which are predicted to contact the membrane only in GTP-H-ras (Gorfe et al, 2007b) (Figure 2A) and determined the FRET vectors of R128A/R135A mutants in GTP and GDP backgrounds. Figure 2C (ii) shows that five Emax values of the eight-tuple FRET vector of GTP-H-ras-R128A/R135A were significantly altered. In contrast, the FRET vector of GDP-H-ras-R128A/R135A was almost unchanged, with only one Emax value being significantly different (Figure 2D (ii)). These data, therefore, strongly suggest a GTP-specific role for residues R128 and R135 in maintaining the membrane organization of H-ras. The GDP-H-ras MD model shows stabilizing membrane contacts of R169 and K170 in the HVR (Figure 2B). Correspondingly, the mutations R169A/K170A significantly changed two Emax values of the GDP-H-ras FRET vector (Figure 2D (iii)). We also observed significant changes to three Emax values of the GTP-H-ras-R169A/K170A FRET vector (Figure 2C (iii)), suggesting that these residues contact the membrane in both GTP- and GDP-bound H-ras. However, these GTP and GDP FRET vectors were not identical, indicating some guanine nucleotide specificity. This interpretation is supported by an analysis of H-ras proteins with all four basic residues converted to alanines. The FRET vector of GDP-H-ras-R128A/R135A/R169A/K170A (Figure 2D (iv)) showed four significant changes to Emax values, whereas only two such changes were found in the FRET vector of GTP-H-ras-R128A/R135A/R169A/K170A (Figure 2C (iv)). Comparison of the latter with the vector of GTP-H-ras-R128A/R135A, reveals that wild-type residues R169 and K170 were required for the five Emax changes in the GTP-H-ras-R128A/R135A FRET vector, suggesting that residues R128/R135 (helix α4) and R169/K170 (HVR) do not function independently (Figure 2C). A similar argument can be advanced for GDP-H-ras, as the FRET vector of GDP-H-ras-R128A/R135A/R169A/K170A showed four significantly different Emax values (Figure 2D (iv)), whereas those of R128A/R135A (Figure 2D (ii)) or R169A/K170A (Figure 2D (iii)) were altered in only one or two Emax values, respectively. Given the strong correlation between the observed FRET vectors and the predicted membrane contacts of basic residues in helix α4 and the HVR, we conclude that residues R128/R135 and R160/K170 exhibit guanine nucleotide-specific functions in regulating H-ras membrane interactions, as suggested by our MD models. However, residues R128/R135 and R169/K170 do not operate independently of each other. As a direct physical interaction between helix α4 and the HVR is not possible, this coupling must occur through conformational changes of H-ras, as indicated by the MD simulations (Figure 2A and B).
A novel C-terminal switch region controls H-ras membrane organization
If H-ras undergoes GTP-induced conformational changes as suggested by MD simulations and the FRET data (Figure 2A and B), we reasoned that structural changes in a membrane-orientation switch region that is physically linked to helix α4 or the HVR might be expected. A comparison of the crystal structures of GTP- and GDP-bound H-ras revealed that in a region adjacent to the HVR residues, D47, E49 in the β2–β3 loop and R161, R164 in helix α5 are arranged differently (Supplementary Figure S2A). The H-ras MD models also showed two different networks of salt bridges formed by residues D47, E49, R161, R164 and K170 (Figure 3A and B). We hypothesized that conformational changes induced by GTP loading would rearrange the β2–β3 loop and helix α5, ultimately leading to a differential engagement of the HVR and helix α4 with the membrane. In support of this hypothesis, the FRET vector of GTP-H-ras-R161A/R164A was highly correlated with that of GTP-H-ras-R128A/R135A (correlation coefficient, r=0.932; Figures 2C (ii) and 3C (i), respectively) reflecting very similar changes of their Emax values. Similarly, alanine substitution of residues D47 and E49 in the β2–β3 loop of GTP-H-ras, which have salt bridges with R161 and R164 (Figure 3A and B), resulted in a FRET vector that was highly correlated with the FRET vectors of GTP-H-ras-R161A/R164A (r=0.827) and GTP-H-ras-R128A/R135A (r=0.871) (Figures 2C and 3C). Analysis of a set of single-point mutations (Figure 3C) suggested that D47A contributes predominantly to the observed perturbations in the GTP-H-ras-D47A/E49A FRET vector, whereas mutation R164A appears central to the changes in the GTP-H-ras-R161A/R164A FRET vector (Figure 3C and E).
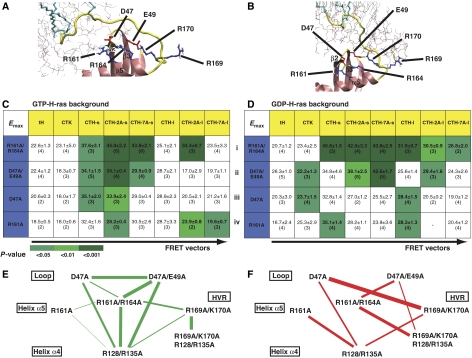
Figure 3.
A membrane-orientation switch region in H-ras. A combination of MD simulation and structural analysis (Supplementary Figure S2A) suggests operation of a membrane-orientation switch region in H-ras that comprises the β2–β3 loop, helix α5 and the HVR. (A, B) The models for GTP- and GDP-H-ras show networks of salt bridges between basic and acidic residues, which we propose are involved in the structural changes that reorient H-ras after GTP loading. Colouring is as in Figure 2A. Lipid molecules are represented as thin sticks. (C, D) The two matrices show FRET vectors of GTP- and GDP-H-ras with alanine substitutions at residues involved in the membrane-orientation switch. Emax values are given±s.e.m., and number of independent experiments in brackets. Emax values of each mutant–marker pair were compared with the corresponding value in the cognate wild-type background (grey FRET vectors in Figure 2C and D) using t-tests, significant differences are shown as green shading. (E, F) Correlation maps of all GTP- and GDP FRET vectors in C, D and Figure 2C and D. In these maps, mutated residues are arranged in a similar spatial layout as they appear in the H-ras structure, mutated residues are connected by lines if the correlation coefficient is >0.6, the thickness of the line is proportional to the correlation coefficient (given in Supplementary Figure S2B). Hence, mutants linked by thicker lines cause similar perturbations in the parent FRET vector. Changes in the correlation maps are suggestive of actual structural rearrangements, possibly linking regions of correlated motion. The network in E clearly shows that residues in the switch region stabilize a conformation that realizes the membrane contact of helix α4. The network in F reveals complex rearrangements of linkages in the switch region that correspond to a different stabilization of the GDP-H-ras conformation; the strongest participation is that between D47 and R169/K170.
Mutation of D47, E49, R161 and R164 in the GDP background also resulted in strong perturbations of the GDP-H-ras FRET vectors (Figure 3D), suggesting that this putative network of salt bridges is important to maintain both GTP- and GDP-H-ras membrane organization. Pairwise comparison of double- and single-point mutants in the GDP background did not allow unambiguous assignment of function to individual residues. Therefore, we devised a novel approach to visualize perturbations of the FRET vectors. We generated GTP- and GDP-specific correlation matrices, which compared the similarity of changes in each FRET vector relative to the parent vector (Supplementary Figure S2B). These matrices were then translated into correlation maps (Figure 3E and F), by placing mutants into a similar spatial layout as they appear in the H-ras structure, and connecting them with lines, the thickness of which is proportional to their correlation coefficient. Thus, correlation maps report on conformational changes in H-ras related to lateral segregation and orientation with respect to the membrane and may indicate correlated motion, for example, of the switch I region and the novel switch region comprising the β2–β3 loop and helix α5 (AA Gorfe, B Grant and JA McCammon, personal communication). The two distinct GTP- and GDP-specific correlation maps (Figure 3E and F, respectively) clearly illustrate a shift in the GTP state from highly correlated mutants D47A, D47A/E49A, R161A/R164A and R128A/R135A, without strong linkage to R169A/K170A (Figure 3E), to a more ramified GDP state correlation map that shows a greater involvement of R161A and a notable strong link between D47A and R169A/K170A (Figure 3F). Such network changes are exactly what would be expected from a switch region that interacts with associated switched structural elements. In fact, we can now distinguish between ‘switched elements' that directly contact the membrane (residues in the HVR and helix α4) and residues comprising the putative ‘switch region' (β2–β3 loop and helix α5) that may be involved in transmitting conformational changes.
The Ras-binding domain of C-Raf recognizes the relative membrane orientation of H-ras
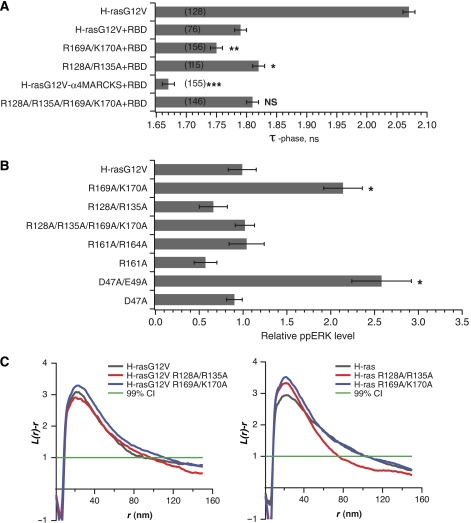
We reasoned that changes in the membrane orientation of H-ras could be manifest in the efficiency of effector interactions. To explore this hypothesis, we used fluorescence lifetime imaging microscopy FRET (FLIM-FRET) to study the interaction of the Ras-binding domain (RBD) of C-Raf (residues 51–131) with certain key GTP-H-ras mutants. The minimal RBD does not have any intrinsic membrane-binding motifs, thus recruitment of the RBD from the cytosol by membrane-anchored GTP-H-ras should report exclusively on the effect of H-ras orientation on RBD binding. Figure 4A shows that coexpression of mGFP-tagged GTP-H-ras with mRFP-RBD results in a strong reduction in the fluorescence lifetime of the mGFP donor due to FRET, reflecting a strong interaction between GTP-H-ras and the RBD. This strong GTP-H-ras RBD interaction was, however, significantly enhanced by R169A/K170A mutations, resulting in a further reduction in mGFP lifetime. In contrast, the FLIM assay indicated that interaction of GTP-H-ras-R128A/R135A with the RBD was significantly reduced compared with control GTP-H-ras (Figure 4A). Mutation of all four basic residues restored the balance of the GTP-H-ras orientation, resulting in the same mGFP lifetime for GTP-H-ras-R128A/R135A/R169A/K170A as for GTP-H-ras (Figure 4A).
Figure 4.
The novel membrane-orientation switch regulates H-ras interaction with C-Raf and MAPK signaling. (A) HEK293 cells transiently expressing mGFP-tagged H-rasG12V mutants with or without an excess of mRFP-RBD were imaged using FLIM. The mean mGFP donor fluorescence lifetime (±s.e.m.) was determined for multiple ROIs and data pooled from three independent experiments. The number of ROIs analysed is given in brackets. Statistically significant differences from the lifetime of mGFP-H-rasG12V coexpressed with mRFP-RBD were assessed using t-tests (*P<0.05, **P<0.01, ***P<0.001). Whereas only the lifetime of mGFP-H-rasG12V in the absence of mRFP-RBD is shown, the lifetimes of all other mGFP-H-rasG12V mutants expressed alone were the same (Supplementary Figure S2C). (B) BHK cells transiently expressing fluorescently tagged H-rasG12V (GTP-H-ras) with the specified mutations were assayed for MAPK activation by quantitative immunoblotting for ppERK. The figure shows mean ppERK levels (±s.e.m.; n=3). *Significant (P<0.05) increase in MAPK activity compared with non-mutated H-rasG12V. (C) Intact 2D plasma membrane sheets prepared from BHK cells expressing mGFP-H-ras (GDP-H-ras) or mGFP-H-rasG12V proteins with the specified mutations were immunogold labelled and imaged by EM. The graphs show a statistical analysis of the resulting immunogold point patterns as weighted mean K-functions calculated from n=16–27 membrane sheets. The gold labelling density for these experiments was 281–478/μm2. Nanoclustering is quantified by the extent of the positive deflection of the L(r)–r curve out of the confidence interval for a random pattern (±1). The radius of the nanoclusters is correlated with the radius at which the L(r)–r deflection is maximum. Using parametric bootstrap tests, we detected no significant differences in either of these nanoclustering parameters from control GTP- or GDP-H-ras point patterns for any of the mutants that were evaluated.
To provide a reference for the GTP-induced reorientation of H-ras, we constructed a new H-ras mutant, where we substituted helix α4 for amino acids 154–171 of the MARCKS protein. This polybasic α-helical sequence normally exerts an effect as a second membrane anchor to attach the MARCKS protein with high affinity to the plasma membrane (McLaughlin and Murray, 2005). Because of the now high membrane affinity of helix α4, we anticipated that α4-MARCKS GTP-H-ras would be quantitatively reoriented with respect to the plasma membrane. The α4-MARCKS chimaera indeed showed the strongest reduction in mGFP lifetime of any GTP-H-ras mutant in the FLIM assay (Figure 4A), a result consistent with complete reorientation of GTP-H-ras leading to the strongest interaction with the RBD. Taken together, these data establish the RBD as an orientation-sensitive interaction module within C-Raf.
Mutations in the novel switch region result in enhanced H-ras signal output
If the orientation-dependent interaction of H-ras with the RBD of C-Raf is functionally relevant, we would expect that perturbation of the novel switched elements that directly contact the cell membrane will result in changes in MAPK signalling. We, therefore, measured ERK activation in BHK cells expressing different GTP-H-ras mutants. Figure 4B shows that ppERK levels were more than two-fold greater in BHK cells expressing GTP-H-ras-R169A/K170A than in cells expressing GTP-H-ras (P<0.001). In contrast, the mutations R128A/R135A impaired the ability of GTP-H-ras-R128A/R135A to activate the MAPK pathway, and completely abrogated the increased potency associated with the R169A/K170A mutations.
To explain these signalling and RBD interaction data, we suggest that both GTP- and GDP-loaded H-ras visit the two conformations identified by the MD simulations (Figure 2A and B) (Gorfe et al, 2007b), but the probability of visiting a given conformation is determined by the bound nucleotide and can be altered by specific mutations. The FRET vector data provide the first evidence for this interpretation by showing that R169 and K170 (Figure 2C (iii)) normally interact with the membrane in GTP-H-ras (Figure 2B). Alanine substitutions at R169 and K170 eliminate the possibility of these residues contacting the plasma membrane and therefore increase the probability that GTP-H-ras-R169A/K170A visits the signalling-competent GTP-H-ras conformation (Figure 2A). This interpretation is strengthened by the RBD FLIM-FRET analysis (Figure 4A), which establishes the RBD as an orientation-sensitive interaction module in C-Raf that translates the altered orientation into different ppERK signalling outputs (Figure 4B).
We next explored if residues in the putative membrane-orientation switch region were similarly involved in the structural rearrangements that lead to the more signalling-competent H-ras conformation. Figure 4B shows that GTP-H-ras-D47A/E49A was also hyperactive, with ppERK levels over 2.5 times higher than that of GTP-H-ras. This involvement of D47/E49 in stabilizing the active GTP-H-ras orientation is consistent with the strong linkage of these residues with helix α4 shown in our correlation map (Figure 3E). Finally, these changes in signalling were not related to the actual formation of nanoclusters, as mutations in the HVR and helix α4 had no significant effect on the univariate clustering parameters measured by electron microscopy (Figure 4C).
Collecting together the MD data in conjunction with our FRET, signalling and EM data, we propose a model in which membrane-anchored H-ras operates similarly to a GTP-modulated balance (Figure 5). In this balance model, GDP-H-ras membrane anchorage is predominantly stabilized through the HVR. GTP loading triggers structural rearrangements in switch regions I and II. This results in reorientation of the β2–β3 loop containing D47 and E49, which subsequently engages differently with residues R161 and R164 reorienting helix α5. The coupling of this region with the adjacent HVR, in turn alters interactions of residues R169 and K170 with membrane lipids, allowing reorientation of helix α4 and its contact with the membrane, effectively ‘tipping over' GTP-H-ras and altering the orientation of the whole G-domain with respect to the plasma membrane (Figure 5).
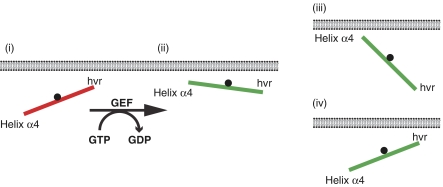
Figure 5.
A balance model identifies another level of regulating Ras signal output. The balance model introduces the orientation of the G-domain with respect to the plasma membrane as an additional determinant of signalling specificity among Ras isoforms. The balance is regulated by the membrane-orientation switch region, which comprises residues in the β2–β3 loop and helix α5, and is indicated by the black fulcrum. MD and FRET analysis of H-ras show that for inactive, GDP-bound Ras (red) contacts of residues in the hypervariable region (HVR) prevail (i) so the balance is shifted to the HVR. Upon GTP loading, Ras is reoriented by interactions of helix α4 with the membrane (ii) so the balance is shifted to helix α4. In the context of this balance model, different Ras isoforms of the Ras subfamily may adopt different preferred orientations with respect to the membrane depending on the precise combination of residues present in the HVR and helix α4 (Supplementary Figure S3). Schemes (iii) and (iv) show examples of the membrane-interaction balance being shifted more and less towards helix α4, respectively. We speculate that such submembrane orientations are an important determinant for the propensity of GTP-loaded Ras proteins to interact with other membrane-associated proteins, such as its activity modulators (e.g. galectins) and other effectors (e.g. PI3 kinase).
The RBD interaction FLIM data further indicate that the G-domain orientation is critical for effector interaction, possibly by facilitating access to the switch I region. We also have preliminary evidence that the interaction of GTP-H-ras with galectin-1, a nanocluster scaffold (Hancock and Parton, 2005), is orientation sensitive and may therefore serve to stabilize the signalling-competent conformation of H-ras. Interestingly, a comparison of the amino-acid sequences of 13 Ras subfamily members reveals that helix α4 is the second most divergent sequence outside of the HVR (Supplementary Figure S3). We speculate that this balance model may also apply to other Ras isoforms and that evolution has assigned to helix α4 a modulating function similar to that of the HVR.
Our previous work has shown that sequence differences in the HVR result in isoform-specific lateral segregation that may underlie the signalling specificity of Ras proteins (Prior and Hancock, 2001; Prior et al, 2003a; Hancock and Parton, 2005). The balance model proposed here now suggests that the interaction of Ras with membrane-associated proteins may be regulated on at least two levels. The first level is lateral segregation, governed by the sum of interactions of the lipid-modified HVR and helix α4. The second level is GTP-regulated orientation of membrane-bound Ras that is stabilized by specific proteo-lipid contacts on helix α4. Such an orientation-based code, or level of regulation, is in good agreement with the various angularities noted for different Ras–effector complexes (Pacold et al, 2000; Herrmann, 2003). More broadly our work suggests that activity of other membrane-associated signalling proteins could be modulated by their relative orientation to the membrane, which, by acting both as a boundary and interaction partner, can spatially constrain possible signalling conformations.
Materials and methods
Molecular cloning
Nanodomain markers tH, tK and CTH were described previously (Rotblat et al, 2004; Abankwa and Vogel, 2007). Other nanodomain marker-targeting sequences were PCR amplified and cloned in frame with the appropriate fluorescent protein cDNA. Point mutations in H-ras were introduced using QuickChange site-directed mutagenesis (Stratagene). The RBD of C-Raf was PCR amplified and cloned in frame with the cDNA of mRFP (Campbell et al, 2002). For the α4-MARCKS chimaera, the cDNA sequence encoding amino acids 121–138 of H-ras in the mGFP-H-rasG12V construct was replaced by the cDNA encoding for amino acids 154–171 of the human MARCKS protein, using ‘splicing by overlap extension'-PCR (Horton et al, 1989). Subcellular localization of all H-ras constructs and nanodomain markers are shown in Supplementary Figure S4.
Cell culture and phospho-ERK analysis
BHK cells were cultured as described (Roy et al, 2005) and transfected using Lipofectamine (Invitrogen) with donor and acceptor constructs 48 h before analysis. For experiments with wild-type H-ras proteins, cells were serum starved overnight. For immunoblotting, cells were harvested 24 h after transfection in lysis buffer (1% NP-40 in 50 mM Tris, 75 mM NaCl, 25 mM NaF, 5 mM MgCl2, 5 mM EGTA, pH 7.5, 1 mM DTT, 330 ng/ml aprotinin, 3.3 μg/ml leupeptin, 100 μM Na3VO4). Western blots were probed with anti-GFP (cat. no. 11814460001; Roche) or anti-phospho-Erk (9106; Cell Signalling) antibodies and quantification was carried out using a LumiImager (Roche) and ImageJ software. To summarize data from repeat experiments, in each repeat raw intensities were normalized using the sum of all phospho-ERK intensities. These normalized values were then averaged and the standard errors were determined. Subsequently, we plotted these normalized and averaged phospho-ERK intensities relative to the average signal of H-rasG12V (which itself kept the error from averaging through error propagation).
FRET analysis by flow cytometry
We used a LSRII flow cytometer (BD Biosciences) to measure fluorescence of cells in donor (405 nm excitation, 450/50 nm emission), acceptor (488 nm excitation, 530/30 nm emission) and FRET channels (405 nm excitation, 560/20 nm emission). Doublet discrimination was implemented to measure signals of single cells. FRET analysis of flow cytometer data was carried out as described (Abankwa and Vogel, 2007) using custom-written algorithms in IgorPro5 (Wavemetrics). Calibration for normalized acceptor surface concentration cA was carried out essentially as described (Abankwa and Vogel, 2007) using cA=NAc/AcellR02, where NAc, the number of mCit fluorophores per cell, Acell is the surface area of a model spherical BHK cell and R0=4.7 nm, the Försters radius of mCFP/mCit, calculated using spectroscopic data. Fluorescent FITC beads (no. 1139; Bangs Laboratories) with defined fluorescein equivalents and size were used to calibrate NAc and Acell. An mCFP–mCit fusion protein was used to calibrate FRET efficiency and donor–acceptor ratio. The FRET efficiency was calculated per cell by using an adapted sensitized acceptor emission method. Only cells with a donor mole fraction xD=0.5±0.1, corresponding to a ∼1:1 donor–acceptor ratio, were analysed. The characteristic Emax value was determined using equation (1), as described (Abankwa and Vogel, 2007).
where A1=0.6322, k1=3.1871, A2=0.3678, k2=0.7515, for Rc/R0=0.7 as in Wolber and Hudson (1979), using Rc, the closest approach distance of the fluorescent proteins (=3.4 nm) (Abankwa and Vogel, 2007). Curve fitting was carried out using IgorPro5 software by iterative minimization of χ2 with the Levenberg–Marquardt algorithm. Standard errors of Emax were calculated based on the residuals. All data were fitted in a batch procedure, with identical starting parameters and constraints. In an exhaustive set of Snedecor's F-tests, we formally evaluated whether the errors associated with fitting Emax values were significantly different from the errors associated with the averaged Emax value from replicated experiments (Figures 2 and 3). In >93% of cases (of the 461 measurements), the variance associated with the averaged Emax value for replicated experiments was greater than, or not significantly different from the variance associated with fitting Emax values to the raw data. Given this result our statistical inferences using the larger sampling variances associated with the replicated Emax values are actually more conservative than had we used the variances associated with fitting the Emax values in individual experiments.
FRET vector correlation coefficients for construction of a linkage map
We define a FRET vector as the set of n Emax values of FRET pairs comprising a given H-ras mutant with n nanodomain markers. The FRET vector of each mutant H-ras protein, m, was converted to a reduced FRET vector Δm by subtracting the cognate GDP or GTP wild-type vector, b (grey fields in Figure 2C and D (i)); Δm=m−b, and all nonsignificant Emax differences were set to zero. Δm therefore contains only the significant Emax changes relative to background. The correlation, rij, of two reduced FRET vectors Δmi and Δmj is defined as rij=Δmi·Δmj/(∣Δmi∣ ∣Δmj∣). The resulting correlation matrices yield high values for mutants that have similar FRET vectors.
FLIM
FLIM-FRET experiments were carried out using a lifetime fluorescence imaging attachment (Lambert Instruments, Leutingwolde, The Netherlands) on an inverted microscope (Olympus IX71). HEK293 cells expressing for 2 days mGFP-H-ras constructs, alone or with mRFP-tagged RBD from C-Raf (acceptor) were excited using a sinusoidally modulated 3 W 470 nm LED at 40 MHz under epi-illumination. Cells were imaged with a × 60 NA 1.45 oil objective using an appropriate GFP filter set. The phase and modulation were determined from a set of 12 phase settings using the manufacturer's software. Fluorescein was used as a lifetime reference standard. Thus, the phase lifetime of the donor (mGFP constructs) was determined for regions of interest containing 1–4 cells, coexpressing indicated constructs. The number of regions of interest assayed for each donor/acceptor combination was ⩾76 (Figure 4A).
Structural models
Models for the GDP- and GTP-bound H-ras structure were derived from previously reported MD simulations (Gorfe et al, 2007b). Snapshots at 25 ns were used here. Note that the simulations provided two ensembles of structures; the predicted models for GDP- and GTP-bound forms predominate in simulations with GDP and GTP, respectively (Gorfe et al, 2007b). About 10–20% of the structures from simulations with GDP dynamically adopted the GTP-bound structure, and vice versa.
Electron microscopy
Apical plasma membrane sheets were prepared, fixed with 4% PFA, 0.1% glutaraldehyde and labelled with affinity-purified anti-GFP antibodies conjugated directly to 5 nm gold as described (Prior et al, 2003b; Plowman and Hancock, 2005). Digital images of the immunogold-labelled plasma membrane sheets were taken at × 100 000 magnification in an electron microscope (Jeol 1011). Intact 1 μm2 areas of the plasma membrane sheet were identified using ImageJ and the (x, y) coordinates of the gold particles were determined as described (Prior et al, 2003b; Plowman and Hancock, 2005).
Statistical analysis
Student's t-tests (two-tailed) were used to assess significant differences between mean values. Statistical analysis of immunogold point patterns using K-functions and parametric bootstrap tests was carried out as described (Diggle et al, 1991, 2000; Prior et al, 2003a; Hancock and Prior, 2005; Plowman et al, 2005).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Dr Tianhai Tian for advice on vector correlation and Ms Annette Lane for technical support. DA is a fellow of the Swiss National Science Foundation (PA00A-111446). This study was supported by grants from the NHMRC (Australia) and NIH. The IMB is a Special Research Centre of the ARC. AAG gratefully acknowledges financial support from Nachwuchsförderungskredit Stiefel-Zangger-Stiftung der Universität Zürich. Work of JAM was supported by grants from NIH, NSF, HHMI, NBCR, CTBP and NSF supercomputer centres and Accelrys.
References
- Abankwa D, Vogel H (2007) A FRET map of membrane anchors suggests distinct microdomains of heterotrimeric G proteins. J Cell Sci 120: 2953–2962 [DOI] [PubMed] [Google Scholar]
- Berney C, Danuser G (2003) FRET or no FRET: a quantitative comparison. Biophys J 84: 3992–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle PJ, Lange N, Benes FM (1991) Analysis of variance for replicated spatial point patterns in clinical neuroanatomy. J Am Stat Assoc 86: 618–625 [Google Scholar]
- Diggle PJ, Mateu J, Clough HE (2000) A comparison between parametric and non-parametric approaches to the analysis of replicated spatial point patterns. Adv Appl Probab 32: 331–343 [Google Scholar]
- Gorfe AA, Babakhani A, McCammon JA (2007a) H-ras protein in a bilayer: interaction and structure perturbation. J Am Chem Soc 129: 12280–12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfe AA, Bayer M-H, Abankwa D, Hancock JF, McCammon JA (2007b) Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem 50: 674–684 [DOI] [PubMed] [Google Scholar]
- Hancock JF (2003) Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol 4: 373–384 [DOI] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57: 1167–1177 [DOI] [PubMed] [Google Scholar]
- Hancock JF, Parton RG (2005) Ras plasma membrane signalling platforms. Biochem J 389: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ (1990) A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63: 133–139 [DOI] [PubMed] [Google Scholar]
- Hancock JF, Prior IA (2005) Electron microscopic imaging of Ras signaling domains. Methods 37: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C (2003) Ras-effector interactions: after one decade. Curr Opin Struct Biol 13: 122–129 [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Kiskowski MA, Kenworthy AK (2007) In silico characterization of resonance energy transfer for disk-shaped membrane domains. Biophys J 92: 3040–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438: 605–611 [DOI] [PubMed] [Google Scholar]
- Nazarov PV, Koehorst RB, Vos WL, Apanasovich VV, Hemminga MA (2006) FRET study of membrane proteins: simulation-based fitting for analysis of membrane protein embedment and association. Biophys J 91: 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov PV, Koehorst RB, Vos WL, Apanasovich VV, Hemminga MA (2007) FRET study of membrane proteins: determination of the tilt and orientation of the N-terminal domain of M13 major coat protein. Biophys J 92: 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL (2000) Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103: 931–943 [DOI] [PubMed] [Google Scholar]
- Plowman SJ, Hancock JF (2005) Ras signaling from plasma membrane and endomembrane microdomains. Biochim Biophys Acta 1746: 274–283 [DOI] [PubMed] [Google Scholar]
- Plowman SJ, Muncke C, Parton RG, Hancock JF (2005) H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA 102: 15500–15505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Hancock JF (2001) Compartmentalization of Ras proteins. J Cell Sci 114: 1603–1608 [DOI] [PubMed] [Google Scholar]
- Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF (2001) GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol 3: 368–375 [DOI] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, Hancock JF (2003a) Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol 160: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Parton RG, Hancock JF (2003b) Observing cell surface signaling domains using electron microscopy. Sci STKE 2003: PL9. [DOI] [PubMed] [Google Scholar]
- Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF (2004) Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol Cell Biol 24: 6799–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF (2005) Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol 25: 6722–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF (2007) Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol 9: 905–914 [DOI] [PubMed] [Google Scholar]
- Wolber PK, Hudson BS (1979) An analytic solution to the Forster energy transfer problem in two dimensions. Biophys J 28: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures