Abstract
p53 is known to prevent tumour formation by restricting the proliferation of damaged or oncogene-expressing cells. In contrast, how the p53 family member p73 suppresses tumour formation remains elusive. Using a step-wise transformation protocol for human cells, we show that, in premalignant stages, expression of the transactivation-competent p73 isoform TAp73 is triggered in response to pRB pathway alterations. TAp73 expression at this stage of transformation results in increased sensitivity to chemotherapeutic drugs and oxidative stress and inhibits proliferation and survival at high cell density. Importantly, TAp73 triggers a transcriptional programme to prevent anchorage-independent growth, which is considered a crucial hallmark of fully transformed cells. An essential suppressor of anchorage-independent growth is KCNK1, which is directly transactivated by TAp73 and commonly downregulated in glioma, melanoma and ovarian cancer. Oncogenic Ras switches p73 expression from TAp73 to the oncogenic ΔNp73 isoform in a phosphatidyl-inositol 3-kinase-dependent manner. Our results implicate TAp73 as a barrier to anchorage-independent growth and indicate that downregulation of TAp73 is a key transforming activity of oncogenic Ras mutants.
Keywords: malignant transformation, p53, p73, tumour suppressor genes
Introduction
p53 is central to an intricate network of pathways that senses various types of cellular stress to coordinate cell fate decisions, such as cell-cycle arrest, senescence, differentiation or apoptosis (Stiewe, 2007). In addition to p53, the p53 gene family comprises p63 and p73, both of which share more than 60% amino-acid identity within the DNA-binding domain of p53 and regulate an overlapping set of target genes (Melino et al, 2002). The TP73 gene encodes two antagonistic classes of p73 proteins that can be distinguished by their transactivation function. The transactivation-competent TAp73 has many p53-like properties of a tumour suppressor, whereas the transactivation-defective, N-terminally truncated ΔNp73 functions as a dominant-negative inhibitor of all p53 family members by competing with DNA binding or by forming transactivation-defective heteromeric complexes (Pozniak et al, 2000; Stiewe et al, 2002a).
In contrast to p53, p73 is not bound and inactivated by transforming viral oncoproteins that are known to target p53 (e.g. simian virus 40 (SV40) large T (LT), HPV16 E6 or adenoviral E1B-55K), which suggests that inactivation of p73 may not be required for transformation (Marin et al, 1998; Roth et al, 1998). However, in the last few years, evidence has accumulated that p73 does have tumour suppressor activity in both mice and men. p73+/− mice show a reduced life span compared with their wild-type littermates and exhibit a spectrum of tumours, including lung adenocarcinomas, thymic lymphomas and hemangiosarcomas (Flores et al, 2005). Compared with p53+/− mice, combined mutation of p73 and p53 (p53+/−;p73+/−) results in an increased tumour burden and a switch to a metastatic tumour phenotype, strongly suggesting that p73 controls pathways limiting tumour aggressiveness and metastasis (Flores et al, 2005).
In cancer patients, p73 function is frequently compromised although the TP73 gene is not directly mutated. Many different studies on multiple tumour entities have described elevated expression of ΔNp73 isoforms in tumour tissues (Casciano et al, 2002; Zaika et al, 2002; Stiewe et al, 2002b, 2004; Concin et al, 2005; Muller et al, 2005; Cam et al, 2006; Dominguez et al, 2006). Importantly, high ΔNp73 expression levels correlate with an aggressive tumour phenotype indicated by lymph node metastasis and vascular invasion, chemotherapeutic failure and poor patient survival prognosis (Casciano et al, 2002; Concin et al, 2005; Muller et al, 2005; Dominguez et al, 2006). Furthermore, ΔNp73 expression proved crucial for the maintenance of the tumorigenic phenotype in childhood solid tumours, such as rhabdomyosarcomas or neuroblastomas (Simoes-Wust et al, 2005; Cam et al, 2006). Experimentally, ΔNp73 has been shown to transform immortalized NIH3T3 fibroblasts to tumorigenicity and to cooperate with H-RasV12 in the malignant transformation of primary mouse embryonic fibroblasts (Stiewe et al, 2002b; Petrenko et al, 2003). A switch in p73 expression from the tumour-suppressive TAp73 to oncogenic ΔNp73 therefore frequently compromises the tumour suppressor activity of the TP73 gene. Although this switch appears to be an important event in tumorigenesis, the underlying molecular mechanisms are yet unclear.
Results
Regulation of p73 expression during experimental transformation
Here, we used a step-wise experimental transformation protocol to investigate the role of p73 in the course of human cell transformation (Hahn et al, 1999). Normal human diploid BJ fibroblasts were rendered tumorigenic by sequential retroviral transduction with the catalytic telomerase subunit hTERT (human telomerase reverse transcriptase), the SV40 early region expressing the LT and small T (ST) antigens, and the activated Ras allele H-RasV12 (Figure 1A). As previously shown, only cells expressing all three genetic elements showed anchorage-independent growth in soft agar (Figure 1B) and were tumorigenic in nude mice (Hahn et al, 1999). Expression of the transgenes was confirmed by telomerase activity assays (data not shown) and western blots (Figure 1D). Interestingly, expression of TAp73 was undetectable in primary and hTERT-immortalized cells but strongly increased in the premalignant BJ-TE stage under conditions of confluent growth (Figure 1C and D). This increase in TAp73 expression could also be observed when BJ-TE cells were analysed under progressively increasing cell densities (Figure 1E). A similar density-dependent increase was observed in HA1E human embryonic kidney cells expressing hTERT and the SV40 early region, indicating that upregulation of TAp73 is not only seen in the fibroblast model but also in epithelial cells (Figure 1E). Upon full transformation by H-RasV12 (BJ-TER stage), a downregulation of TAp73 expression and a concomitant increase in ΔNp73 levels was observed (Figure 1C and D). Activated Ras signalling therefore switches the TAp73/ΔNp73 ratio in favour of ΔNp73, the constellation frequently observed in cancer patients.
Figure 1.
p73 expression changes during the course of human cell transformation. (A) Experimental transformation protocol. BJ fibroblasts were sequentially transduced with retroviral vectors expressing hTERT, the SV40 early region and the H-RasV12 oncogene. The individual stages of transformation are labelled as BJ, BJ-T, BJ-TE and BJ-TER. (B) Soft agar growth of BJ cells in the various stages of transformation. Scale bar, 200 μm. (C) Semiquantitative RT–PCR for expression of TAp73, ΔNp73, SV40 LT and GAPDH. BJ cells in the indicated stages of transformation were harvested for RNA extraction under subconfluent or confluent conditions. NTC, no template control. (D) Western blot analysis of BJ cells for the ectopically expressed SV40 LT antigen and H-RasV12 as well as the endogenous levels of p53, TAp73 and ΔNp73 under subconfluent and confluent conditions. To enhance detection of TAp73, we immunoprecipitated total p73 protein followed by a western blot with an antibody specifically detecting TAp73. In the case of ΔNp73, specific (◂) and nonspecific (*) bands are labelled. β-actin expression is shown as a control. (E) Semiquantitative RT–PCR for TAp73 expression in BJ-TE or HA1E cells harvested at increasing cell densities.
TAp73 expression is induced in confluent cells with pRB pathway alterations
To mechanistically understand induction of TAp73 in premalignant BJ-TE cells, we first investigated the required gene products of the SV40 early region. Expression of LT on its own induced TAp73 to similar extent as the complete early region whereas expression of ST had no impact on TAp73 levels (Figure 2A). As LT is a complex oncoprotein with multiple transforming functions, we analysed LT mutants that are specifically defective for single activities (Figure 2B; Rangarajan et al, 2004). Importantly, the Δ434–444 mutant, which is unable to bind to and inactivate p53, induced TAp73 to similar extent as the wild type or the p53-binding competent M528S mutant. This indicates that TAp73 induction was not a consequence of p53 inactivation. Of all the mutants tested, only the E107K mutant was completely defective for TAp73 induction (Figure 2C and D). E107K is a point mutation in the LXCXE motif known to be essential for binding and inactivating the ‘pocket proteins' pRB, p107 and p130. Similarly, the adenoviral oncoprotein E1A, which also binds to the pocket proteins, induced TAp73 in a LXCXE motif-dependent manner (Figure 2E). Mutation of the J domain (Y34A), which has accessory functions for inhibition of the pocket proteins resulted in reduced TAp73 induction. These findings are consistent with regulation of the TAp73 promoter by the E2F-RB axis (Irwin et al, 2000; Stiewe and Putzer, 2000; Zaika et al, 2001; Urist et al, 2004) and suggest that alterations of the pRB pathway are the underlying cause for TAp73 induction in this early stage of transformation independent of the p53 status.
Figure 2.
TAp73 expression is induced by pRB pathway alterations. (A) Semiquantitative RT–PCR for expression of SV40 LT, ST, TAp73 and GAPDH in BJ-T cells transduced with lentiviral vectors expressing LT or ST or a combination of both (early region). (B) Domain structure of SV40 LT. The bipartite p53-binding region (aa 351–450 and 533–626), the LXCXE motif (aa 103–107) crucial for pocket protein binding and the N-terminal J domain (aa 1–82) are marked. The mutations Y34A, E107K, Δ434–444 and M528S are labelled (▿). (C) Western blot analysis for expression of TAp73 in BJ-T cells infected with recombinant adenoviral vectors for wild-type LT and the point mutants E107K, Y34A and M528S. Expression of LT and β-actin are shown as controls. (D) Semiquantitative RT–PCR for TAp73 expression in BJ-T cells infected with lentiviral vectors expressing LT wt and the mutants E107K and Δ434–444. Expression of LT is shown by western blot. (E) Semiquantitative RT–PCR for TAp73 expression in BJ-T cells infected with lentiviral vectors expressing E1A and the pocket protein-binding defective pm47/124 point mutant of E1A. (F) TAp73 mRNA expression level in BJ-TE cells transfected with dominant-negative E2F1 (dnE2F) under subconfluent and confluent conditions. Mock-transfected cells and cells treated for 8 h with 0.2 μM adriamycin are shown for comparison. AU, arbitrary units. (G) Western blot showing E2F1 expression in BJ-TE cells depending on cell confluence (50 versus 100%) and DNA damage treatment (0.2 μM adriamycin for 8 h). (H) Chromatin immunoprecipitation (ChIP) for E2F1 recruitment on the proximal TAp73 promoter. BJ-TE cells were analysed under both subconfluent (50%) and confluent (100%) conditions. ChIPs were carried out with a polyclonal E2F1 antiserum (C-20) and as a no-antibody control. PCR detection of the proximal TAp73 promoter was performed with the minimal amount of cycles to allow quantification within the linear amplification range. 1% input DNA was amplified as a control.
Importantly, upregulation of TAp73 was only observed under conditions of confluent growth (Figures 1C, D and 2F). This upregulation was comparable to the previously described E2F1-dependent activation in response to adriamycin-induced DNA damage (Figure 2F), suggesting a causal role of E2F1 in this process (Pediconi et al, 2003). In support of this hypothesis, a dominant-negative inhibitor of E2F (Maehara et al, 2005) completely abrogated upregulation of TAp73 in confluent cells (Figure 2F). E2F1 levels were low in cells proliferating at low cell density, whereas increased expression of E2F1 was observed both in response to DNA damage signals and in cells grown to confluence (Figure 2G). Furthermore, E2F1 binding to the TAp73 promoter was increased in cells harvested under confluent but not under subconfluent conditions (Figure 2H). These data clearly indicate that high cell density triggers an E2F1-dependent upregulation of TAp73 expression in cells with defects in pocket protein function.
Antiproliferative function of TAp73 in the premalignant stage of transformation
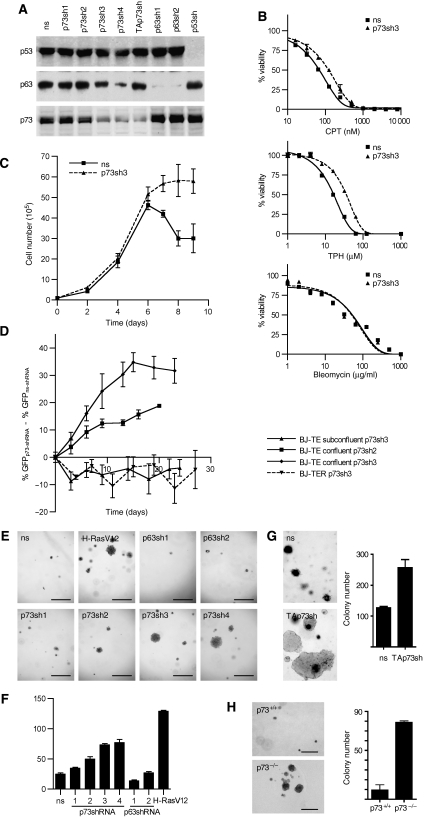
To interrogate the functional relevance of TAp73 upregulation, we carefully designed short hairpin RNA (shRNA) constructs to downregulate p73 expression. Owing to high sequence homology between the p53 family members, we tested all constructs for their inhibitory activity on p53, p63 and p73 in western blots (Figure 3A). All shRNAs were able to clearly discriminate between p53 and p63/p73. Although some shRNAs depleted both p63 and p73 (e.g. p73sh4), p73-selective shRNAs (p73sh2 and p73sh3) were identified. p63-selective shRNAs (p63sh1 and p63sh2) were used to exclude p63-related effects. As a compromise between knockdown efficiency and target specificity, most studies were performed with p73sh3, which strongly reduced p73 levels but only marginally affected p63 expression.
Figure 3.
p73 knockdown enables anchorage-independent growth. (A) Knockdown efficiency and specificity of various p53, p63 and p73 shRNA. BJ-TE cells were transfected with indicated shRNAs and grown to confluence before harvesting for western blot analysis of p53, p63 and p73 expression. p73 detection was performed by first immunoprecipitating total p73 with a polyclonal p73 antiserum followed by a western blot for TAp73. (B) Cell viability assay. BJ-TE cells stably transduced with the p73sh3 construct or a non-silencing shRNA (ns) as a control were treated for 24 h with the indicated doses of camptothecin (CPT), tert-butyl hydroperoxide (TBH) and bleomycin. Cell viability was determined with the ATP-based CellTiter Glo assay. Shown are the mean and standard deviation of triplicate measurements. (C) Growth curves of BJ-TE cells stably expressing the p73sh3 construct or a ns shRNA as a control. Cells were seeded at a density of 1 × 105 cells per 60-mm-dish and counted in triplicates at the indicated time points. Shown is the mean cell number±s.d. (D) Competitive coculture experiment. BJ-TE (or BJ-TER) cells were infected with lentiviral vectors coexpressing GFP and a shRNA construct (ns, p73sh2, p73sh3) as indicated. The percentage of GFP-expressing cells was adjusted to approximately 30% with parental cells. The percentage of GFP-positive cells was determined by flow cytometry over a time course of 3–4 weeks. The growth advantage of p73-depleted cells is presented as the difference in the percentage of GFP-positive cells in the p73-shRNA-expressing population and the ns-shRNA population. The cell cultures were grown under conditions of high (confluent) or low (subconfluent) cell density. Shown is the mean of a triplicate experiment. (E–H) Anchorage-independent growth assay. (E–G) BJ-TE cells were stably transduced with lentiviral vectors expressing H-RasV12 or one of the indicated shRNAs. (H) Mouse embryonic fibroblasts were isolated from p73+/+ and p73−/− embryos, immortalized with LT antigen and tested for anchorage-independent growth. Shown is the clonogenic growth in soft agar medium and the mean number of colonies±s.d. Scale bars, 200 μm.
p73-depleted BJ-TE cells showed increased resistance to the DNA-damaging agent camptothecin, which was previously shown to mediate apoptosis in a p73-dependent manner (Figure 3B; Urist et al, 2004). Furthermore, p73 depletion conferred increased resistance to oxidative stress but not to the radiomimetic agent bleomycin (Figure 3B). Together these findings indicate that upregulated TAp73 is functionally active in BJ-TE cells and increases sensitivity to p73-mediated cell death.
Because p73 is known to inhibit cell proliferation, we tested whether p73 depletion confers a growth advantage to BJ-TE cells. However, in initial experiments, p73-depleted cells proliferated equally fast as the controls (data not shown). Considering that TAp73 expression was triggered in confluent cell cultures only, we repeated the experiment with special focus on high-density cultures (Figure 3C). Here, p73-depleted cells continued to grow and remained viable at the time when the control cells became confluent and eventually started to die. This suggests that TAp73 contributes to contact inhibition by limiting cell growth and survival under conditions of confluent growth. To investigate whether p73 knockdown at this stage can lead to a long-term growth advantage, we measured the proliferation of p73-depleted BJ-TE cells in a competitive coculture experiment (Figure 3D). For this, BJ-TE cells were infected with lentiviral vectors coexpressing green fluorescent protein (GFP) and either a non-silencing or a p73-directed shRNA. The percentage of the GFP/shRNA-expressing cells was adjusted to approximately 30% by mixing with parental, non-infected cells. The mixed populations were cultured under conditions of low or high cell density for several passages and the percentage of GFP-positive cells was measured by flow cytometry at 2–4 days intervals. To control for nonspecific effects of viral infection on cell growth, the percentage of GFP-positive cells in the non-silencing shRNA population was subtracted from the percentage of GFP-positive cells in the p73-depleted population. This difference serves as an indicator for the p73-shRNA-induced growth advantage under competitive growth conditions. We observed a strong growth advantage conferred by two different p73-knockdown constructs (p73sh2 and p73sh3) when the cells were cultured under conditions of high cell density. Importantly, under subconfluent conditions, no growth advantage was observed. Similarly, no growth advantage was seen in BJ-TER cells, which express only low levels of TAp73 due to activated Ras signalling. This provides further evidence that the growth advantage conferred by the p73-knockdown constructs is a specific effect of TAp73 depletion.
As contact inhibition is a major barrier to anchorage-independent growth as one of the hallmarks of transformed cells, we tested the effect of p73 depletion on growth in soft agar (Figure 3E and F). All p73-knockdown constructs enabled anchorage-independent growth of BJ-TE cells. The colony numbers and size correlated directly with the p73-knockdown efficiency (Figure 3A, E and F). Similar to total p73 depletion, the specific knockdown of TAp73 also induced anchorage-independent growth properties (Figure 3G). However, although p73 depletion allowed anchorage-independent growth comparable to oncogenic Ras, only H-RasV12-expressing cells were tumorigenic in nude mice.
Likewise, LT-expressing mouse embryo fibroblasts isolated from genetically defined p73-knockout embryos showed anchorage-independent growth properties that were not apparent in the wild type (Figure 3H). Despite this clear difference in soft agar growth between wild-type and knockout cells, the murine cells proliferated slower and the colonies rarely reached the size seen in the human transformation model.
We conclude that p73 loss is sufficient to enable anchorage-independent growth. The knockdown of p63 does not confer anchorage-independent growth properties, although we cannot formally exclude that a p63 knockdown enhances the anchorage-independent phenotype of p73-depleted cells. Together these results indicate that increased expression of TAp73 due to aberrant proliferation control limits cell growth and survival under conditions of high cell density and presents a barrier to anchorage-independent growth.
Identification of a TAp73-regulated transcriptional programme
To identify p73-regulated pathways, we compared genome-wide expression profiles of p73-depleted BJ-TE cells with control cells under both subconfluent and confluent conditions (Figure 4A). TAp73 expression was low in both subconfluent cell populations and was specifically induced in the confluent control cells (Figure 4B). This induction was effectively prevented by the p73-knockdown construct (Figure 4B). Expression of a total of 50 probe sets was changed by p73 depletion by more than twofold. This number appears small compared with the number of changes commonly seen upon p53 depletion, but it is similar to the number of genes that are differentially expressed between p73-knockout and wild-type MEFs (data not shown). Importantly, 28 of the 50 (56%) p73-dependent probe sets were regulated in a density-dependent manner and are shown in the heatmap in Figure 4A (Supplementary Table 1). Six of these genes were expressed in a manner that paralleled expression of TAp73 (Figure 4B) and were therefore chosen for further analysis. Independent experiments confirmed p73- and density-dependent regulation (Figure 4B). Furthermore, downregulation of these six genes by an shRNA specific for the TAp73 isoform demonstrated that these genes are regulated by TAp73 and not ΔNp73 (Figure 4B). As TAp73 is induced by LT in a LXCXE-dependent manner, we also investigated regulation by LT and the E107K mutant. Of the six genes, FAM38B, IGSF3 and KCNK1 were induced by LT but not by E107K in BJ-T cells (Figure 4C).
Figure 4.
Identification of a TAp73-induced transcriptional programme. (A) BJ-TE cells expressing a non-silencing or p73-depleting shRNA were analysed under subconfluent and confluent conditions with Affymetrix GeneChip Human Genome U133A 2.0 Arrays. Shown is a heatmap depicting the expression level of the 28 probe sets that were both density- and p73-regulated. The box contains the seven probe sets with a similar expression pattern as TAp73. (B, C) Semiquantitative RT–PCR for expression of six genes identified by gene expression profiling as putative mediators of TAp73 function. (B) BJ-TE cells transduced with a non-silencing (ns), total p73-depleting (p73sh3) or TAp73-depleting (TAp73sh) shRNA were analysed under subconfluent and confluent conditions. (C) BJ-T cells were analysed after infection with adenoviral vectors expressing wild-type or mutant LT. (D, E) BJ-TE cells were infected with lentiviral vectors coexpressing GFP and shRNAs directed against FAM38B, IGSF3 or KCNK1. Shown is the growth of these cells in soft agar medium by bright field and fluorescence microscopy (D). Scale bars: 200 μm. The knockdown efficiency of the shRNAs was determined by the psiCHECK system (E). The ratio of Renilla (R) and firefly (F) luciferase activity was normalized to the mock-transfected control cells. Shown is the mean normalized R/F ratio±s.d. An inactive shRNA results in a normalized R/F ratio of 1.0, whereas an efficient knockdown is characterized by a normalized R/F ratio lower than 0.3.
To investigate the requirement of these genes for TAp73-mediated suppression of anchorage-independent growth, we designed multiple shRNAs to knockdown expression of FAM38B, IGSF3 and KCNK1 (Figure 4D and E). In the absence of suitable antibodies for detection of the FAM38B, IGSF3 and KCNK1 proteins, we determined the knockdown efficiency using the psiCHECK system. For this, the targeted mRNA sequence was cloned into the 3′UTR of a Renilla luciferase expression cassette and cotransfected with the shRNA construct. Efficient knockdown by mRNA degradation or translational inhibition resulted in suppression of Renilla luciferase activity. A firefly luciferase expression cassette not regulated by any of the analysed shRNAs was used for normalization (Figure 4E). All shRNAs reduced the target expression to less than 30% of the control. Importantly, knockdown of either FAM38B or KCNK1 mimicked the effect of a p73 knockdown in BJ-TE cells and enabled anchorage-independent growth (Figure 4D). Despite efficient knockdown, the IGSF3 shRNAs did not enable soft agar growth, which further confirmed the specificity of the FAM38B- and KCNK1-knockdown phenotype.
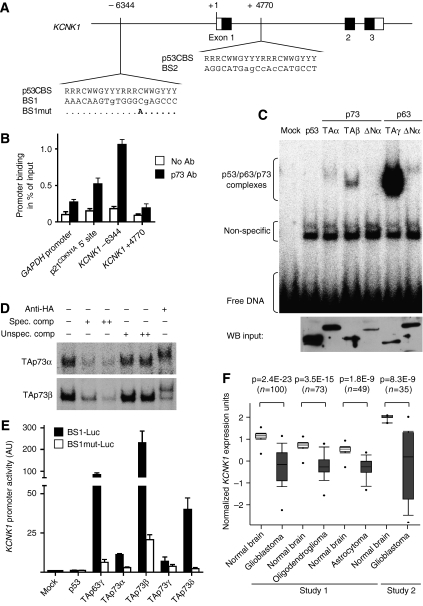
To investigate regulation of these two genes by p73, we scanned the genomic region surrounding the transcriptional start site for binding sites by bioinformatics and chromatin immunoprecipitation. In the case of FAM38B, which encodes a potential multipass membrane protein with yet unknown function, no binding site for p73 could be identified by either method, suggesting that FAM38B is indirectly regulated in response to p73 activation. In the KCNK1 gene, which encodes a ubiquitously expressed potassium channel protein (Lesage et al, 1996), MatInspector located two potential binding sites with four or less mismatches to the consensus p53-binding sequence (Figure 5A). Binding site 1 (BS1) is located 6344 bp upstream of the transcription start, whereas the putative binding site 2 (BS2) is located 4770 bp downstream in the first intron. Chromatin immunoprecipitation revealed binding of p73 to KCNK1 BS1 in vivo, which was even stronger than binding to the established 5′ binding site in the p53 family target gene p21CDKN1A (Figure 5B). No significant association with KCNK1 BS2 was detected. Electrophoretic mobility shift assays confirmed binding of in vitro-translated recombinant TAp73 and also p63 to BS1 (Figure 5C), but not BS2 (data not shown). Interestingly, although the in vitro-translated p53 protein efficiently bound p21 promoter sequences (data not shown), p53 was unable to bind to the KCNK1 BS1 sequence, indicating that this binding site is able to discriminate between p53 and p63/p73. The specificity of the observed DNA–protein complexes was confirmed by competition and supershift analysis (Figure 5D). A luciferase reporter plasmid containing 530 bp surrounding BS1 linked to a minimal TATA box promoter was strongly transactivated by TAp73β (Figure 5E). A single point mutation in BS1 reduced transactivation by TAp73β from 230-fold to only 20-fold. Although TAp63γ showed strong DNA binding in vitro, TAp73β was a 2.7-fold better transactivator. Together these data establish that KCNK1 is a bona fide target of p73.
Figure 5.
KCNK1 as an effector of TAp73-mediated tumour suppression. (A) Location of two potential binding sites for p53 family members as determined by bioinformatics (MatInspector). Shown is the p53 consensus binding sequence (p53CBS) in comparison to binding sites BS1 and BS2 in the promoter region and intron 1, respectively. (B) Chromatin immunoprecipitation (ChIP). BJ-T cells were transfected with TAp73 and chromatin was isolated after cross-linking with formaldehyde. p73 was precipitated and the associated DNA fragments were quantified by qPCR for the presence of BS1 and BS2 in the KCNK1 gene, the 5′ p53-binding site in the p21CDKN1A promoter and the GAPDH promoter as a negative control. No-antibody control immunoprecipitations serve as specificity controls. Promoter binding was normalized to the amount of input DNA. Shown is the mean promoter binding±s.d. of three independent chromatin immunoprecipitation experiments. (C, D) Electrophoretic mobility shift assays showing binding of in vitro-translated HA-tagged p53, TAp73α, TAp73β, ΔNp73α, TAp63γ and ΔNp63α to a 30 bp double-stranded, 32P-labelled oligonucleotide containing the KCNK1 BS1-binding site (5 nM). (D) A 50- and 200-fold molar excess of a p53 consensus binding site without [32P] was used as a specific competitor, a scrambled oligonucleotide as an unspecific competitor. A 200 ng portion of anti-HA antibody was added for supershift analysis as indicated. (C, lower panel) A western blot demonstrates successful in vitro translation of all proteins. (E) Luciferase reporter assay. The 530 bp surrounding KCNK1 BS1 were cloned upstream of a minimal TATA box promoter into the luciferase expression plasmid pGL3. A single-point mutation was introduced into BS1 by site-directed mutagenesis. Shown is the promoter activity 48 h following cotransfection of 200 ng BS1-Luc (or BS1mut-Luc) with 100 ng of the indicated p53, p63 or p73 expression plasmids. Shown is the mean±s.d. of at least two independent experiments performed in duplicates each. (F) KCNK1 is downregulated in brain tumours relative to normal brain. The data are taken from two independent studies (Bredel et al, 2005; Sun et al, 2006) and were obtained from www.oncomine.org.
As the transactivation function of TAp73 is frequently disabled in various cancer types by mutant p53 or overexpression of the dominant-negative p73 inhibitor ΔNp73 (Marin et al, 2000; Tuve et al, 2004; Concin et al, 2005; Wager et al, 2006), downstream effectors of TAp73, such as KCNK1 or FAM38B, would be expected to be downregulated in cancer tissues. Data obtained from a publicly available database of tumour expression profiles (www.oncomine.org; Rhodes et al, 2004) showed that downregulation of KCNK1 relative to normal tissue occurs in tumour entities such as glioma (glioblastoma multiforme, astrocytoma and oligodendroglioma; Figure 5F) and melanoma (Supplementary Figure 1A). In addition, KCNK1 is expressed in a grade- and stage-specific manner in ovarian cancer (Supplementary Figure 1C and D), with expression being lowest in advanced tumour stages. Likewise, expression of FAM38B is also significantly reduced in most glioblastomas (Supplementary Figure 1B). Together the gene expression profiling and RNA interference knockdown experiments identified KCNK1 (and FAM38B), which are downregulated in human cancer types with frequent loss of TAp73 activity, as essential inhibitors of anchorage-independent growth.
Control of p73 expression by activated Ras
The central question arising from these findings is how this TAp73-dependent barrier to full transformation is eventually overcome during tumorigenesis. The initial experiment in BJ fibroblasts indicated that activated Ras downregulates TAp73 expression and concomitantly enhances expression of the antagonistic ΔNp73 isoform (Figure 1C and D). When TAp73 expression was restored or when ΔNp73 was depleted, BJ-TER cells lost their anchorage-independent growth properties (Figure 6A–C). Both downregulation of TAp73 and upregulation of ΔNp73 are therefore important for the transforming activity of H-RasV12.
Figure 6.
Activated Ras regulates the TAp73/ΔNp73 ratio through PI3K-signalling. (A) Restoring TAp73 expression inhibits anchorage-independent growth of BJ-TER cells. BJ-TER cells were transfected with TAp73 or GFP as a control and tested for anchorage-independent growth in soft agar. Shown is the clonogenic growth in soft agar medium and the mean number of colonies±s.d. Scale bars, 200 μm. (B, C) Knockdown of ΔNp73 inhibits soft agar growth of BJ-TER cells. (B) ΔNp73 was detected by a western blot with a polyclonal antiserum raised against the ΔNp73-specific epitope encoded by the alternative exon 3′. ΔNp73-specific (◂) and non-specific (*) bands are indicated. ΔNp73 was detected under both basal and p53-stimulated conditions following expression of a non-silencing control (ns) or ΔNp73-directed shRNA. (C) BJ-TER cells were transduced with the lentiviral shRNA constructs as indicated and tested for growth in soft agar. Shown are representative images of soft agar colonies and the mean number of colonies±s.d. Scale bars, 200 μm. (D, E) Semiquantitative RT–PCR for TAp73 and ΔNp73 expression and western blot for TAp73 in H-RasV12-expressing BJ-TER cells treated with the MEK-inhibitor U0126 or the PI3K inhibitor LY294002 for 24 h. Non-transformed BJ-TE cells are shown for comparison. (F) BJ-TER cells were treated with LY294002 or DMSO as a control and tested for growth in soft agar. Shown are colony morphology and the mean number of colonies±s.d. (G) PI3K pathway activation switches the TAp73/ΔNp73 ratio in BJ-TE cells. BJ-TE cells were transduced with retroviral vectors encoding H-RasV12WT and the effector loop mutants H-RasV12T35S, H-RasV12E37G and H-RasV12Y40C. Shown are western blots for PI3K-AKT and p44/42 MAPK pathway activation. For specific detection of TAp73 and ΔNp73, we first immunoprecipitated total p73 with a polyclonal p73 antiserum before western blot detection with antibodies specific for TAp73 and ΔNp73 epitopes. (H) HCT116 cells were transduced with a non-silencing (ns) or a K-RasD13-directed shRNA with or without coexpression of H-RasV12WT or H-RasV12Y40C. PI3K-AKT and p44/42 MAPK pathway activation, TAp73 and ΔNp73 expression were analysed by western blot in HCT116 cells grown to confluence.
The H-RasV12-induced switch in p73 expression from TAp73 to ΔNp73 involves phosphatidylinositol-3 kinase (PI3K)-mediated signalling as the PI3K inhibitor LY294002, but not the MEK inhibitor U0126, switches the TAp73/ΔNp73 ratio back to TAp73 (Figure 6D and E). Consistently, we observed reduced colony formation in soft agar when BJ-TER cells were treated with LY294002 (Figure 6F). This pharmacological result was further confirmed genetically using effector-loop mutants of oncogenic Ras. These mutants are single amino-acid substitutions in the effector loop domain of oncogenic H-RasV12 that cause each of them to preferentially activate only one of the major effector pathways. Thus, the H-RasV12T35S mutant binds to and activates Raf, but not PI3K or Ral-GEF, while the H-RasV12E37G and H-RasV12Y40C mutants bind to and activate only Ral–GEFs and PI3K, respectively (Rodriguez-Viciana et al, 1997). As these mutants are less active than wild-type H-RasV12 (H-RasV12WT), they were expressed at higher levels to obtain comparable pathway activation. This was controlled by measuring phosphorylation of AKT and p44/42 MAPK (Figure 6G). Only the PI3K-activating mutant H-RasV12Y40C was able to switch the p73 isoform expression similar to H-RasV12WT providing genetic evidence that PI3K activation is necessary and sufficient for switching TAp73 expression to ΔNp73 (Figure 6G).
To investigate regulation of p73 expression by Ras in tumour cells that have developed naturally in a cancer patient, we used HCT116 colon cancer cells carrying an activating G13D mutation in one of the two K-Ras alleles. As previously described (Brummelkamp et al, 2002), specific depletion of the oncogenic K-Ras allele abolishes soft agar growth and tumorgenicity in nude mice. Although total K-Ras levels were only reduced marginally, we observed a significant downregulation of PI3K pathway activation as assessed by Thr308 phosphorylation of AKT (Figure 6H). As a consequence, TAp73 levels were strongly induced, but restored back to normal following PI3K activation by ectopic expression of either H-RasV12WT or H-RasV12Y40C (Figure 6H). As previously seen in BJ-TE cells, TAp73 was only induced under conditions of high cell density (data not shown). In contrast to the profound regulation of ΔNp73 by Ras in BJ cells, Ras depletion had only little inhibitory effect on ΔNp73 levels in HCT116 cells indicating the presence of additional factors activating ΔNp73 expression in HCT116 cells. Taken together, these results demonstrate that activated Ras controls TAp73 expression and thus the TAp73/ΔNp73 ratio through PI3K signalling not only in an experimental model of human cell transformation but also in tumour cells from a cancer patient.
Discussion
In summary, our data provide an explanation of how p73 can function as a tumour suppressor independent of its famous family member p53. We show that the transactivating p73 isoform TAp73 is specifically induced when cells with pRB pathway alterations due to expression of LT or E1A are grown to high cell density (Figure 7). In contrast, p53 activity occurs only in actively growing but not in contact-inhibited high-density cultures (Deffie et al, 1995).
Figure 7.
Model of p73 expression changes during the course of experimental transformation.
Regulation of the TAp73 promoter by the E2F-RB axis has been reported previously (Irwin et al, 2000; Stiewe and Putzer, 2000; Zaika et al, 2001; Urist et al, 2004). Physiologically, the TAp73 promoter was shown to be repressed by E2F4–p130–HDAC complexes (Pediconi et al, 2003), suggesting that LT or E1A derepress TAp73 transcription by binding and inactivating the pocket protein family member p130. However, inactivation of pocket proteins was not sufficient to activate TAp73 expression in our setting. In the premalignant BJ-TE cells, TAp73 was only induced when the cells were grown to high cell density, which stimulated E2F1 expression and binding to the TAp73 core promoter (Figure 2G and H). It has previously been demonstrated that E2F1 needs to be activated by acetylation for recruitment on the TA73 promoter following DNA damage (Pediconi et al, 2003). In the BJ cells, high cell density might provide such an activating signal to E2F1 that is already present in many cancer cell lines due to constitutively active DNA damage signalling induced by oncogenic Ras or c-Met (Di Micco et al, 2006). This might explain why high cell density has not been identified as an essential factor for TAp73 promoter activation in other studies that were mostly performed in fully transformed cancer cell lines with activated DNA damage signalling pathways (Zaika et al, 2001).
Binding to pocket proteins is known to require the LXCXE motif present in both LT and E1A (Ali and DeCaprio, 2001). Interference with the pRB-related proteins p107 and p130 further requires the aminoterminal J domain of LT (Stubdal et al, 1997; Ali and DeCaprio, 2001). In line with this, we identified both the LXCXE motif and the J domain of LT to be involved in TAp73 induction (Figure 2C). The LT protein, which binds to and inactivates p53, therefore, not only spares p73 from inhibition, but also even causes upregulation of TAp73 expression. LT, therefore, discriminates between the p53 family members and regulates them in an opposite manner. p53 is inactivated, whereas TAp73 is activated and unfolds a transcriptional programme that limits cell survival under conditions of high cell density and impairs anchorage-independent growth as one of the major hallmarks of fully transformed cells. Thus, TAp73 expression may not be detrimental in the context of viral replication, but as our data indicate, it does pose a barrier to malignant transformation.
The role for p73 in preventing anchorage-independent growth provides an intriguing explanation for the poorly understood enhanced metastasis and tumour aggressiveness that was observed upon loss of p73 function in p53+/− mice and the poor prognosis of cancer patients with altered p73 expression (Tannapfel et al, 1999; Concin et al, 2005; Flores et al, 2005; Muller et al, 2005; Dominguez et al, 2006). Interestingly, KCNK1 and FAM38B, the two targets of TAp73 that we found to be essential for suppression of anchorage-independent growth, are frequently downregulated in patients with melanoma, glioma and ovarian cancer (Figure 5F; Supplementary Figure 1A and B). Moreover, expression of these genes in ovarian cancer is further decreased in more advanced tumour stages (Supplementary Figure 1C and D). In all these cancer entities, overexpression of ΔNp73 as a potent inhibitor of TAp73 has been documented and in many cases linked to poor prognostic markers (Tuve et al, 2004; Concin et al, 2005; Wager et al, 2006). It will be interesting to see whether KCNK1 and FAM38B are also downregulated in the highly aggressive tumours from p53/p73 compound heterozygous mice (Flores et al, 2005). What remains unclear at present is how these two genes affect anchorage-independent growth in molecular terms. Both genes encode membrane surface proteins: FAM38B, a potential multipass membrane protein with yet unknown function and KCNK1 (TWIK-1) a member of the two-pore-domain potassium leak channel family, which determines the resting membrane potential (Lesage et al, 1996). Although rather speculative, it could be conceived that these cell-surface proteins fulfill functions in cell adhesion or transmembrane signalling through intracellular adapter proteins or modulation of the membrane potential in response to cell contacts, which might be instrumental in integrating cell density with cell-cycle progression. Much more experiments will certainly be required to better understand how FAM38B and KCNK1 affect cell proliferation and viability in response to changes in cell density and how this eventually contributes to tumour suppression.
Several lines of evidence hint at an accessory role of p63 in anchorage-independent growth control, which would be consistent with its role in regulating cell adhesion and migration (Carroll et al, 2006). First, the p73-directed shRNA, which also targets p63 (p73sh4) due to sequence homology, is the most effective inducer of soft agar growth. Second, KCNK1 is not only transactivated by TAp73 but also by TAp63. It is therefore conceivable that an additional loss of TAp63 function might further enhance the anchorage-independent growth of p73-depleted cells. However, depletion of only p63 did not enable soft agar growth, whereas the selective ablation of p73 with the highly specific TAp73-shRNA (Figure 3A and G) and with a genetically defined p73-knockout (Figure 3H) was sufficient to promote anchorage-independence. We, therefore, conclude that TAp73 is the major barrier to anchorage-independent growth in this transformation model.
How is the TAp73-imposed barrier to full transformation overcome in malignant tumours? One answer is provided by our previous findings that the TAp73-inhibitory p73 isoform ΔNp73 renders NIH3T3 cells anchorage-independent and tumorigenic (Stiewe et al, 2002a). A similar effect was observed with oncogenic Ras, which suggested that Ras signalling might control p73 activity. In fact, our results from the experimental transformation model and also from patient-derived tumour cell lines indicate that activating mutations in the Ras genes allows cells to overcome this barrier by downregulating TAp73 expression and enhancing expression of the antagonistic p73 isoform ΔNp73 (Figures 6D, G and 7). Both alterations are crucial for the transforming activity of Ras in this model system, as restoration of TAp73 and knockdown of ΔNp73 prevented soft agar growth (Figure 6A–C). Interestingly, the Ras-induced switch of the TAp73/ΔNp73 ratio in favour of ΔNp73 has been frequently observed in many different cancer types and been shown to correlate with advanced tumour stage, lymph node metastasis, vascular invasion, unsatisfactory response to chemo- and radiotherapy, and poor overall patient survival (Casciano et al, 2002; Stiewe et al, 2004; Concin et al, 2005; Muller et al, 2005; Dominguez et al, 2006). Whether the poor prognosis of patients with an altered TAp73/ΔNp73 ratio is caused by ΔNp73 overexpression, loss of TAp73 expression or both remains to be investigated.
Although the effects of Ras on ΔNp73 levels were only observed in BJ cells, the inhibitory effects of Ras-induced PI3K signalling on TAp73 transcription were seen in both the BJ model system and the patient-derived HCT116 cell line. In contrast, in UR61 pheochromocytoma and SH-SY5Y neuroblastoma cells, but not H1299 cells, Ras-induced MAP kinase signalling was shown to stabilize the TAp73 protein (Fernandez-Garcia et al, 2007). The antagonistic effects of PI3K signalling on TAp73 transcription and of MAPK signalling on TAp73 protein stability indicate that the control of p73 by Ras is complex and occurs in a tissue- and context-specific manner. Therefore, much more work is needed to better understand how expression of the prognostically relevant TAp73/ΔNp73 ratio is regulated and how it can be therapeutically modulated. Nevertheless, our results clearly demonstrate that TP73 encodes tumour suppressor functions that are distinct from p53 and are specifically targeted by activated Ras signalling. Consequently, p73 is not a simple backup for p53, but has tumour-suppressor activities on its own.
Materials and methods
Cell lines and cell culture
Primary human diploid BJ fibroblasts, HA1E (Hahn et al, 1999), H1299, HCT116, 293, 293T and PT67 cells were cultured in DMEM (Sigma) supplemented with 10% fetal bovine serum (Sigma), 2 mM L-glutamine and penicillin-streptomycin (Sigma). When cells were grown to confluence the medium was replaced with fresh medium every 1–2 days. Transfections were performed using Escort V reagent (Sigma). Viral infections were performed as described previously (Wiznerowicz and Trono, 2003; Cam et al, 2006). In brief, retroviruses were produced by transient transfection of PT67 cells on 60-mm dishes with 5 μg vector plasmid. Retrovirus containing supernatant was collected between 48 and 72 h following transfection, passed through a 0.45-μm-pore filter and supplemented with polybrene to a final concentration of 8 μg/ml. Cells were spin-infected at 500 g and 37°C for 1 h. Transduced target cells were selected with appropriate selection antibiotics. Infection efficiency was controlled by GFP-fluorescence. Adenoviruses were produced using the AdEasy-system (Stratagene). Cell viability was measured 48 h after drug treatment with the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions on a FLUOstar Optima (BMG). For luciferase assays, H1299 cells were cotransfected with 200 ng of luciferase reporter plasmid (BS1-Luc or BS1mut-Luc) and 100 ng of p53, p63 or p73 expression plasmid and collected for luciferase activity measurement with the Luciferase Assay System (Promega) at 48 h later according to the manufacturer's protocol. For assays of soft agar growth, 2 × 104 cells were seeded on a six-well plate in DMEM containing 10% fetal bovine serum and 0.4% agarose on top of a layer of medium containing 0.8% agarose and overlaid with medium containing 10% fetal calf serum, which was changed every 3–4 days. Colonies were counted under a fluorescence microscope approximately 2 weeks after seeding. For each construct, three independent experiments were performed and at least 10 microscopic images per experiment were used for measurement of colony sizes with the cellF software (Olympus). In all experiments, a cutoff diameter of at least 90 μm was used to distinguish colonies from background debris. All diagrams report the mean number of colonies±s.d.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were performed as described previously (Stiewe et al, 2002a). The following double-stranded oligonucleotides (p53-binding sites underlined) were used: p53 consensus sequence (specific competitor) 5′-GGG TAG ACA TGC CTA GAC ATG CCT AAG CTC CC-3′, p53 consensus scrambled (unspecific competitor) 5′-GGG CCA GCT AGC AGG CAG CAT CAG TAC TTC CC-3′, KCNK1 BS1 5′-GGA GCA AAC AAG TGT GGG CGA GCC CAT TCC-3′.
Western blot and RT–PCR
For immunoblotting, cells were lysed in RIPA buffer (50 mM Tris-Cl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS)). Samples (100 μg per lane) were separated by SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Amersham) for immunodetection. RNA isolation and reverse transcription were performed using the RNeasy Mini Kit (Qiagen) and Omniscript Reverse Transcriptase (Qiagen), according to the manufacturer's instructions. cDNA was amplified with HotStar Taq (Qiagen), and the number of amplification cycles was adjusted to remain within the linear amplification range. Primer sequences are available upon request.
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed as described previously (Cam et al, 2006). Immunoprecipitated DNA fragments were analysed by semiquantitative PCR (TAp73 promoter) or by quantitative PCR (p21CDKN1A, KCNK1, GAPDH). Primer sequences are available upon request. qPCR data from triplicate measurements were expressed as promoter occupancy in percentage of input DNA.
Gene expression profiling
BJ-TE cells expressing a p73-directed (p73sh3) or non-silencing shRNA as a control were analysed under subconfluent and confluent conditions. Total RNA was extracted using the RNeasy Mini Kit (Qiagen). The samples were analysed for gene expression using Affymetrix GeneChip Human Genome U133A 2.0 Arrays and further analysed with GeneSpring 7.0 (Silicon Genetics). For normalization, data measurements less than 0.01 were set to 0.01, each measurement was divided by the 50th percentile of all measurements in that sample, and each gene was divided by the median of its measurements in all samples. To avoid noise from genes expressed at levels close to the detection limit, a raw expression threshold level of 70 was applied. A probe set was considered to be p73-induced when expression in both p73-shRNA samples was more than twofold lower than in the non-silencing samples. Vice versa, a probe set was considered to be p73-repressed when expression was more than twofold higher in both p73-shRNA samples compared to the non-silencing samples. Density-dependent regulation was assumed for probe sets that changed more than 1.5-fold between the two non-silencing samples. The complete set of microarray data has been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE7201.
Statistical analysis
All measurements are presented as the mean±s.d.
Supplementary Material
Supplementary Table 1
Supplementary Figure 1
Supplementary Information
Acknowledgments
We thank William C Hahn, Robert Weinberg, Scott Lowe, Eiji Hara, Bert Vogelstein, Hans van Bokhoven, Frank McKeon and Didier Trono for providing expression plasmids, cell lines and mice. AR is supported by the Interdisciplinary Center for Clinical Research (IZKF), University of Würzburg. This work was funded by grants from the Deutsche Forschungsgemeinschaft (Transregio TR17 ‘Ras-dependent pathways in human cancer', Forschungszentrum FZ82).
References
- Ali SH, DeCaprio JA (2001) Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol 11: 15–23 [DOI] [PubMed] [Google Scholar]
- Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI (2005) Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res 65: 8679–8689 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247 [DOI] [PubMed] [Google Scholar]
- Cam H, Griesmann H, Beitzinger M, Hofmann L, Beinoraviciute-Kellner R, Sauer M, Huttinger-Kirchhof N, Oswald C, Friedl P, Gattenlohner S, Burek C, Rosenwald A, Stiewe T (2006) p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell 10: 281–293 [DOI] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW (2006) p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 8: 551–561 [DOI] [PubMed] [Google Scholar]
- Casciano I, Mazzocco K, Boni L, Pagnan G, Banelli B, Allemanni G, Ponzoni M, Tonini GP, Romani M (2002) Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ 9: 246–251 [DOI] [PubMed] [Google Scholar]
- Concin N, Hofstetter G, Berger A, Gehmacher A, Reimer D, Watrowski R, Tong D, Schuster E, Hefler L, Heim K, Mueller-Holzner E, Marth C, Moll UM, Zeimet AG, Zeillinger R (2005) Clinical relevance of dominant-negative p73 isoforms for responsiveness to chemotherapy and survival in ovarian cancer: evidence for a crucial p53-p73 cross-talk in vivo. Clin Cancer Res 11: 8372–8383 [DOI] [PubMed] [Google Scholar]
- Deffie A, Hao M, Montes de Oca Luna R, Hulboy DL, Lozano G (1995) Cyclin E restores p53 activity in contact-inhibited cells. Mol Cell Biol 15: 3926–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Dominguez G, Garcia JM, Pena C, Silva J, Garcia V, Martinez L, Maximiano C, Gomez ME, Rivera JA, Garcia-Andrade C, Bonilla F (2006) DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol 24: 805–815 [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia B, Vaque JP, Herreros-Villanueva M, Marques-Garcia F, Castrillo F, Fernandez-Medarde A, Leon J, Marin MC (2007) p73 cooperates with Ras in the activation of MAP kinase signaling cascade. Cell Death Differ 14: 254–265 [DOI] [PubMed] [Google Scholar]
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7: 363–373 [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA (1999) Creation of human tumour cells with defined genetic elements. Nature 400: 464–468 [DOI] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG Jr (2000) Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407: 645–648 [DOI] [PubMed] [Google Scholar]
- Lesage F, Guillemare E, Fink M, Duprat F, Lazdunski M, Romey G, Barhanin J (1996) TWIK-1, a ubiquitous human weakly inward rectifying K+ channel with a novel structure. EMBO J 15: 1004–1011 [PMC free article] [PubMed] [Google Scholar]
- Maehara K, Yamakoshi K, Ohtani N, Kubo Y, Takahashi A, Arase S, Jones N, Hara E (2005) Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional pRB and p53. J Cell Biol 168: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, Vousden KH, Allday MJ, Gusterson B, Ikawa S, Hinds PW, Crook T, Kaelin WG Jr (2000) A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 25: 47–54 [DOI] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Irwin MS, DeCaprio JA, Caput D, Kaelin WG Jr (1998) Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol 18: 6316–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G, De Laurenzi V, Vousden KH (2002) p73: friend or foe in tumorigenesis. Nat Rev Cancer 2: 605–615 [DOI] [PubMed] [Google Scholar]
- Muller M, Schilling T, Sayan AE, Kairat A, Lorenz K, Schulze-Bergkamen H, Oren M, Koch A, Tannapfel A, Stremmel W, Melino G, Krammer PH (2005) TAp73/DeltaNp73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ 12: 1564–1577 [DOI] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, Gulino A, Levrero M (2003) Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 5: 552–558 [DOI] [PubMed] [Google Scholar]
- Petrenko O, Zaika A, Moll UM (2003) deltaNp73 facilitates cell immortalization and cooperates with oncogenic Ras in cellular transformation in vivo. Mol Cell Biol 23: 5540–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD (2000) An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 289: 304–306 [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA (2004) Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6: 171–183 [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J (1997) Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89: 457–467 [DOI] [PubMed] [Google Scholar]
- Roth J, Konig C, Wienzek S, Weigel S, Ristea S, Dobbelstein M (1998) Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol 72: 8510–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Wust AP, Sigrist B, Belyanskaya L, Hopkins Donaldson S, Stahel RA, Zangemeister-Wittke U (2005) DeltaNp73 antisense activates PUMA and induces apoptosis in neuroblastoma cells. J Neurooncol 72: 29–34 [DOI] [PubMed] [Google Scholar]
- Stiewe T (2007) The p53 family in differentiation and tumorigenesis. Nat Rev Cancer 7: 164–168 [DOI] [PubMed] [Google Scholar]
- Stiewe T, Putzer BM (2000) Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet 26: 464–469 [DOI] [PubMed] [Google Scholar]
- Stiewe T, Theseling CC, Putzer BM (2002a) Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem 277: 14177–14185 [DOI] [PubMed] [Google Scholar]
- Stiewe T, Tuve S, Peter M, Tannapfel A, Elmaagacli AH, Putzer BM (2004) Quantitative TP73 transcript analysis in hepatocellular carcinomas. Clin Cancer Res 10: 626–633 [DOI] [PubMed] [Google Scholar]
- Stiewe T, Zimmermann S, Frilling A, Esche H, Putzer BM (2002b) Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res 62: 3598–3602 [PubMed] [Google Scholar]
- Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA (1997) Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol 17: 4979–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA (2006) Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9: 287–300 [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Wasner M, Krause K, Geissler F, Katalinic A, Hauss J, Mossner J, Engeland K, Wittekind C (1999) Expression of p73 and its relation to histopathology and prognosis in hepatocellular carcinoma. J Natl Cancer Inst 91: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Tuve S, Wagner SN, Schittek B, Putzer BM (2004) Alterations of DeltaTA-p 73 splice transcripts during melanoma development and progression. Int J Cancer 108: 162–166 [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C (2004) p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 18: 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager M, Guilhot J, Blanc JL, Ferrand S, Milin S, Bataille B, Lapierre F, Denis S, Chantereau T, Larsen CJ, Karayan-Tapon L (2006) Prognostic value of increase in transcript levels of Tp73 DeltaEx2-3 isoforms in low-grade glioma patients. Br J Cancer 95: 1062–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D (2003) Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 77: 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika A, Irwin M, Sansome C, Moll UM (2001) Oncogenes induce and activate endogenous p73 protein. J Biol Chem 276: 11310–11316 [DOI] [PubMed] [Google Scholar]
- Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM (2002) DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med 196: 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Figure 1
Supplementary Information